Abstract
We summarize findings of SARS-CoV infections in several animal models each of which support viral replication in lungs accompanied by histopathological changes and/or clinical signs of illness to varying degrees. New findings are reported on SARS-CoV replication and associated pathology in two additional strains (C57BL/6 and 129S6) of aged mice. We also provide new comparative data on viral replication and associated pathology following infection of golden Syrian hamsters with various SARS-CoV strains and report the levels of neutralizing antibody titers following these infections and the cross-protective efficacy of infection with these strains in protecting against heterologous challenge. Finally, we summarize findings of a variety of vaccine approaches and discuss the available in vitro and in vivo data addressing the potential for disease enhancement following re-infection in animals previously vaccinated against or infected with SARS-CoV.
Keywords: SARS coronavirus, BALB/c, C57BL/6, 129S6 mice, Hamsters, Ferrets, Non-human primates, Vaccines, FIPV, Enhanced disease, Urbani, HKU-39849, Frankfurt 1, GD03T0013
1. Introduction
In studies of pathogenesis, prophylaxis, and treatment, animal models that mimic human disease and protection from disease are invaluable. Since the outbreak of severe acute respiratory syndrome (SARS) in late 2002 and identification of the etiological agent as a novel coronavirus (SARS-CoV) in 2003, several animal models have been identified for use in such evaluations. Although each model reviewed in this article has some utility in the study of SARS disease and prevention, the kinetics of viral replication and resolution of disease are much more rapid in animal models compared to human infections, and no model fully reflects the spectrum of clinical illness (morbidity and mortality), associated pathology, and viral replication observed in human cases of SARS. Of the animal models that have been employed for evaluating SARS-CoV replication and disease, those models that have been most fully characterized and which offer the best potential for evaluation of prevention and therapeutic strategies are discussed here and include inbred mice, hamsters, ferrets and non-human primates. We also reference other, less well-characterized animal models.
A number of approaches for the development of SARS vaccines have been explored, including inactivated whole-virus, protein subunit, live virus-vectored, DNA-vectored, and combination vaccines. We review the data available on the evaluation of these candidate vaccines.
Lastly, we address the potential for enhanced disease following exposure to SARS-CoV in previously vaccinated animals. The potential for this phenomenon is given special consideration because immunization of kittens with a type-1 coronavirus vaccine against feline infectious peritonitis virus (FIPV) led to exacerbated disease following challenge with FIPV. This has led to concern and speculation about the potential for disease enhancement following use of SARS vaccines.
2. Animal models
2.1. Inbred mice
Several inbred mouse species (BALB/c, C57BL/6 (B6), 129S) have been shown to support SARS-CoV replication and to demonstrate pneumonitis (129S) and clinical signs of SARS disease (aged BALB/c) (Glass et al., 2004, Hogan et al., 2004, Roberts et al., 2005a, Subbarao et al., 2004). The inbred mouse model has several advantages including small size, cost, availability in large enough numbers for statistical evaluation, and the ability to be manipulated at a genetic level (i.e. to develop gene knock-outs and knock-ins). In addition, immunological reagents are available for studying elements of pathogenesis in many inbred mouse strains.
Following intranasal inoculation under light anesthesia, SARS-CoV replicates in the epithelium of nasal turbinates and lungs of mice as early as 1 day post-infection (p.i.). The peak titer in lungs occurs at days 2–3 p.i., and virus is cleared in most mice by days 5–7 p.i. In young mice, SARS-CoV replication has not been associated with overt signs of clinical illness or pronounced pathology. However, 129S6 mice seem to be slightly more susceptible to disease associated with SARS-CoV infection than are BALB/c or B6 mice, as evidenced by an approximate 8% loss of body weight, with a nadir at days 5–7 p.i., and by the presence of mild interstitial pneumonitis at day 2 p.i. (Hogan et al., 2004; authors’ unpublished data). Interstitial pneumonitis following SARS-CoV infection has not been previously reported in BALB/c mice when evaluated at days 2, 4 and 9 p.i. (Subbarao et al., 2004). However, slightly elevated levels of the proinflammatory cytokine TNF-alpha were observed in young BALB/c mice at day 3 p.i. (authors’ unpublished data), and upon recent evaluation, young BALB/c mice displayed moderate interstitial pneumonitis on day 3 p.i., comparable to that seen in B6 and Rag1−/− mice (Glass et al., 2004, Roberts et al., 2007). This pneumonitis is quickly resolved and is not observed on day 4 p.i. This transient inflammation coincides with viral clearance since little viral antigen is observed at or after day 3 p.i., and virus titers in lungs also steadily decline from day 3 p.i. onwards (Subbarao et al., 2004).
Aged BALB/c mice develop more severe disease than young BALB/c mice in a pattern of age-related severity that mimics observations in the SARS outbreak in humans, where age was a predictor of severe disease and mortality (Roberts et al., 2005a). In addition to serving as a model of age-related susceptibility to disease, aged BALB/c mice consistently demonstrate several features of an ideal animal model for SARS-CoV studies. In addition to supporting viral replication in respiratory tissues, the aged BALB/c mouse (BALB/cAnNHsd, Harlan) demonstrates clinical signs of illness including weight loss, dehydration, and ruffled fur, and several histopathological findings, i.e. diffuse alveolar damage including edema, hyaline membrane formation, and pneumonitis, that correlate with those reported in autopsies of SARS patients (Roberts et al., 2005a).
We also examined the susceptibility of aged C57BL/6J (The Jackson Laboratory, B6) and 129S6/SvEvTac (Taconic, 129S6) mice to SARS-CoV infection. Following intranasal inoculation with 105 TCID50 of SARS-CoV (Urbani), both B6 and 129S6 aged mice (12–14 months old) had weight loss (∼7% at the nadir) from day 3 through day 5 p.i. that was comparable to that observed in aged BALB/c mice (Roberts et al., 2005a). The level and kinetics of viral replication in lungs of aged B6 and BALB/c mice following inoculation were also similar. At days 5 and 6 p.i., SARS-CoV-infected B6 mice had 10–70-fold lower titers of virus than BALB/c mice (Fig. 1 ). In contrast, aged 129S6 mice did not support prolonged replication of SARS-CoV as virus was cleared from lungs by day 5 p.i. (data not shown). Nevertheless, early histopathological features in the lungs of aged B6 and 129S6 mice were similar to those seen in aged BALB/c mice following SARS-CoV infection. Changes observed in the lungs at day 3 p.i included perivascular and peribronchiolar mononuclear inflammatory infiltrates, intraluminal necrotic debris in bronchioles comprised of dead respiratory epithelium and inflammatory cells, and small foci of interstitial pneumonitis (Fig. 2A, C, and E). Viral antigens were detected by immunohistochemical (IHC) staining in the respiratory epithelial cells of bronchioles and in alveolar pneumocytes of all mouse strains at day 3 p.i., but appeared most abundant in the bronchioles of 129S6 mice at this time point (Fig. 2B, C and F). Varying amounts of mononuclear inflammatory infiltrates persisted around bronchioles and small pulmonary vessels through day 9 p.i.; however, no viral antigens were detected by IHC at this time point. As previously reported, the aged BALB/c mouse has features that would make it an excellent model for evaluation of pathogenesis and vaccine efficacy studies. Based on weight loss and histopathological analyses, we believe that aged B6 and 129S6 mice may provide additional models for the study of age-dependent susceptibility to SARS-CoV. However, the drawback to widespread utility of the aged mouse model is the difficulty in procuring large numbers of mice that are over 12 months of age.
Fig. 1.
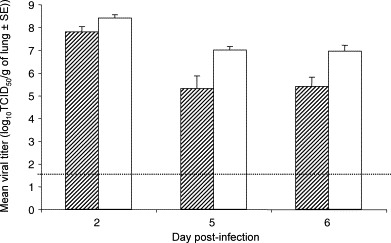
Replication of SARS-CoV in lungs of aged B6 and BALB/c mice. B6 and BALB/c mice (N = 15 per strain, aged 12–14 months) were intranasally infected with SARS-CoV (105 TCID50/mouse) at day 0. Five mice per strain were sacrificed at days 2, 5 and 6 post-infection. Bars (hatched = B6; open = BALB/c) indicate the mean titer of virus detected in 10% (w/v) lung homogenates by 50% tissue culture infectious dose (TCID50) assays on Vero cell monolayers. Error bars indicate standard error. Dashed line indicates limit of detection, 101.5 TCID50/g lung.
Fig. 2.
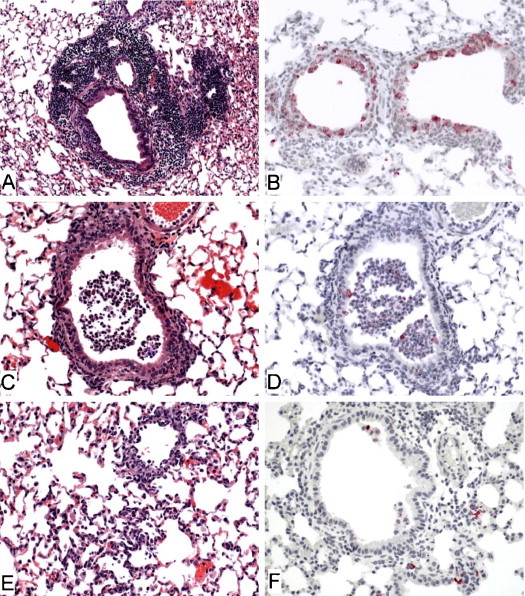
Histopathologic features of infection with SARS-CoV in the lungs of aged 129S6, B6 and BALB/c mice at 3 days post-infection. (A) Florid perivascular and peribronchiolar inflammatory cell infiltrates comprised predominantly of mononuclear cells (strain 129S6, original magnification 25×). (B) Abundant SARS-CoV antigens (red) in the cytoplasm of bronchiolar epithelial cells (strain 129S6, original magnification 50×). (C) Necrotic respiratory epithelium and inflammatory cells in the lumen of a small pulmonary bronchiole (strain BL6, original magnification 50×). (D) Immunohistochemical staining of the same section in C, showing scattered SARS-CoV antigens in the intraluminal debris. (E) Focus of interstitial inflammatory cell infiltrate (strain BALB/c, original magnification 50×). (F) SARS-CoV antigen in alveolar pneumocytes adjacent to a bronchiole (strain BALB/c, original magnification 50×). Hematoxylin and eosin stain (A, C, E); immunoalkaline phosphatase with naphthol fast-red substrate and hematoxylin counterstain (B, D, F).
Although a few strains of mice with targeted deletions of genes have been evaluated following SARS-CoV infection including Rag1−/−, CD1−/−, and Beige mice (Glass et al., 2004), the only strain with a notable clinical course or histopathological outcome different from wild-type strains of mice are STAT1−/− mice in the 129S background (Hogan et al., 2004; authors’ unpublished data). These mice support prolonged viral replication in the lungs with accompanying pathology, viremia, and dissemination of virus to the liver and spleen. These mice are suitable for pathogenesis studies and evaluation of antiviral agents. Other wild-type strains of inbred mice are suitable for studies of immunopathology, immunogenicity and efficacy of vaccine candidates.
2.2. Golden Syrian hamsters
The golden Syrian hamster (strain LVG, Charles River Laboratories; http://www.criver.com/research_models_and_services/research_models/LVG_Hamsters.html) is an excellent model for SARS-CoV infection because viral replication is accompanied by pathological changes in the lungs including pneumonitis and consolidation (Roberts et al., 2005b). Following intranasal inoculation with 103 TCID50 SARS-CoV, hamsters support viral replication in the nasal turbinates and lungs. Peak viral replication in the lungs occurs at day 3 p.i. and is cleared by day 7 p.i. Virus titers in nasal turbinates are about 10–100-fold lower than in lungs, but are detectable through day 14 p.i. Viremia is detected 2–3 days p.i.; and virus can be recovered from the spleen and liver. Histopathological changes in the lungs accompany SARS-CoV infection of hamsters. By day 3 p.i., focal areas of interstitial inflammation and consolidation are visible. At day 5 p.i., inflammation is more pronounced and consolidation is more widespread. Consolidation continues through day 7 p.i. and involves 30–40% of the surface area of the lung. By day 21 p.i., pathological changes in the lung have resolved (Roberts et al., 2005b).
Although histopathological signs of disease are pronounced, clinical symptoms of disease have been difficult to identify in hamsters. However, use of exercise wheels to evaluate overnight activity demonstrates that hamsters infected with SARS-CoV are less active from day 2 through day 7 p.i. than they were prior to infection or compared to uninfected or mock-infected animals (Table 1 ). This is the first objective evidence of clinical illness in hamsters.
Table 1.
Reduced activity in SARS-CoV-infected hamsters
| Inoculation group | Average no. of revolutions per hour ± S.E. |
||
|---|---|---|---|
| Prior to infection | Days 2–4 p.i. | Days 9–10 p.i. | |
| SARS-CoV Urbani | 865 ± 25 | 61 ± 23 | 770 ± 5 |
| Mock-infected | 946 ± 21 | 726 ± 46 | |
Golden Syrian hamsters, 5–10 weeks old, were allowed to run overnight in an ABSL3 animal containment facility under a biosafety cabinet on a Nalgene activity wheel for rodents equipped with a magnetic switch with LCD counter that counts whole revolutions (Fisher Scientific). Two to five nights prior to inoculation and one to ten nights following inoculation, hamsters were allowed to run and the number of revolutions was recorded and reported as an average of revolutions per hour; average run time 14 h/night. Each activity wheel was in a separate cage; one hamster per cage; water and food were available ad libitum. Hamsters were rested on alternate nights.
2.3. Evaluation of various strains of SARS-CoV in hamsters
Several strains of SARS-CoV have been evaluated in the hamster model, including Urbani (GenBank no. AY27874), HKU-39849 (HK-39; GenBank no. AY278491), Frankfurt 1 (Frk-1; GenBank no. AY291315) and a recombinant SARS-CoV infectious clone (ic) expressing the spike protein of GD03T0013 (GenBank no. AY525636) (icGD03; Deming et al., 2006). Primary intranasal infection with 103 TCID50/hamster with SARS-CoV (Urbani, HK-39 or Frk-1) results in virus titers of >107 TCID50/g of lung by day 2 p.i. (Fig. 3A–C), whereas primary infection with icGD03 results in significantly lower titers (104 TCID50/g on day 2 p.i.) (Fig. 3D). Our findings suggest that compared with the Urbani strain, icGD03 is slightly attenuated in the hamster model with a 1000-fold reduction in peak virus titers in the lungs following infection, and that SARS-CoV Frk-1 may be slightly more virulent than other strains. Although dose-dependent mortality was not observed, 3 of 20 hamsters died following infection with SARS-CoV Frk-1, and peak virus titers in lungs are higher than peak virus titers following infection with Urbani or HK-39. However, the difference in peak virus titer did not achieve statistical significance (Fig. 3A–C). Histopathological findings in lungs following infection with SARS-CoV (HK-39 or Frk-1) are indistinguishable from those reported following infection with SARS-CoV (Urbani) (Fig. 4 ). The spike proteins of the SARS-CoV Urbani and HK-39 isolates are identical in amino acid sequence, and the Frk-1 spike protein sequence differs from these by only one amino acid in the S2 domain (L1148F). Antibodies detected in hamster sera following primary SARS-CoV infections cross-neutralize the various virus strains and homologous viruses to comparable degrees (Urbani, Frk-1 and HK-39: ∼1:500 to 1:1000), and hamsters infected with each of these strains of SARS-CoV are protected from re-infection with the homologous virus or heterologous viruses (Fig. 3A–D).
Fig. 3.
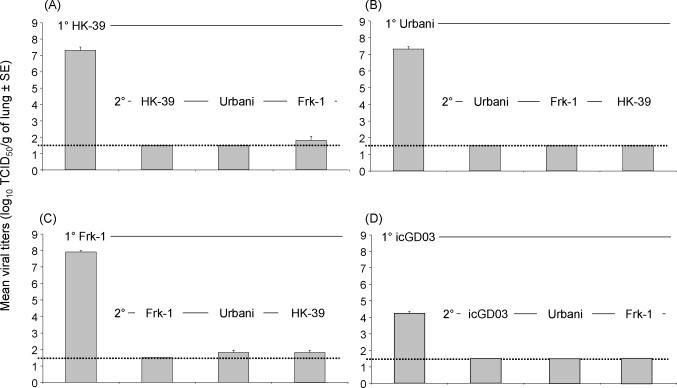
Primary replication and replication after challenge of SARS-CoV in hamster lungs. Golden Syrian hamsters (50–55 days old) were intranasally inoculated with one of four SARS-CoV strains (A) HK-39 (N = 16), (B) Urbani (N = 16), (C) Frk-1 (N = 16), and (D) icGD03 (N = 12) of SARS-CoV (103 TCID50/hamster). For evaluating viral replication following primary (1°) infection, four hamsters per strain (three hamsters for icGD03) were sacrificed at day 2 post-infection. For evaluation of protection from secondary (2°) infection, the remaining hamsters were challenged 28 days after 1° infection with the indicated homologous or heterologous strain of SARS-CoV (N = 4 per group; or N = 3/group for icGD03; 103 TCID50/hamster) and sacrificed 2 days later. Bars indicate the mean titer of virus detected in 10% (w/v) lung homogenates by TCID50 assay on Vero cells (1° = day 2 after initial infection; 2° = day 2 after challenge). Error bars indicate standard error. Dashed line indicates limit of detection, 101.5 TCID50/g lung.
Fig. 4.
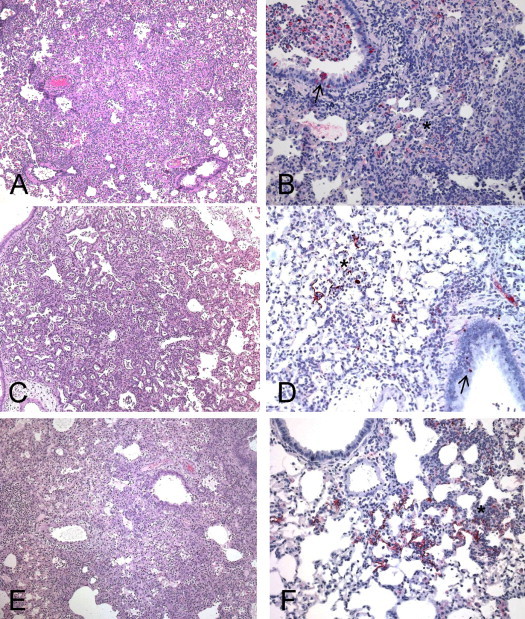
Pathology in lungs of golden Syrian hamsters following SARS-CoV infection. Lungs from SARS-CoV-infected (Urbani, A and B; Frankfurt-1, C and D; HKU-39849, E and D) hamsters at day 5 p.i. show confluent pneumonic consolidation (hematoxylin-and-eosin stain, A, C, and E), and SARS-CoV antigens (IHC assays, antigen in red; B, D, and F) in consolidated areas (asterisk) and bronchial epithelium (arrows). Original magnifications: 25× (A, C and E); 50× (B, D and F).
Although hamsters are not an inbred species, the level of genetic variability among golden Syrian hamsters is limited because they have been a closed, outbred colony since 1951. Furthermore, since hamsters demonstrate high levels of viral replication, pronounced and widespread histopathological findings in the lungs, and some clinical evidence of infection (Table 1), they are suitable for vaccine efficacy, immunoprophylaxis and treatment studies. Although the availability of immunological reagents for the hamster are limited, the use of real-time RT-PCR to study cytokine and chemokine responses following infection may reveal mechanisms of pathogenesis in this model and will lead to a better understanding of the pathogenesis of SARS in humans.
2.4. Ferrets
Ferrets (Mustela furo) support SARS-CoV replication and develop multifocal pulmonary lesions involving 5–10% of the surface area of the lung (ter Meulen et al., 2004). Ferrets infected with 103 or 104 TCID50 sustain virus replication at 106 TCID50/mL at day 4 p.i. (Martina et al., 2003, ter Meulen et al., 2004) Two groups of investigators have utilized the ferret model with somewhat different findings (Martina et al., 2003, ter Meulen et al., 2004, Weingartl et al., 2004b), suggesting that this model needs further characterization for vaccine efficacy and immunoprophylaxis studies. Clinical symptoms following SARS-CoV infection were reported in one study (Martina et al., 2003) but not in two others (ter Meulen et al., 2004, Weingartl et al., 2004b). Additional studies would help define the biological variability of this outbred species and the potential contribution of co-pathogens to the variability observed between the different studies. Finally although the ferret model has been used successfully for immunotherapy and prophylaxis studies (ter Meulen et al., 2004), further evaluation is needed to understand the poor efficacy and immunogenicity seen in MVA-vectored, SARS-vaccinated ferrets (Weingartl et al., 2004b) compared to that observed in other animals immunized with a similar vaccine (Bisht et al., 2004, Chen et al., 2005).
2.5. Non-human primates (NHP)
NHPs support viral replication and pneumonitis with variable clinical symptoms and pathology, depending upon the species. Various strains of SARS-CoV, including Urbani, PUMC-01 and HKU-39849, have been evaluated in rhesus macaques (McAuliffe et al., 2004, Qin et al., 2005, Rowe et al., 2004), cynomolgus macaques (Fouchier et al., 2003, Kuiken et al., 2003, Lawler et al., 2006, McAuliffe et al., 2004, Qin et al., 2006, Rowe et al., 2004), common marmosets (Greenough et al., 2005b), African green monkeys (McAuliffe et al., 2004), squirrel monkeys and mustached tamarins. Squirrel monkeys and mustached tamarins were not susceptible to SARS-CoV Urbani infection (authors’ unpublished data). Virus was not detected in the respiratory tract following inoculation and monkeys did not develop antibodies to SARS-CoV. All other NHP species studied were susceptible to infection to variable degrees. Of these susceptible species, no single model is preferred to any other. Each support viral replication in lungs (Table 2 ) and viral shedding in respiratory secretions and most develop some histopathological changes consistent with human infections including infection of type I and type II pneumocytes and diffuse alveolar damage. Few vaccine candidates have been evaluated in NHPs. The vaccines that were evaluated in NHPs were efficacious in reducing titers of wild-type SARS-CoV challenge virus replication and/or histopathological changes in immunized animals compared to mock-immunized controls. Due to individual variation in responses of outbred animals, small groups of monkeys (N = 4 or 5) are not large enough to evaluate statistical outcomes in these studies. Therefore, special considerations should be given before conducting vaccine efficacy studies in NHPs. Challenge studies should be conducted in large numbers (N = 10–12 per group), and consideration should also be given to the history of the animals used, utilizing the highest quality of monkeys (avoiding those that have had free-range periods in life or those that are known to be infected with co-pathogens) available (Roberts et al., 2006b). In vaccine efficacy studies, immunological responses should also be evaluated. Due to the lack of available immunological reagents (including micro-array assays) for other NHP species, rhesus and cynomolgus macaques are likely to be the most informative of the non-human primate models for continued SARS research. Despite the limitations of NHP models, evaluation of vaccine candidates in NHPs may be necessary to proceed to clinical studies. The cost for large studies that will provide the needed sample size and the other special considerations mentioned will limit these vaccine efficacy studies to the most promising candidates.
Table 2.
Pulmonary replication of SARS-CoV in various animal models
| Model (species) | Input virus (dose) | Viral titer (per gram) achieved in lung | Peak virus titer (day p.i.) | Reference(s) |
|---|---|---|---|---|
| Mouse (BALB/c) | Urbani (105 TCID50) | 107 TCID50 | 2 | Roberts et al. (2005a), Subbarao et al. (2004) |
| Mouse (129SvEv) | Tor2 (107 TCID50) | 106 TCID50 | 3 | Hogan et al. (2004) |
| Urbani (105 TCID50) | 106 TCID50 | 3 | Authors’ unpublished data | |
| Mouse (C57BL6) | Urbani (104 TCID50) | 107 TCID50 | 3 | Glass et al. (2004) |
| Hamster (golden Syrian) | Urbani | 107 TCID50 | 2 | Roberts et al. (2005b) and authors’ unpublished data |
| HKU-39849 | 107 TCID50 | |||
| Frankfurt 1 | 108 TCID50 | |||
| icGD03 (103 TCID50) | 104 TCID50 | |||
| Ferret | HKU-39849 (103–106 TCID50) | 106 TCID50a | 4 and 7 | Martina et al. (2003), ter Meulen et al. (2004) |
| NHP (cynomolgus) | HKU-39849 (106 TCID50) | 105.5 TCID50a,b | 6 | Kuiken et al. (2003)c |
| NHP (AGM) | Urbani (106 TCID50) | 107.2 TCID50a,d | 2 and 4 | McAuliffe et al. (2004) |
| NHP (rhesus) | PUMC01 (105 TCID50) | NRa,e | 2, 5, and 7 | Qin et al. (2005) |
| NHP (marmoset) | Urbani (106 TCID50) | NDf | 2 and 4 | Greenough et al. (2005b) |
Time corresponding to peak virus titer has not been determined; titers, if given, are for indicated day in bold-type, other days sampled are also given.
Two of four macaques had undetectable levels of virus by viral titration assays; one had a titer of 105 TCID50 and one a titer of 106 TCID50/ml.
Additional studies: Fouchier et al. (2003) reported virus in lung tissue by tissue culture (actual titer not reported) in one of two infected macaques, euthanized at day 6 p.i.; McAuliffe et al. (2004) reported peak virus titers in tracheal lavage samples at 101.1 TCID50/g on day 2 p.i. in one of four macaques; Lawler et al. (2006) reported detection of virus genome by RT/PCR in pharyngeal swabs intermittently at days 2–10 p.i. in eight of eight macaques tested.
Peak titers were detected in right lobes of the lung and were much higher that titers from tracheal lavage samples collected on the same day.
NR: not reported; five of eight macaques had virus isolated from combined nose and throat swabs, but actual titers are not reported.
ND: none detected; virus was not isolated but viral genome was detected in lung homogenates by RT/PCR at the indicated times.
2.6. Summary of animal models
Although additional reports exist on other animal models such as farmed masked palm civets, guinea pigs, and voles (Chepurnov et al., 2004, Gao et al., 2005, Wu et al., 2005), there are insufficient data available on these models to recommend their use in the evaluation of SARS-CoV vaccines and antiviral drugs. Use of these models in such evaluations would necessitate a thorough characterization of the model, including viral replication data and histopathological analysis of SARS-CoV- and mock-infected animals. Different species may prove useful for studying different aspects of SARS-CoV, and the study of pathogenesis may best be addressed in several species for which tools are available for immunological analysis, whereas vaccines and antivirals may be studied in models that allow large enough numbers for statistical evaluation.
The best animal model would be one that mimics human disease including features such as comparable levels of mortality to that seen in humans (∼10%) and increased susceptibility in older animals. It would likely be possible to develop such a model by increasing virulence of SARS-CoVs for various species, as has been done for influenza A, influenza B and Ebola viruses. Such efforts are underway in mice and NHPs.
3. Vaccines and immunotherapy for SARS
Although all the correlates of protection from SARS associated disease have not been identified in human infections, neutralizing antibodies directed at SARS-CoV spike (S) protein are present in convalescent human serum (J.S. Zhang et al., 2005) and have been detected in experimentally infected animals (Fouchier et al., 2003, Martina et al., 2003, McAuliffe et al., 2004, Qin et al., 2005, Roberts et al., 2005a, Roberts et al., 2005b, Rowe et al., 2004, Subbarao et al., 2004, Weingartl et al., 2004b, Wu et al., 2005). In animal studies, neutralizing antibody provides protection from re-infection with SARS-CoV (Subbarao et al., 2004).
3.1. Immunoprophylaxis and therapy
Prophylactically administered monoclonal antibodies specific to the SARS spike protein and passive transfer of SARS-CoV hyper-immune sera to naive mice (Greenough et al., 2005a, Subbarao et al., 2004, Sui et al., 2004, Traggiai et al., 2004, X. Wang et al., 2005), hamsters (Roberts et al., 2006a) and ferrets (ter Meulen et al., 2004) prevent or reduce SARS-CoV replication and associated disease following challenge. Monoclonal antibodies specific to the SARS spike protein administered therapeutically (i.e. after the onset of infection) prevent further increase in viral burden and reduce associated disease (e.g. consolidation) in hamsters (Roberts et al., 2006a). Sera from mice immunized with recombinant spike protein, inactivated whole SARS-CoV, live-viral vectored vaccines expressing the spike protein (VSV-S or MVA-S) or DNA vaccines (expressing SARS-S and -M proteins) have been shown to contain neutralizing antibodies to SARS-CoV (Table 3, Table 4 ). Passive transfer of these immune sera to naive mice prevents infection with SARS-CoV (Bisht et al., 2004, Kapadia et al., 2005, Stadler et al., 2005, Yang et al., 2004). Although cell-mediated immunity may have a protective role in viral clearance or resolution of disease in the human population, antibody alone has been shown to have both prophylactic and therapeutic benefits in animal models. The data from several studies strongly suggests that an effective SARS-CoV vaccine will be one that induces high and sustained levels of neutralizing antibodies and that administration of neutralizing MAbs specific to the SARS-CoV spike protein may be a useful strategy in post-exposure treatment and prophylaxis in at risk populations.
Table 3.
Neutralizing titers achieved following vaccination with
| Vaccine administered | Reciprocal neutralizing titer (log2) |
||
|---|---|---|---|
| Pre-bleed and Post-dose 1 | Post-dose 2 | Post-dose 3 | |
| Recombinant protein vaccine (Bisht et al., 2005) | |||
| Spikea + adjuvantb | <3 | 8.3 | 10.0 |
| L1Rc + adjuvantb | <3 | <3 | <3 |
| Inactivated, whole-virus vaccine (Stadler et al., 2005) | |||
| BPL-SARSd + saline | <3 | <3 | 6.0 |
| BPL-SARSd + adjuvante | <3 | 6.5 | 9.3 |
| BPL-Flud + adjuvante | <3 | <3 | <3 |
10 μg/dose, s.c.
Adjuvant = QS21 (saponin).
L1R = vaccinia virus L1R protein.
5 μg total protein/dose, s.c.
Adjuvant = MF59 (oil-in-water emulsion).
Table 4.
Summary of SARS-CoV vaccine candidates that have demonstrated immunogenicity or efficacy
| Animal model | Immunogena | Vaccine type | Evaluation | Challenge virus used in efficacy studies | References |
|---|---|---|---|---|---|
| Mouse | Spike | Recombinant protein (baculovirus) | Efficacy | Urbani | Bisht et al. (2005) |
| ±QS21 | |||||
| ±MPL/TDMb | |||||
| BPL(BJ01) | Inactivated WVc | Neutralizing Abs | ND | Tang et al. (2004) | |
| ±Al(OH)3 | |||||
| BPL-(FRA) | Inactivated WVc | Efficacy | Urbani | Stadler et al. (2005) | |
| ±MF59 | |||||
| BPL-(Tor2) | Inactivated WVc | Efficacy | Tor2 | See et al. (2006) | |
| Formaldehyde (GZ50) | Inactivated WV | Neutralizing Abs | ND | Qu et al. (2005) | |
| ±alumd | |||||
| ±CpG | |||||
| ±CTBe | |||||
| Formaldehyde (F69) | Inactivated WVc | Neutralizing Abs | ND | Xiong et al. (2004), C.H. Zhang et al. (2005) | |
| ±Freund's | |||||
| ±Al(OH)3 | |||||
| ±CpG | |||||
| UV (HKU-39849) | Inactivated WVc | Neutralizing Abs | ND | Takasuka et al. (2004) | |
| ±alumd | |||||
| Formaldehyde + UV (Utah) | Inactivated WVc | Efficacy | Utah | Spruth et al. (2006) | |
| ±Al(OH)3 | |||||
| DNA-Spike | DNA | Efficacy | Urbani | Yang et al. (2004) | |
| DNA-spike | Combination | Neutralizing Abs | ND | Woo et al. (2005) | |
| ±S peptide | |||||
| DNA-Spike | Combination | Neutralizing Abs | ND | Zakhartchouk et al. (2005) | |
| BPL(Tor2) + alumd | |||||
| VSV-S | Live-vectored | Efficacy | Urbani | Kapadia et al. (2005) | |
| MVA-S | Efficacy Neutralizing Abs | Urbani | Bisht et al. (2004) | ||
| MVA-S | Neutralizing Abs Efficacy | ND | Chen et al. (2005) | ||
| RV-S | ND | Faber et al. (2005) | |||
| AdS/N (Tor2) | Tor2 | See et al. (2006) | |||
| Hamster | Spike-(HKU-39849) ± Al(OH)3 | Recombinant protein | Efficacy | Urbani | Kam et al. (2007) |
| BHPIV3-S | Live-vectored | Efficacy | Urbani | Buchholz et al. (2004) | |
| Rabbit | DNA-spike | DNA | Neutralizing Abs | ND | S. Wang et al. (2005) |
| Ferret | rMVA-S (Tor2) | Live-vectored | Efficacy | Tor2 | Weingartl et al. (2004a) |
| NHPf | |||||
| Cyno. | BPL(BJ01) ± Al(OH)3 | Inactivated WVc | Efficacy | BJ01 | Qin et al. (2006) |
| Rhesus | BPL(BJ01) ± alumd | Inactivated WVc | Efficacy | GD01 | Qin et al. (2006) |
| Rhesus | Formaldehyde (ZJ01) | Inactivated WVc | Efficacy | NS1 | Zhou et al. (2005) |
| Rhesus | MVA-S | Live-vectored | Efficacy | PUMC01 | Chen et al. (2005) |
| Rhesus | AdS/N/M | Live-vectored | Neutralizing Abs | ND | Gao et al. (2003) |
| AGM | BHPIV3-S | Live-vectored | Efficacy | Urbani | Bukreyev et al. (2004) |
The SARS-CoV strain that was used as the immunogen is designated in parentheses. If strain is not designated it is the Urbani strain. Urbani (AY278741); Tor2 (AY274119); FRA (AY310120); Utah (AY714217); HKU-39849 (AY278491); BJ01 (AY278488); NS1 (AY508724); ZJ01 (AY297028); GD01 (AY278489); GZ50 (AY304495).
Sigma–Aldrich #M6536: 0.5 mg monophosphoryl lipid A (detoxified endotoxin) from S. minnesota (MPL) and 0.5 mg synthetic trehalose dicorynomycolate (TDM) in 2% oil (squalene)–Tween 80–water.
WV: whole-virus.
alum: Al2(SO4)3.
CTB: cholera toxin B subunit.
NHP: non-human primate; Cyno.: cynomolgus macaque (Macaca fascicularis), Rhesus: rhesus macaque (Macaca mulatta), AGM: African green monkey (Cercopithecus aethiops or Chlorocebus sabaeus).
3.2. Recombinant protein and inactivated whole-virus vaccines
The safety of vaccines should always be of paramount consideration and must always be empirically determined. Recombinant protein vaccines are well accepted as being among the safest vaccines although they are not always cost-effective. Recombinant protein vaccines are excellent at eliciting sterilizing immunity that can last up to several years depending upon the immunogenicity of the protein. These vaccines do not carry the risks of incomplete inactivation, genetic recombination with circulating viruses, or reversion to virulent phenotypes. Recombinant proteins are often also very stable. Furthermore, adjuvants such as aluminum salts (Al(OH)3 and Al2(SO4)3 or alum) or a squalene emulsion (MF59) (licensed for use in the US and Europe, respectively) may be added to the formulation of protein-based vaccines to increase immunogenicity.
The SARS-CoV spike (S) protein has been identified as the major target of neutralizing antibodies (Buchholz et al., 2004, Pang et al., 2004, Sui et al., 2005, S. Wang et al., 2005, Z.Y. Yang et al., 2005, H. Zhang et al., 2004, Y. Zhang et al., 2004, Zhao et al., 2004, Zhou et al., 2004). One study demonstrated neutralizing antibodies specific for the membrane (M) protein (Pang et al., 2004). The role of antibodies specific to the SARS-S protein in conferring protection has also been well established (Greenough et al., 2005a, Roberts et al., 2006a, Subbarao et al., 2004, Sui et al., 2005, ter Meulen et al., 2004, Traggiai et al., 2004, X. Wang et al., 2005). As such, vaccines containing recombinant SARS spike proteins or neutralizing epitopes of the spike protein should be fairly attractive. In fact, spike protein and trimers of spike proteins have been expressed from baculovirus or semliki-virus-defective particle expression systems, respectively, and were evaluated for their safety, immunogenicity and efficacy in murine and hamster models (Bisht et al., 2005, Kam et al., 2007). These proteins have demonstrated excellent immunogenicity (Table 3, Table 4) and efficacy in protection from challenge in animal models.
Inactivated, whole-virus vaccines have also been shown to be immunogenic and efficacious (Table 3, Table 4). Like recombinant protein vaccines, these vaccines contain substantial amounts of the spike protein, and they elicit neutralizing antibodies at levels similar to those elicited by recombinant protein vaccines (Table 3) that confer protection from subsequent challenge in vaccinated animals. Like recombinant protein vaccines, inactivated, whole-virus vaccines would likely have a relatively stable formulation. Potential benefits of inactivated, whole-virus vaccines over recombinant protein vaccines are that the conformation of the spike protein may be maintained and that antibodies to other viral proteins may contribute to protective immunity. Major disadvantages of inactivated, whole-virus vaccines would be the need to work with whole infectious virus in the preparation of the vaccine and the potential for incomplete inactivation in the processing and manufacturing of the vaccine.
3.3. Vectored vaccines
Although vaccines that elicit both humoral and cellular immunity may provide theoretical advantages, vaccines that produce a neutralizing antibody response may be sufficient for protection from SARS-CoV infection. Convalescent human sera had a mean plaque reduction neutralization titer of 1:61 with a normal distribution of titers around the mean (J.S. Zhang et al., 2005) but the titer of neutralizing antibodies necessary to achieve protection in a human population exposed to SARS-CoV is not known. A recombinant-, DNA-, or live-vectored vaccine, or a combination of these, have the advantage that laboratory workers are not exposed to live SARS-CoV during vaccine production.
Although DNA vectors have potential to be safe, stable and relatively inexpensive vaccines, they have not been proven to be highly efficacious or immunogenic in humans. When DNA vaccines expressing the SARS-S protein were evaluated in mice, they stimulated both humoral and cell-mediated immunity (Z. Wang et al., 2005, Yang et al., 2004) and were efficacious. Antibodies to the S gene were responsible for protection (Yang et al., 2004). SARS-CoV DNA vaccines have demonstrated greater potential in prime-boost regimens in species in which robust immunity is not induced by a DNA vector alone (Table 4). However, such combinations may lessen the economic benefit of a DNA-vector approach and necessitate licensure of more than one vaccine for human use.
Several live viruses have been tested as vectors to express SARS-CoV proteins, including BHPIV3, MVA, VSV and adenovirus (Table 4). Not all vectors were equally immunogenic, and efficacy data were not available from all the studies. Nevertheless, those vectors that elicited neutralizing antibodies and protective immunity were those that expressed the spike protein of SARS-CoV. Vectors expressing other SARS-CoV proteins including M and N did not confer protection from challenge with SARS-CoV, and the addition of the M and N genes did not enhance the immunogenicity or efficacy or the vaccine encoding the spike protein alone (Buchholz et al., 2004). In senescent mice, immunization with a vectored vaccine expressing the SARS-CoV N protein alone, or with co-administered spike protein. Enhanced pathology was seen following challenge (Deming et al., 2006). A summary of the vectored vaccines tested, their immunogenicity and protective efficacy and the animal models in which they were tested are listed in Table 4. Of all the live-vectored vaccines tested to date, only one construct (MVA-S) was variably immunogenic and variably efficacious. In ferrets, vaccination with this vector did not confer protection and may have led to formation of hepatic lesions following challenge with SARS-CoV. These findings contrast with other data reported using a MVA-S-vectored vaccine in mice and rhesus macaques where they were significantly immunogenic and efficacious. It is possible that the immunogenicity of MVA-vectored vaccines, like DNA vaccines varies in different species.
3.4. Combination vaccines
Finally, combination vaccines may prove to be more immunogenic than DNA vaccines, inactivated vaccines, or vectored vaccines alone. A thorough study of a combination of DNA and peptide vaccines demonstrated that such a regimen greatly enhanced the neutralizing antibody response to the SARS-S protein (Woo et al., 2005). However, the complexity of developing two systems and requiring a prime/boost regimen are disadvantages in comparison to a single immunization with a highly immunogenic vaccine. One may question whether a more complex regimen is warranted if a single vaccine administered once can produce sterilizing immunity.
4. The potential for enhanced disease following use of SARS vaccines
4.1. Enhancement of disease in vivo with the type-1 coronavirus FIPV and other viruses
Enhanced disease and mortality have been observed in kittens immunized against feline infectious peritonitis virus (FIPV), a type-I coronavirus, when they were subsequently exposed to FIPV (Vennema et al., 1990, Weiss and Scott, 1981). Enhanced disease associated with FIPV is not well understood but apparently is mediated by sub-neutralizing antibodies that facilitate viral entry into macrophages via an Fc-receptor-mediated mechanism (Corapi et al., 1992, Corapi et al., 1995, Hohdatsu et al., 1991, Hohdatsu et al., 1993, Olsen et al., 1992, Olsen et al., 1993). In the absence of serotype-specific protective antibodies or in the presence of low levels or sub-neutralizing levels of virus specific antibody, other human viruses including respiratory syncytial virus (RSV) and dengue virus demonstrate enhanced disease upon re-infection (Connors et al., 1992a, Connors et al., 1992b, Halstead, 2003, Kakuk et al., 1993, Murphy et al., 1990, Sullivan, 2001). Disease enhancement in cases of RSV occurs in the absence of protective titers of antibodies and is likely mediated by cellular immune responses that lead to immunopathology. Antibody dependent entry observed in dengue virus infections is similar to that which occurs in FIPV infection. In dengue virus infections, antibody enhanced entry occurs after previously infected individuals are re-infected with a second serotype of dengue virus. The antibodies to the first serotype of dengue virus are not protective against the second serotype, and virus entry into susceptible cells is facilitated through Fc-receptor-mediated mechanisms by these non-protective antibodies.
Although SARS-CoV antigen and nucleic acid have been detected in pulmonary macrophages in lungs from infected humans and experimentally infected animals, it is unclear whether this is due to phagocytosis of virions or to antibody- or receptor-mediated viral entry (Li et al., 2003, Yilla et al., 2005). Macrophages infected ex vivo do not support replication of SARS-CoV, a type-II coronavirus. However, as discussed below, there are some in vitro data to suggest that enhanced entry of SARS-CoV into susceptible cells may occur in the presence of non-neutralizing or sub-neutralizing levels of antibodies.
4.2. Enhancement of viral entry in vitro
Pseudo-typed lentiviruses, expressing the spike protein from a SARS-CoV isolated from civet cats, have demonstrated enhanced entry into a human renal adenocarcinoma cell line 786-O when incubated with antibodies to human SARS-CoV isolates (Z.Y. Yang et al., 2005). Enhancement was not observed with pseudo-typed viruses expressing the spike protein of a human SARS-CoV isolate. Enhancement has only been demonstrated at the level of virus entry and not in virus replication, and it is yet to be determined if enhanced replication occurs in the presence of sub-neutralizing levels of antibodies. Furthermore, these in vitro findings have not been linked to any known component of human disease or infection in vivo in animal models; this phenomenon has only been reported with pseudo-typed viruses and has not been observed with human isolates of SARS-CoV; and these observations in 786-O cells were not reproducible according to He et al. (2006). Further in vitro experiments examining productive or enhanced SARS-CoV (HK-39) replication in differentiated human macrophages in the presence or absence of SARS-specific antibodies demonstrated that SARS-CoV was able to enter these cells and that entry was not enhanced by antibody to SARS-CoV. Viable virus was not recovered from these macrophage cultures two days after viral entry, and qRT-PCR analysis showed little to no viral replication in these cells (ter Meulen et al., 2006). Ex vivo experiments examining SARS-CoV (Urbani and mouse-adapted strain MA15, Roberts et al., 2007) replication in primary murine (BALB/c, BL/6 and 129S6 strains) macrophages, isolated from the lungs, spleen and peritoneum, failed to demonstrate productive infection as well (Roberts et al., unpublished data).
4.3. Enhancement of disease in vivo with SARS-CoV
Several experiments have evaluated SARS-CoV infection in various animal models in the presence of neutralizing and sub-neutralizing levels of SARS-antisera or MAbs specific to SARS-CoV S protein. Evidence of disease enhancement has not been seen in any of the studies where appreciable levels of neutralizing antibodies were achieved following vaccination (See references in Table 4). Furthermore, enhanced disease has not been reported in respiratory tissues or in GI tissues, which are the primary sites of viral replication in SARS-CoV infection. In a single set of experiments (Weingartl et al., 2004a), ferrets were immunized with MVA-SARS-S via interperitoneal and subcutaneous routes with 108 pfu of vaccine per ferret at day 0, boosted with the same regimen at day 14, and challenged intranasally with 106 pfu/ferret of SARS-CoV (Tor2) at day 28. MVA-SARS-S vaccinated ferrets demonstrated low levels of neutralizing antibodies to SARS-CoV 1 week after the booster immunization (i.e. day 21; titer 1:40 or less). Strangely, neutralizing antibodies in this group were not detectable at day 28 (<1:20), prior to challenge and no significant difference was observed in the level of virus detected in pharyngeal swabs from animals vaccinated with MVA-SARS-S and control animals following challenge with SARS-CoV. Foci of hepatic necrosis were observed in MVA-SARS-S-vaccinated animals and to a lesser extent, in animals immunized with the MVA vector alone or PBS. It is unclear why the MVA-SARS-S vaccine was so poorly immunogenic (indicated by low level and transient detection of neutralizing antibodies) in ferrets since a similar vaccine was immunogenic and efficacious in mice and NHPs. It is also unclear why a memory antibody response was observed in MVA-SARS-S-immunized animals but was unable to neutralize virus or clear virus from these ferrets any faster than virus was cleared from mock-immunized ferrets. The possibility of co-pathogens as a cause for hepatitis was not examined, and it is possible that co-pathogens might have played a role in the observations reported in these ferrets. It is also possible that the observed phenomenon may be limited to a specific vector (MVA) or animal model (ferrets). Finally, although elevated levels of liver enzymes following SARS-CoV infection have been reported in human cases of SARS, hepatitis was rarely if ever reported in SARS patients (Z. Yang et al., 2005). Therefore, it is difficult to interpret the findings of hepatitis in ferrets and to determine if these findings have any relevance to the possibility of disease enhancement in SARS-vaccinated animals. Although unconfirmed, it may be possible that hepatic lesions observed in this study occur through cell-mediated mechanisms similar to that observed in the lungs in RSV infection following the use of a formalin inactivated vaccine. The finding of hepatitis following use of this MVA-vectored vaccine in ferrets merits further evaluation.
5. Summary
Although all of the correlates of immunity are not known for human cases of SARS or for current SARS animal models, much is now known about generating protection against SARS-CoV infection. Vaccines and monoclonal antibodies specific to SARS-CoV spike protein are highly efficacious in prophylaxis. Although concerns regarding potential enhancement of disease in previously vaccinated animals are being addressed experimentally, the body of literature supporting complete and partial protection in several animal models following a variety of vaccine strategies suggests that a successful SARS vaccine can be made.
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, NIAID. We acknowledge Dr. Marisa St. Claire and staff at Bioqual and Dr. Wun Ju Shieh from CDC, IDPA for participation and evaluation of SARS-CoV infection in squirrel monkeys and mustached tamarin monkeys.
References
- Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. U.S.A. 2004;101(17):6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht H., Roberts A., Vogel L., Subbarao K., Moss B. Neutralizing antibody and protective immunity to SARS coronavirus infection of mice induced by a soluble recombinant polypeptide containing an N-terminal segment of the spike glycoprotein. Virology. 2005;334(2):160–165. doi: 10.1016/j.virol.2005.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz U.J., Bukreyev A., Yang L., Lamirande E.W., Murphy B.R., Subbarao K., Collins P.L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. U.S.A. 2004;101(26):9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukreyev A., Lamirande E.W., Buchholz U.J., Vogel L.N., Elkins W.R., St Claire M., Murphy B.R., Subbarao K., Collins P.L. Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363(9427):2122–2127. doi: 10.1016/S0140-6736(04)16501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhang L., Qin C., Ba L., Yi C.E., Zhang F., Wei Q., He T., Yu W., Yu J., Gao H., Tu X., Gettie A., Farzan M., Yuen K.Y., Ho D.D. Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J. Virol. 2005;79(5):2678–2688. doi: 10.1128/JVI.79.5.2678-2688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepurnov A.A., Dadaeva A.A., Malkova E.M., Kolesnikov S.I., Sandakhchiev L.S. Symptoms of infection caused by SARS coronavirus in laboratory mice and guinea pigs. Dokl. Biol. Sci. 2004;397:310–313. doi: 10.1023/B:DOBS.0000039701.92375.b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors M., Collins P.L., Firestone C.Y., Sotnikov A.V., Waitze A., Davis A.R., Hung P.P., Chanock R.M., Murphy B.R. Cotton rats previously immunized with a chimeric RSV FG glycoprotein develop enhanced pulmonary pathology when infected with RSV, a phenomenon not encountered following immunization with vaccinia—RSV recombinants or RSV. Vaccine. 1992;10(7):475–484. doi: 10.1016/0264-410x(92)90397-3. [DOI] [PubMed] [Google Scholar]
- Connors M., Kulkarni A.B., Firestone C.Y., Holmes K.L., Morse H.C., III, Sotnikov A.V., Murphy B.R. Pulmonary histopathology induced by respiratory syncytial virus (RSV) challenge of formalin-inactivated RSV-immunized BALB/c mice is abrogated by depletion of CD4+ T cells. J. Virol. 1992;66(12):7444–7451. doi: 10.1128/jvi.66.12.7444-7451.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corapi W.V., Olsen C.W., Scott F.W. Monoclonal antibody analysis of neutralization and antibody-dependent enhancement of feline infectious peritonitis virus. J. Virol. 1992;66(11):6695–6705. doi: 10.1128/jvi.66.11.6695-6705.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corapi W.V., Darteil R.J., Audonnet J.C., Chappuis G.E. Localization of antigenic sites of the S glycoprotein of feline infectious peritonitis virus involved in neutralization and antibody-dependent enhancement. J. Virol. 1995;69(5):2858–2862. doi: 10.1128/jvi.69.5.2858-2862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming D., Sheahan T., Heise M., Yount B., Davis N., Sims A., Suthar M., Harkema J., Whitmore A., Pickles R., West A., Donaldson E., Curtis K., Johnston R., Baric R. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 2006;3(12):e525. doi: 10.1371/journal.pmed.0030525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber M., Lamirande E.W., Roberts A., Rice A.B., Koprowski H., Dietzschold B., Schnell M.J. A single immunization with a rhabdovirus-based vector expressing severe acute respiratory syndrome coronavirus (SARS-CoV) S protein results in the production of high levels of SARS-CoV-neutralizing antibodies. J. Gen. Virol. 2005;86(Pt 5):1435–1440. doi: 10.1099/vir.0.80844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423(6937):240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Tamin A., Soloff A., D’Aiuto L., Nwanegbo E., Robbins P.D., Bellini W.J., Barratt-Boyes S., Gambotto A. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362(9399):1895–1896. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Peng J.P., Deng W., Shi D., Bao L., Wang D., Zhang B., Qin C., Zhang Z. Infection of SARS-CoV on juvenile and adult Brandt's vole. Chinese Sci. Bull. 2005;50(12):1199–1204. doi: 10.1007/BF03183693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass W.G., Subbarao K., Murphy B., Murphy P.M. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 2004;173(6):4030–4039. doi: 10.4049/jimmunol.173.6.4030. [DOI] [PubMed] [Google Scholar]
- Greenough T.C., Babcock G.J., Roberts A., Hernandez H.J., Thomas W.D., Jr., Coccia J.A., Graziano R.F., Srinivasan M., Lowy I., Finberg R.W., Subbarao K., Vogel L., Somasundaran M., Luzuriaga K., Sullivan J.L., Ambrosino D.M. Development and characterization of a severe acute respiratory syndrome-associated coronavirus-neutralizing human monoclonal antibody that provides effective immunoprophylaxis in mice. J. Infect. Dis. 2005;191(4):507–514. doi: 10.1086/427242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenough T.C., Carville A., Coderre J., Somasundaran M., Sullivan J.L., Luzuriaga K., Mansfield K. Pneumonitis and multi-organ system disease in common marmosets (Callithrix jacchus) infected with the severe acute respiratory syndrome-associated coronavirus. Am. J. Pathol. 2005;167(2):455–463. doi: 10.1016/S0002-9440(10)62989-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead S.B. Neutralization and antibody-dependent enhancement of dengue viruses. Adv. Virus. Res. 2003;60:421–467. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- He Y., Li J., Li W., Lustigman S., Farzan M., Jiang S. Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein. J. Immunol. 2006;176(10):6085–6092. doi: 10.4049/jimmunol.176.10.6085. [DOI] [PubMed] [Google Scholar]
- Hogan R.J., Gao G., Rowe T., Bell P., Flieder D., Paragas J., Kobinger G.P., Wivel N.A., Crystal R.G., Boyer J., Feldmann H., Voss T.G., Wilson J.M. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J. Virol. 2004;78(20):11416–11421. doi: 10.1128/JVI.78.20.11416-11421.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohdatsu T., Nakamura M., Ishizuka Y., Yamada H., Koyama H. A study on the mechanism of antibody-dependent enhancement of feline infectious peritonitis virus infection in feline macrophages by monoclonal antibodies. Arch. Virol. 1991;120(3–4):207–217. doi: 10.1007/BF01310476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohdatsu T., Yamada H., Ishizuka Y., Koyama H. Enhancement and neutralization of feline infectious peritonitis virus infection in feline macrophages by neutralizing monoclonal antibodies recognizing different epitopes. Microbiol. Immunol. 1993;37(6):499–504. doi: 10.1111/j.1348-0421.1993.tb03242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuk T.J., Soike K., Brideau R.J., Zaya R.M., Cole S.L., Zhang J.Y., Roberts E.D., Wells P.A., Wathen M.W. A human respiratory syncytial virus (RSV) primate model of enhanced pulmonary pathology induced with a formalin-inactivated RSV vaccine but not a recombinant FG subunit vaccine. J. Infect. Dis. 1993;167(3):553–561. doi: 10.1093/infdis/167.3.553. [DOI] [PubMed] [Google Scholar]
- Kam Y.W., Kien F., Roberts A., Cheung Y.C., Lamirande E.W., Vogel L., Chu S.L., Tse J., Guarner J., Zaki S.R., Subbarao K., Peiris M., Nal B., Altmeyer R. Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcgammaRII-dependent entry into B cells in vitro. Vaccine. 2007;25(4):729–740. doi: 10.1016/j.vaccine.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia S.U., Rose J.K., Lamirande E., Vogel L., Subbarao K., Roberts A. Long-term protection from SARS coronavirus infection conferred by a single immunization with an attenuated VSV-based vaccine. Virology. 2005;340(2):174–182. doi: 10.1016/j.virol.2005.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D., Laman J.D., de Jong T., van Doornum G., Lim W., Ling A.E., Chan P.K., Tam J.S., Zambon M.C., Gopal R., Drosten C., van der Werf S., Escriou N., Manuguerra J.C., Stohr K., Peiris J.S., Osterhaus A.D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362(9380):263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler J.V., Endy T.P., Hensley L.E., Garrison A., Fritz E.A., Lesar M., Baric R.S., Kulesh D.A., Norwood D.A., Wasieloski L.P., Ulrich M.P., Slezak T.R., Vitalis E., Huggins J.W., Jahrling P.B., Paragas J. Cynomolgus macaque as an animal model for severe acute respiratory syndrome. PLoS Med. 2006;3(5):e149. doi: 10.1371/journal.pmed.0030149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wo J., Shao J., Zhu H., Wu N., Li M., Yao H., Hu M., Dennin R.H. SARS-coronavirus replicates in mononuclear cells of peripheral blood (PBMCs) from SARS patients. J. Clin. Virol. 2003;28(3):239–244. doi: 10.1016/S1386-6532(03)00195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina B.E., Haagmans B.L., Kuiken T., Fouchier R.A., Rimmelzwaan G.F., Van Amerongen G., Peiris J.S., Lim W., Osterhaus A.D. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425(6961):915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe J., Vogel L., Roberts A., Fahle G., Fischer S., Shieh W.J., Butler E., Zaki S., St Claire M., Murphy B., Subbarao K. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology. 2004;330(1):8–15. doi: 10.1016/j.virol.2004.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy B.R., Sotnikov A.V., Lawrence L.A., Banks S.M., Prince G.A. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3–6 months after immunization. Vaccine. 1990;8(5):497–502. doi: 10.1016/0264-410x(90)90253-i. [DOI] [PubMed] [Google Scholar]
- Olsen C.W., Corapi W.V., Ngichabe C.K., Baines J.D., Scott F.W. Monoclonal antibodies to the spike protein of feline infectious peritonitis virus mediate antibody-dependent enhancement of infection of feline macrophages. J. Virol. 1992;66(2):956–965. doi: 10.1128/jvi.66.2.956-965.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen C.W., Corapi W.V., Jacobson R.H., Simkins R.A., Saif L.J., Scott F.W. Identification of antigenic sites mediating antibody-dependent enhancement of feline infectious peritonitis virus infectivity. J. Gen. Virol. 1993;74(Pt 4):745–749. doi: 10.1099/0022-1317-74-4-745. [DOI] [PubMed] [Google Scholar]
- Pang H., Liu Y., Han X., Xu Y., Jiang F., Wu D., Kong X., Bartlam M., Rao Z. Protective humoral responses to severe acute respiratory syndrome-associated coronavirus: implications for the design of an effective protein-based vaccine. J. Gen. Virol. 2004;85(Pt 10):3109–3113. doi: 10.1099/vir.0.80111-0. [DOI] [PubMed] [Google Scholar]
- Qin C., Wang J., Wei Q., She M., Marasco W.A., Jiang H., Tu X., Zhu H., Ren L., Gao H., Guo L., Huang L., Yang R., Cong Z., Guo L., Wang Y., Liu Y., Sun Y., Duan S., Qu J., Chen L., Tong W., Ruan L., Liu P., Zhang H., Zhang J., Zhang H., Liu D., Liu Q., Hong T., He W. An animal model of SARS produced by infection of Macaca mulatta with SARS coronavirus. J. Pathol. 2005;206(3):251–259. doi: 10.1002/path.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin E., Shi H., Tang L., Wang C., Chang G., Ding Z., Zhao K., Wang J., Chen Z., Yu M., Si B., Liu J., Wu D., Cheng X., Yang B., Peng W., Meng Q., Liu B., Han W., Yin X., Duan H., Zhan D., Tian L., Li S., Wu J., Tan G., Li Y., Li Y., Liu Y., Liu H., Lv F., Zhang Y., Kong X., Fan B., Jiang T., Xu S., Wang X., Li C., Wu X., Deng Y., Zhao M., Zhu Q. Immunogenicity and protective efficacy in monkeys of purified inactivated Vero-cell SARS vaccine. Vaccine. 2006;24(7):1028–1034. doi: 10.1016/j.vaccine.2005.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu D., Zheng B., Yao X., Guan Y., Yuan Z.H., Zhong N.S., Lu L.W., Xie J.P., Wen Y.M. Intranasal immunization with inactivated SARS-CoV (SARS-associated coronavirus) induced local and serum antibodies in mice. Vaccine. 2005;23(7):924–931. doi: 10.1016/j.vaccine.2004.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Paddock C., Vogel L., Butler E., Zaki S., Subbarao K. Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. J. Virol. 2005;79(9):5833–5838. doi: 10.1128/JVI.79.9.5833-5838.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Vogel L., Guarner J., Hayes N., Murphy B., Zaki S., Subbarao K. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J. Virol. 2005;79(1):503–511. doi: 10.1128/JVI.79.1.503-511.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Thomas W.D., Guarner J., Lamirande E.W., Babcock G.J., Greenough T.C., Vogel L., Hayes N., Sullivan J.L., Zaki S., Subbarao K., Ambrosino D.M. Therapy with a severe acute respiratory syndrome-associated coronavirus-neutralizing human monoclonal antibody reduces disease severity and viral burden in golden Syrian hamsters. J. Infect. Dis. 2006;193(5):685–692. doi: 10.1086/500143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Wood J., Subbarao K., Ferguson M., Wood D., Cherian T. Animal models and antibody assays for evaluating candidate SARS vaccines: summary of a technical meeting, August 25–26, 2005, London, UK. Vaccine. 2006;24(49–50):7056–7065. doi: 10.1016/j.vaccine.2006.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Deming D., Paddock C.D., Cheng A., Yount B., Vogel L., Herman B.D., Sheahan T., Heise M., Genrich G.L., Zaki S.R., Baric R., Subbarao K.A. Mouse-Adapted SARS-Coronavirus Causes Disease and Mortality in BALB/c Mice. PLoS Pathog. 2007;3(1):e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe T., Gao G., Hogan R.J., Crystal R.G., Voss T.G., Grant R.L., Bell P., Kobinger G.P., Wivel N.A., Wilson J.M. Macaque model for severe acute respiratory syndrome. J. Virol. 2004;78(20):11401–11404. doi: 10.1128/JVI.78.20.11401-11404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See R.H., Zakhartchouk A.N., Petric M., Lawrence D.J., Mok C.P., Hogan R.J., Rowe T., Zitzow L.A., Karunakaran K.P., Hitt M.M., Graham F.L., Prevec L., Mahony J.B., Sharon C., Auperin T.C., Rini J.M., Tingle A.J., Scheifele D.W., Skowronski D.M., Patrick D.M., Voss T.G., Babiuk L.A., Gauldie J., Roper R.L., Brunham R.C., Finlay B.B. Comparative evaluation of two severe acute respiratory syndrome (SARS) vaccine candidates in mice challenged with SARS coronavirus. J. Gen. Virol. 2006;87(Pt 3):641–650. doi: 10.1099/vir.0.81579-0. [DOI] [PubMed] [Google Scholar]
- Spruth M., Kistner O., Savidis-Dacho H., Hitter E., Crowe B., Gerencer M., Bruhl P., Grillberger L., Reiter M., Tauer C., Mundt W., Barrett P.N. A double-inactivated whole virus candidate SARS coronavirus vaccine stimulates neutralising and protective antibody responses. Vaccine. 2006;24(5):652–661. doi: 10.1016/j.vaccine.2005.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler K., Roberts A., Becker S., Vogel L., Eickmann M., Kolesnikova L., Klenk H.D., Murphy B., Rappuoli R., Abrignani S., Subbarao K. SARS vaccine protective in mice. Emerg. Infect. Dis. 2005;11(8):1312–1314. doi: 10.3201/eid1108.041003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W.J., Zaki S., Murphy B. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 2004;78(7):3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U.S.A. 2004;101(8):2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Li W., Roberts A., Matthews L.J., Murakami A., Vogel L., Wong S.K., Subbarao K., Farzan M., Marasco W.A. Evaluation of human monoclonal antibody 80R for immunoprophylaxis of severe acute respiratory syndrome by an animal study, epitope mapping, and analysis of spike variants. J. Virol. 2005;79(10):5900–5906. doi: 10.1128/JVI.79.10.5900-5906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N.J. Antibody-mediated enhancement of viral disease. Curr. Top. Microbiol. Immunol. 2001;260:145–169. doi: 10.1007/978-3-662-05783-4_8. [DOI] [PubMed] [Google Scholar]
- Takasuka N., Fujii H., Takahashi Y., Kasai M., Morikawa S., Itamura S., Ishii K., Sakaguchi M., Ohnishi K., Ohshima M., Hashimoto S., Odagiri T., Tashiro M., Yoshikura H., Takemori T., Tsunetsugu-Yokota Y. A subcutaneously injected UV-inactivated SARS coronavirus vaccine elicits systemic humoral immunity in mice. Int. Immunol. 2004;16(10):1423–1430. doi: 10.1093/intimm/dxh143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Zhu Q., Qin E., Yu M., Ding Z., Shi H., Cheng X., Wang C., Chang G., Fang F., Chang H., Li S., Zhang X., Chen X., Yu J., Wang J., Chen Z. Inactivated SARS-CoV vaccine prepared from whole virus induces a high level of neutralizing antibodies in BALB/c mice. DNA Cell Biol. 2004;23(6):391–394. doi: 10.1089/104454904323145272. [DOI] [PubMed] [Google Scholar]
- ter Meulen J., Bakker A.B., van den Brink E.N., Weverling G.J., Martina B.E., Haagmans B.L., Kuiken T., de Kruif J., Preiser W., Spaan W., Gelderblom H.R., Goudsmit J., Osterhaus A.D. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363(9427):2139–2141. doi: 10.1016/S0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E., Preiser W., Doerr H.W., Chow V.T., de Kruif J., Peiris J.S., Goudsmit J. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3(7):e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traggiai E., Becker S., Subbarao K., Kolesnikova L., Uematsu Y., Gismondo M.R., Murphy B.R., Rappuoli R., Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10(8):871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., de Groot R.J., Harbour D.A., Dalderup M., Gruffydd-Jones T., Horzinek M.C., Spaan W.J. Early death after feline infectious peritonitis virus challenge due to recombinant vaccinia virus immunization. J. Virol. 1990;64(3):1407–1409. doi: 10.1128/jvi.64.3.1407-1409.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Chou T.H., Sakhatskyy P.V., Huang S., Lawrence J.M., Cao H., Huang X., Lu S. Identification of two neutralizing regions on the severe acute respiratory syndrome coronavirus spike glycoprotein produced from the mammalian expression system. J. Virol. 2005;79(3):1906–1910. doi: 10.1128/JVI.79.3.1906-1910.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ni B., Du X., Zhao G., Gao W., Shi X., Zhang S., Zhang L., Wang D., Luo D., Xing L., Jiang H., Li W., Jiang M., Mao L., He Y., Xiao Y., Wu Y. Protection of mammalian cells from severe acute respiratory syndrome coronavirus infection by equine neutralizing antibody. Antivir. Ther. 2005;10(5):681–690. [PubMed] [Google Scholar]
- Wang Z., Yuan Z., Matsumoto M., Hengge U.R., Chang Y.F. Immune responses with DNA vaccines encoded different gene fragments of severe acute respiratory syndrome coronavirus in BALB/c mice. Biochem. Biophys. Res. Commun. 2005;327(1):130–135. doi: 10.1016/j.bbrc.2004.11.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl H., Czub M., Czub S., Neufeld J., Marszal P., Gren J., Smith G., Jones S., Proulx R., Deschambault Y., Grudeski E., Andonov A., He R., Li Y., Copps J., Grolla A., Dick D., Berry J., Ganske S., Manning L., Cao J. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J. Virol. 2004;78(22):12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl H.M., Copps J., Drebot M.A., Marszal P., Smith G., Gren J., Andova M., Pasick J., Kitching P., Czub M. Susceptibility of pigs and chickens to SARS coronavirus. Emerg. Infect. Dis. 2004;10(2):179–184. doi: 10.3201/eid1002.030677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R.C., Scott F.W. Pathogenesis of feline infetious peritonitis: pathologic changes and immunofluorescence. Am. J. Vet. Res. 1981;42(12):2036–2048. [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Tsoi H.W., Chen Z.W., Wong B.H., Zhang L., Chan J.K., Wong L.P., He W., Ma C., Chan K.H., Ho D.D., Yuen K.Y. SARS coronavirus spike polypeptide DNA vaccine priming with recombinant spike polypeptide from Escherichia coli as booster induces high titer of neutralizing antibody against SARS coronavirus. Vaccine. 2005;23(42):4959–4968. doi: 10.1016/j.vaccine.2005.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Tu C., Xin C., Xuan H., Meng Q., Liu Y., Yu Y., Guan Y., Jiang Y., Yin X., Crameri G., Wang M., Li C., Liu S., Liao M., Feng L., Xiang H., Sun J., Chen J., Sun Y., Gu S., Liu N., Fu D., Eaton B.T., Wang L.F., Kong X. Civets are equally susceptible to experimental infection by two different severe acute respiratory syndrome coronavirus isolates. J. Virol. 2005;79(4):2620–2625. doi: 10.1128/JVI.79.4.2620-2625.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong S., Wang Y.F., Zhang M.Y., Liu X.J., Zhang C.H., Liu S.S., Qian C.W., Li J.X., Lu J.H., Wan Z.Y., Zheng H.Y., Yan X.G., Meng M.J., Fan J.L. Immunogenicity of SARS inactivated vaccine in BALB/c mice. Immunol. Lett. 2004;95(2):139–143. doi: 10.1016/j.imlet.2004.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428(6982):561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Xu M., Yi J.Q., Jia W.D. Clinical characteristics and mechanism of liver damage in patients with severe acute respiratory syndrome. Hepatobiliary Pancreat. Dis. Int. 2005;4(1):60–63. [PubMed] [Google Scholar]
- Yang Z.Y., Werner H.C., Kong W.P., Leung K., Traggiai E., Lanzavecchia A., Nabel G.J. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc. Natl. Acad. Sci. U.S.A. 2005;102(3):797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilla M., Harcourt B.H., Hickman C.J., McGrew M., Tamin A., Goldsmith C.S., Bellini W.J., Anderson L.J. SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res. 2005;107(1):93–101. doi: 10.1016/j.virusres.2004.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhartchouk A.N., Liu Q., Petric M., Babiuk L.A. Augmentation of immune responses to SARS coronavirus by a combination of DNA and whole killed virus vaccines. Vaccine. 2005;23(35):4385–4391. doi: 10.1016/j.vaccine.2005.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wang G., Li J., Nie Y., Shi X., Lian G., Wang W., Yin X., Zhao Y., Qu X., Ding M., Deng H. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J. Virol. 2004;78(13):6938–6945. doi: 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Yang, F., Li, Y.H., Li, W.H., Tu, X.M., Wei, Q., Zhu, H., Liu, L., Wang, H., Qin, C., Yuan, G.Y., He, W., Wang, S.H., 2004. [Potent neutralization antibody elicited in mice by SARS-associated coronavirus spike protein S1 domain]. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 18 (3), 258–260. [PubMed]
- Zhang C.H., Lu J.H., Wang Y.F., Zheng H.Y., Xiong S., Zhang M.Y., Liu X.J., Li J.X., Wan Z.Y., Yan X.G., Qi S.Y., Cui Z., Zhang B. Immune responses in BALB/c mice induced by a candidate SARS-CoV inactivated vaccine prepared from F69 strain. Vaccine. 2005;23(24):3196–3201. doi: 10.1016/j.vaccine.2004.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.S., Chen J.T., Liu Y.X., Zhang Z.S., Gao H., Liu Y., Wang X., Ning Y., Liu Y.F., Gao Q., Xu J.G., Qin C., Dong X.P., Yin W.D. A serological survey on neutralizing antibody titer of SARS convalescent sera. J. Med. Virol. 2005;77(2):147–150. doi: 10.1002/jmv.20431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Ke J.S., Qin Z.L., Ren H., Zhao L.J., Yu J.G., Gao J., Zhu S.Y., Qi Z.T. DNA vaccine of SARS-CoV S gene induces antibody response in mice. Acta. Biochim. Biophys. Sin. (Shanghai) 2004;36(1):37–41. doi: 10.1093/abbs/36.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Wang H., Luo D., Rowe T., Wang Z., Hogan R.J., Qiu S., Bunzel R.J., Huang G., Mishra V., Voss T.G., Kimberly R., Luo M. An exposed domain in the severe acute respiratory syndrome coronavirus spike protein induces neutralizing antibodies. J. Virol. 2004;78(13):7217–7226. doi: 10.1128/JVI.78.13.7217-7226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang W., Zhong Q., Hou W., Yang Z., Xiao S.Y., Zhu R., Tang Z., Wang Y., Xian Q., Tang H., Wen L. Immunogenicity, safety, and protective efficacy of an inactivated SARS-associated coronavirus vaccine in rhesus monkeys. Vaccine. 2005;23(24):3202–3209. doi: 10.1016/j.vaccine.2004.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]


