Summary
Ubiquilin/PLIC proteins belong to the family of UBL-UBA proteins implicated in the regulation of the ubiquitin-dependent proteasomal degradation of cellular proteins. A human presenilin-interacting protein, ubiquilin-1 has been suggested as potential therapeutic target for treating Huntington’s disease. Ubiquilin’s interactions with mono- and polyubiquitins are mediated by its UBA domain which is one of the tightest ubiquitin binders among known ubiquitin-binding domains. Here we report the three-dimensional structure of the UBA domain of ubiquilin-1 (UQ1-UBA) free in solution and in complex with ubiquitin. UQ1-UBA forms a compact three-helix bundle structurally similar to other known UBAs, and binds to the hydrophobic patch on ubiquitin with a Kd of 20 μM. To gain structural insights into UQ1-UBA’s interactions with polyubiquitin chains, we have mapped the binding interface between UQ1-UBA and Lys48- and Lys63-linked di-ubiquitins and characterized the strength of UQ1-UBA binding to these chains. Our NMR data show that UQ1-UBA interacts with the individual ubiquitin units in both chains in a mode similar to its interaction with mono-ubiquitin, though with an improved binding affinity for the chains. Our results indicate that, in contrast to UBA2 of hHR23A that has strong binding preference for Lys48-linked chains, UQ1-UBA shows little or no binding selectivity toward a particular chain linkage or between the two ubiquitin moieties in the same chain. The structural data obtained in this study provide insights into the possible structural reasons for the diversity of polyubiquitin chain recognition by UBA domains.
Keywords: ubiquilin, ubiquitin, polyubiquitin, UBA domain, protein-protein interaction
Introduction
Ubiquitination is a post-translational modification involved in the regulation of diverse cellular processes 1. It is defined as the covalent attachment of one or more ubiquitin (Ub) molecules to a surface lysine of a target protein via an isopeptide linkage. So formed single or multiple mono-Ub signals are involved in endocytic sorting as well as transcriptional regulation and DNA repair 2; 3; 4. Alternatively, the attachment could be in the form of a polyubiquitin (polyUb) chain comprising several Ub molecules. Depending on the length of the chain and the lysine residue involved in the isopeptide linkage between two successive Ubs, polyUb signal can be interpreted differently in the cell. For instance, K48-linked polyUb (with n ≥ 4) attached to a substrate protein acts as an efficient signal for proteasomal degradation 5, while K63-linked chains play regulatory role in DNA repair and signal transduction 6; 7; 8; 9. Although all seven lysines of Ub can serve as polyUb assembly sites in vivo 10, the functional role and significance of the other chains than K48- and K63-linked is still obscure.
Recent identification of ubiquitin-binding domains (UBD) was a major turning point in the understanding of the mechanisms of the recognition of Ub signals in the cell 11. So far, at least 16 different UBD classes have been identified 12; 13; 14. UBDs are small independently folded domains of larger proteins and are commonly capable of interacting with Ub although with broad spectrum of affinities (from 3 μM to 2 mM). Most of the UBDs interact with the hydrophobic patch on Ub surface, formed by L8, I44, and V70 15; 16. One of the well-characterized UBDs is the ubiquitin-associated (UBA) domain, a 40–50 amino acid motif, first identified bioinformatically 17 due to its occurrence in certain proteins that participate in protein ubiquitination. Deletion analyses have demonstrated that the Ub-binding acitivity of UBA domains has functional importance for their host proteins 18. In vitro binding studies have further proven that some UBA domains interact with polyUb chains in preference to monoUb 19; 20 and in accordance with their degenerate character (in terms of sequence variability) they can reveal diverse Ub and polyUb-binding properties 21.
The best studied UBA-domain containing protein is the human homolog of budding yeast Rad23 (hHR23A) which functions in nucleotide excision repair 22 and in proteasomal degradation of cellular proteins 23; 24; 25; 26. It consists of an N-terminal UBL domain and one internal and one C-terminal UBA domains (the latter are referred to here as hHR23A-UBA1 and hHR23A-UBA2, respectively). UBL-UBA proteins are described as Ub receptors of the proteasome that build the final contact between Ub-protein conjugates and the 26S-proteasome by their simultaneous interaction with proteasome, via UBL domain, and with ubiquitinated proteins, via UBA domain 11; 27. NMR studies revealed that each UBA domain of hHR23A folds into a compact three-helix bundle with a hydrophobic surface 28; 29 built up of a conserved MGF motif in loop 1 and several C-terminal residues of helix 3. A similar hydrophobic surface is present in all UBA domains and contributes to their interaction with Ub 18. Further characterization of the UBA domains of hHR23A revealed its preferred interaction with K48-linked polyUb chains 20. Structural details of this specificity and the mode of the UBA2’s interaction with K48-linked di-ubiquitin (Ub2) have been documented 30. These include the hydrophobic pocket formed by the L8-I44-V70 patches on both Ubs and extended through the isopeptide linker, combined with the conformational flexibility that allows Ub2 to wrap around and make hydrophobic contacts with both the front (MGF, helix 3) and the back (helix 2) sides of hHR23A-UBA2.
Human ubiquilin-1 (UQ1), also known as hPLIC1, is a presenilin-interacting protein 31 consisting of an N-terminal UBL and a C-terminal UBA domain. The UBA domain of UQ1 interacts with presenilin, and overexpression of UQ1 results in accumulation of presenilin level in cells 31; 32. PLIC proteins are also involved in the regulation of ubiquitin-dependent proteasomal degradation of cellular proteins 33; 34. For instance, hPLIC1 and hPLIC2 physically associate with both proteasomes and E3 ligases, and overexpression of these hPLIC proteins was shown to interfere with Ub-dependent degradation of p53 and IκBα. Furthermore, the UBA domain of hPLIC1 was reported to be responsible for the interaction of this protein with K7 protein of Kaposi’s sarcoma-associated herpesvirus. This interaction inhibits PLIC1 activity, resulting in rapid degradation of IκBα and p53 and, therefore, inhibition of p53-dependent apoptosis, potentially providing a favorable environment for viral reproduction 35. The exact mechanism of this effect, however, remains to be clarified. As another example, hPLIC1 was identified as an HCV NS5B (hepatitis C virus RNA-dependent RNA polymerase)-binding protein 36. This interaction occurs through the UBA domain of hPLIC1 and is required for the degradation of NS5B 36. In the case of hHR23A, another UBL-UBA protein, RNA-interference experiments agreed with its stimulatory effect on the proteolysis of p53 protein 37.
Recent pull-down experiments have demonstrated that, in contrast to hHR23A-UBA2, the UBA domain of ubiquilin-1 (UQ1-UBA) interacts with K48-Ub4, K63-Ub4, and a mixed Ub4 of K29/K6, equally well 21. In addition, the binding studies based on quantitative SPR analyses 21 revealed strong interaction of UQ1-UBA with monoUb with a Kd of 27 μM which is similar to that of the UBA domain of its yeast orthologue Dsk2 (15 μM) 38. Although UQ1-UBA interacts with the same hydrophobic surface patch of Ub as the UBA2 domain of hHR23A, preliminary data suggested that the strength of the interaction in each case is localized to different residues of the patch 21. Knowledge of UQ1-UBA’s structure and the atomic details of its interactions with mono- and polyUb are required to rationalize these findings.
Here we use NMR to understand the structural basis for the different binding properties of UQ1-UBA compared to hHR23A-UBA2. We present the three-dimensional structures of the UQ1-UBA domain and its complex with monomeric Ub. We also examine the mode of UQ1-UBA binding to K48- and K63-linked di-Ub chains and compare it to the binding of hHR23A-UBA2, in order to reveal the structural details of the diversity of these UBA domains.
Results
Solution structure of UQ1-UBA domain
The UQ1-UBA construct used in this study consists of 52 amino acid residues. The first ten residues and the last one do not belong to the UBA domain (see Materials and Methods). In total, 48 backbone amide signals are well resolved in the 1H-15N HSQC spectrum (Fig. 1). NMR signals of most of spin systems were assigned except for the first three residues (GSP) in the N-terminal extension. Solution structure of UQ1-UBA was determined based on the distance, torsional, and orientational constraints, as detailed in the Methods section. The ensemble of 10 lowest-energy structures is shown in Fig. 1; the secondary structure backbone RMSD within the ensemble is 0.30±0.10 Å. The statistics for the experimental constraints and for the derived ensemble of NMR structures are presented in Table 1.
Figure 1.
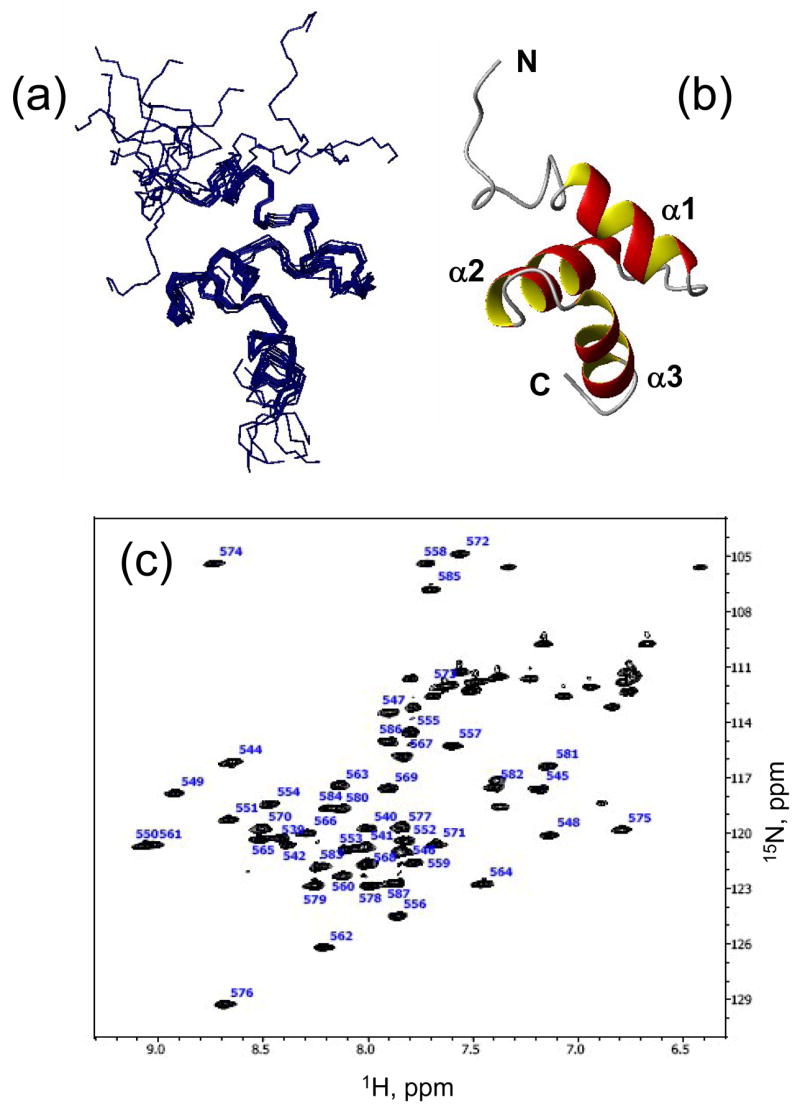
NMR-derived structures of the Ubiquilin1 UBA domain. (a) The ensemble of 10 solution NMR structures of UQ1-UBA, and (b) a ribbon representation of one of the structures. (c) The 1H-15N HSQC spectrum of UQ1-UBA domain. All molecular drawings in this paper were made using MolMol 69.
Table 1.
Statistics of the experimental constraints and the results of structure calculation for UQ1-UBA domain and its complex with monoUb
| UQ1-UBA | UQ1-UBA/Ub complex | ||
|---|---|---|---|
| Constraints | |||
| distance restraints | 1014 | ambiguous intermolecular | 19 active |
| restraints from CSP mapping | 16 passive | ||
| dihedral angle restraints | 66 | unambiguous distance restraints (from NOEs and PREs) | 14 |
| hydrogen bonds | 40 | RDC restraints (secondary structure) | 52 |
| RDC restraints | 48 | ||
| RMSDa (Å) | |||
| backbone, secondary struct. | 0.28 ± 0.07 | at the interface | 0.45 ± 0.15 |
| heavy atoms, secondary struct. | 0.76 ± 0.07 | UBA+Ub backbone all residues | 0.72 ± 0.35 |
| backbone, all residues | 2.19 ± 1.07 | UBA backbone all residues | 0.90 ± 0.35 |
| secondary structure residues | 0.32 ± 0.07 | ||
| heavy atoms, all residues | 2.36 ± 0.91 | Ub backbone all residues | 0.59 ± 0.28 |
| secondary structure residues | 0.46 ± 0.13 | ||
| PROCHECKa | |||
| most favored regions | 82.3 ± 2.81% | most favored regions | 89.1 ± 1.8% |
| allowed regions | 13.2 ± 3.84% | allowed regions | 8.7 ± 1.6% |
| generously allowed regions | 1.82 ± 2.34% | generously allowed regions | 0.8 ± 0.5% |
| disallowed regions | 2.72 ± 1.77% | disallowed regions | 1.3 ± 0.5% |
| G-factor overall | −0.051 ± 0.039 | G-factor overall | 0.29 ± 0.02 |
The RMSD and PROCHECK 69 analyses were performed for the ensemble of ten final lowest-energy structures.
The three-dimensional structure of UQ1-UBA is a compact three-helix bundle comprising helices α1 (residues Q548 to A556), α2 (R562-A571), and α3 (I576-L584) connected by loops 1 and 2 (Fig. 1). Side chains of several residues, L554, F559, L569, I580, and L583, contribute to the formation of the hydrophobic core of the protein; close contacts between these residues are evident from the NOESY spectra (not shown). This 3D fold is typical for UBA domains 18. The first 10 N-terminal residues in the construct studied here are structurally not well defined in the calculated ensemble due to their flexibility (see below). The agreement between the measured and back-calculated RDCs is extremely good, characterized by the Pearson’s correlation coefficient r of 0.997 and the quality R-factor 39 R=0.056 for the secondary structure residues (Supplementary Fig. 1). When all residues are included, the corresponding numbers are r=0.97 and R=0.17.
Mapping the UQ1-UBA/Ub binding interface
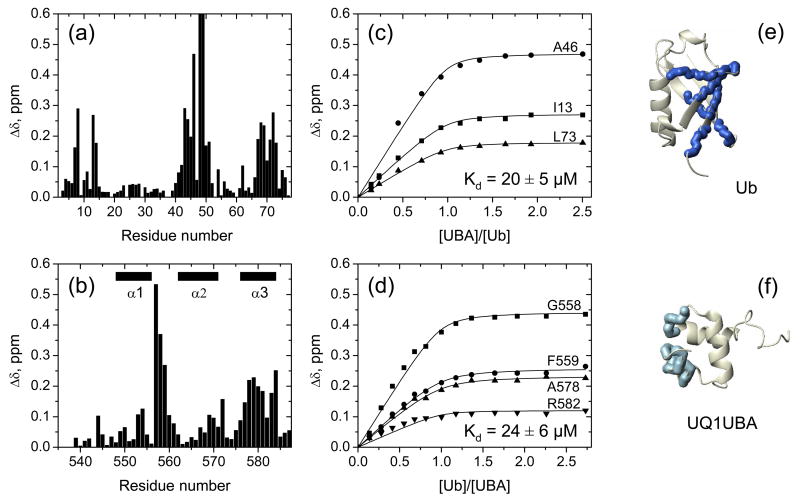
We then used NMR to characterize UQ1-UBA’s interaction with monoUb. Chemical shift perturbations (CSPs) provide a sensitive tool for identifying individual sites in a protein involved in ligand binding by monitoring shifts of the corresponding NMR signals upon ligand titration 40. The CSP profiles observed in both UQ1-UBA and Ub (Fig. 2) suggest a highly specific interaction between the two proteins, mediated predominantly by the hydrophobic contacts.
Figure 2.
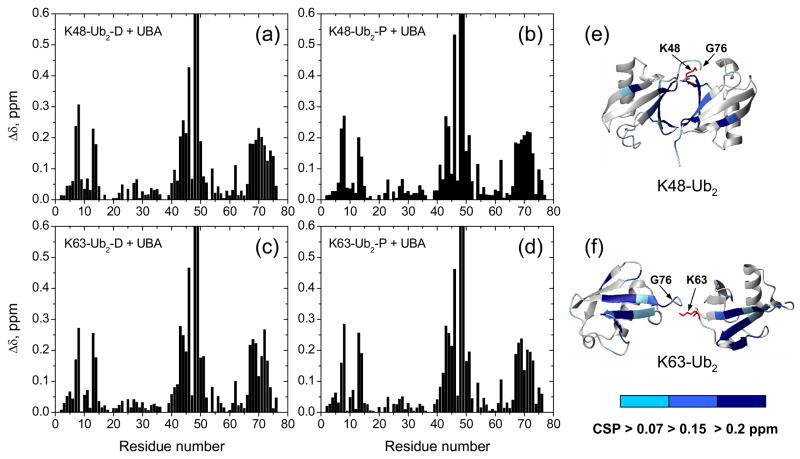
Chemical shift perturbation mapping of the interaction between UQ1-UBA and Ub. Amide CSPs in (a) monoUb and (b) UQ1-UBA at the endpoint of titration as a function of residue number. Panels (c) and (d) show representative titration curves for the perturbed residues in Ub and UQ1-UBA, respectively. These data depict the CSPs as a function of the molar ratio of the titrant and the 15N-labeled protein under the observation. The curves were obtained by fitting the CSPs to a 1:1 binding model (Methods). Panels (e) and (f) show a ribbon representation of the 3D structures of Ub and UQ1-UBA, with the residues exhibiting strong CSPs (Δδ >0.15ppm) shown in the CPK mode and colored blue and light blue, respectively. Note that the Δδ values for K48 (>1 ppm) and Q49 (~0.7 ppm) exceed the vertical scale used in panel (a).
Based on our CSP data, the binding surface on UQ1-UBA comprises residues M557-F559 in loop 1 and A578-L584 of α3. In addition, M557 and G558 also experienced strong signal attenuation (HSQC cross-peaks disappeared in the course of titration and then reappeared at the end), indicative of a slow exchange regime on the chemical shift time scale. Residues in helices 1 and 2, as well as in the N-terminal extension exhibited noticeably weaker CSPs, thus indicating that these elements of the UQ1-UBA construct are not involved in Ub binding.
The UQ1-UBA-binding surface on Ub includes three structural regions, involving residues T7, L8, I13, T14 (β1/β2 loop and β2 strand), L43-E51 (β3 and β4), and L67-L73 (β5), consistent with the role of the L8-I44-V70 hydrophobic patch 15; 16 as a target for UBA and other UBDs 12; 14. In addition, strong signal attenuations were observed in L8, I44, A46, K48, Q49, and H68, indicative of slow exchange on the chemical shift timescale, in agreement with the previous report 21. It is worth mentioning considerable CSPs in L50-E51 and strong signal attenuation in Q49; these residues located at the edge of the I44-centered hydrophobic patch were usually found negligibly perturbed in previous studies of UBA/Ub interactions.
Quantitative analysis of the CSPs in 15N-UQ1-UBA upon titration with monoUb (Fig. 2b,d) yields the dissociation constant (Kd) of 24±6 μM (Table 2), in agreement with 20±5 μM from the titration of 15N-Ub with UQ1-UBA (Fig. 2a,c and Table 2) and 27 μM from the surface plasmon resonance measurements 21.
Table 2.
Equilibrium dissociation constants (Kd) determined from the chemical shift perturbations detected in the ligand titration assays at pH6.8
| Sample | Titrant | Kd, μMa |
|---|---|---|
| 15N-UQ1-UBA | Ub (D77) | 24±6 |
| 15N-Ub (D77) | UQ1-UBA | 20±5 |
| 15N -UQ1-UBA | K48-Ub2 | 7±6 |
| K48-Ub2 (Proximal Ub) | UQ1-UBA | 4±5 |
| K48-Ub2 (Distal Ub) | UQ1-UBA | 12±10 |
| K63-Ub2 (Proximal Ub) | UQ1-UBA | 18±18 |
| K63-Ub2 (Distal Ub) | UQ1-UBA | 32±23 |
The Kd values reported here (mean±SD) were averaged over several residues that showed strong CSPs upon binding. The residues included in this analysis are: in Ub, T7, I13, T14, L43, I44, L50, L67, L69, L71 and L73; in UQ1-UBA, M557-L560, N577-L584. In the case of the 2:1 binding model the Kd values represent average microscopic values of the dissociation constant.
Intermolecular distance information from paramagnetic spin labeling
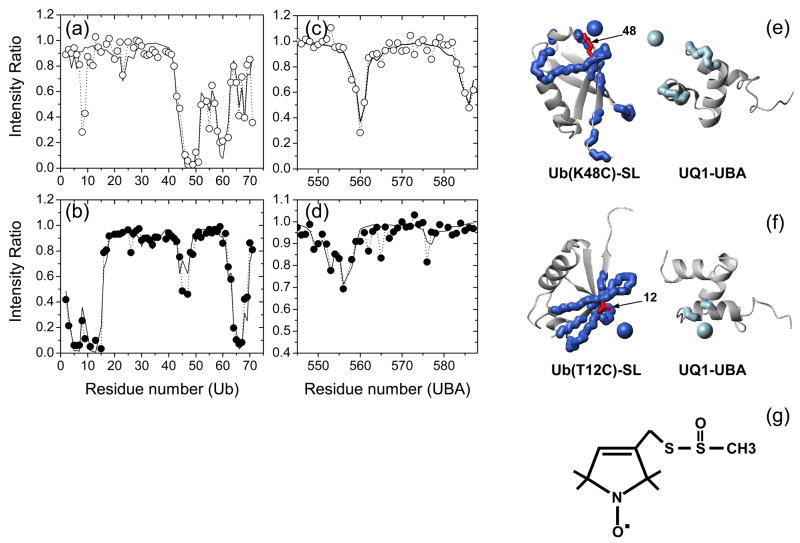
Paramagnetic relaxation enhancement (PRE) measurements were performed to determine the relative orientation of UQ1-UBA and Ub in the complex. The paramagnetic spin label (SL) causes strong signal attenuation in nuclei that are close in space; the magnitude of the PRE effect is inversely proportional to the sixth power of the distance from the unpaired electron.
Figure 3 depicts signal attenuations in both Ub and UQ1-UBA in the complex when the SL was attached to C48 on Ub (K48C). In Ub, the presence of SL clearly decreased the signal of C48 and the neighboring residues, as well as of the β1/β2 loop (T9, G10) and strand β5 (T66, H68, and R71) (Fig. 3a). Although the magnitude of signal attenuations in UQ1-UBA is smaller than in Ub residues around the SL’s attachment site, reflecting UBA’s larger distance from SL, the results clearly show the PRE effect on K560 (loop 1) and G585 (C-terminus of α3) (Fig. 3c). Based on the magnitude of the PREs and the location of the affected residues, we reconstructed the position of the SL with respect to Ub and UQ1-UBA (Fig. 3e), which was further utilized to derive intermolecular distance restraints (Materials and Methods) for refining the relative position and orientation of the two proteins in the complex (see below).
Figure 3.
Paramagnetic relaxation enhancement effect in the UQ1-UBA/Ub complex induced by the spin label. The top and bottom rows correspond to SL attached to Ub sites C48 and C12, respectively. (a, b) and (c, d) depict experimental PREs (circles) observed in SL-Ub and UQ1-UBA, respectively. The solid lines represent PREs back-calculated for the fitted SL position. Panels (e) and (f) show cartoon representations of the relative orientation of Ub and UQ1-UBA (similar to that in the calculated structure) with the reconstructed position of the unpaired electron of the SL shown as a sphere and the residues with significant PREs marked with thick backbone. The PRE-affected residues and the reconstructed spheres derived from measurements on SL-Ub are colored blue, those from UQ1-UBA measurements (in the presence of SL-Ub) are colored light blue. The side chains of K48 and T12 (mutated to a Cys for spin labeling) are shown in red sticks, as indicated. The reconstruction placed the unpaired electron of the SL at a distance of 7.2 Å from the Cα atom of K48 and at 8.8 Å from Cα of T12, the corresponding distances from Cβ atoms were 6.2 Å and 7.8 Å, respectively. Flexible unstructured C-terminal residues 72-76 in Ub and the 10 N-terminal residues in UQ1-UBA were excluded from the fits shown here; including these residues in the analysis had little effect on the SL positions. Also shown (panel (g)) is the chemical structure of the spin label used in this study.
Similar measurements were also conducted with the SL attached to C12 in Ub (T12C), which provided additional unambiguous intermolecular distance restraints for calculating the structure of the UQ1-UBA/Ub complex (Fig. 3b, d, and f).
It is worth pointing out that the observation of PREs in UQ1-UBA induced by a Ub-attached SL serves as an independent evidence of its binding to Ub, and the fact that these PREs are site-specific indicates a specific complex between Ub and UQ1-UBA. Note also that the attachment of the SL to Ub did not affect its binding to UQ1-UBA, as judged from the comparison of the NMR spectra of 15N-UQ1-UBA in the presence of Ub and SL-Ub (Supplementary Fig. 5).
Structure of the UQ1-UBA/monoUb complex
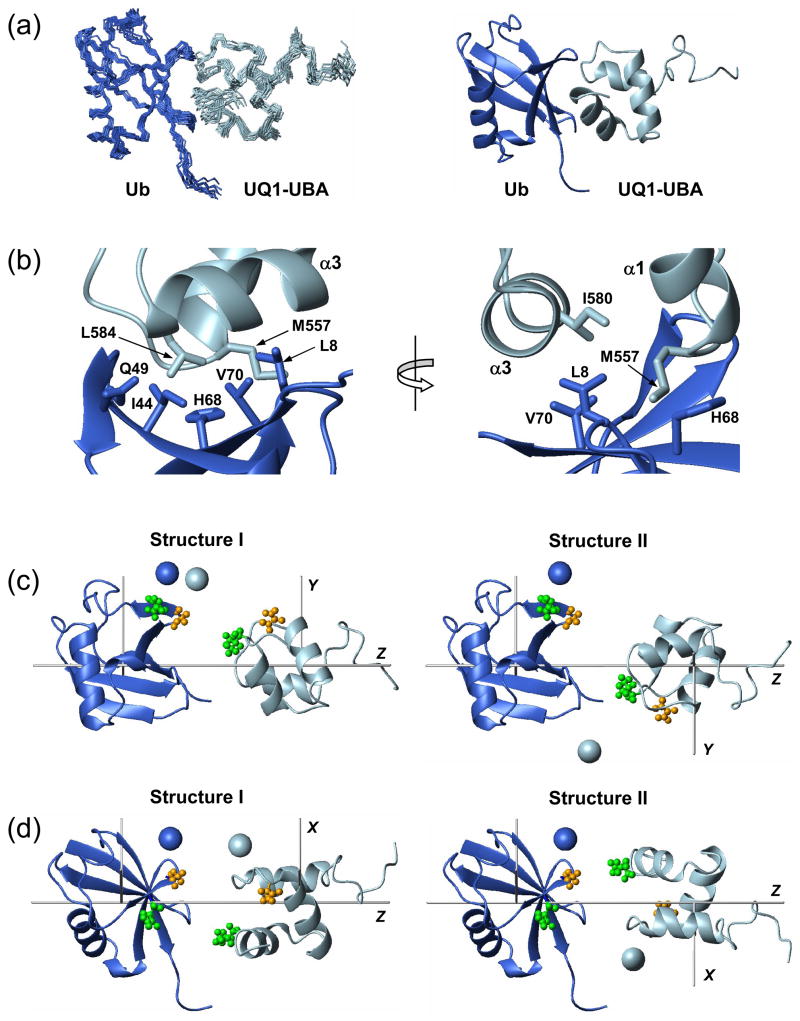
Structure of the UQ1-UBA/monoUb complex was obtained using protein-protein docking approach (HADDOCK41) based on the binding interface mapping data discussed above (Fig. 2). The relatively small magnitudes of the observed CSPs (Fig. 2) in both UQ1-UBA and Ub and the overall similarity of the NMR spectra in the free and bound states indicate that the gross overall fold of these proteins is not significantly perturbed by their binding. This justified the docking approach for determining the structure of the complex as well as the utilization of the structures of these proteins in the unbound state as starting material for docking. The ambiguous intermolecular constraints derived from the CSPs were complemented by long-range orientational constraints from residual dipolar couplings (RDCs) measured for the complex (see Methods). These measurements yielded 54 RDCs for Ub and 46 for UQ1-UBA. Only RDCs belonging to the elements of secondary structure were included as restraints in HADDOCK calculation of the complex. Only a few intermolecular NOEs could be reliably identified from the 15N-filtered NOESY spectrum of the complex of 2H-Ub with 15N-UQ1-UBA, due to signal overlap and the scarcity of NOE contacts between side-chain protons of UBA and amide protons of Ub. The unambiguously assigned NOEs (Q49/L584, G47/N561) were included, together with the PREs (see above) as unambiguous intermolecular distance restraints into the calculation. The details of these calculations are provided below. The average RMSD for the final ensemble of 10 structures of the UQ1-UBA/Ub complex was 1.33 ± 0.36 Å for the backbone atoms in the elements of secondary structure and 1.11 Å for the interface residues. The relevant statistics of the constraints and the calculated structural ensemble, as well as the evaluation of the calculated structures are shown in Table 1.
In the resulting UQ1-UBA/Ub complex (Fig. 4a), the two proteins are in a close contact mediated primarily by hydrophobic interactions. Residues M557 and G558 of loop 1 of UQ1-UBA interact with L8 and H68 of Ub. Helix α3 of UQ1-UBA which includes I580, E581, and L584, is in close contact with the hydrophobic patch residues (L8, I44, and V70), as well as Q49 (Fig. 4b). These contacts between the two proteins are in good agreement with the results of the CSP mapping (Fig. 2).
Figure 4.
The structure of the monoUb:UQ1-UBA complex. (a) The final ensemble of 10 lowest-energy NMR structures and a cartoon representation of the representative structure of the complex. (b) A close-up view of some intermolecular contacts at the Ub:UQ1-UBA binding interface. (c) Two possible relative orientations of Ub and UQ1-UBA agree with the RDC data. These conformations differ by a 180° rotation of one of the domains (in this case, UQ1-UBA) about the Z-axis of the alignment tensor; the grey rods indicate the principal axes of the tensor. Spheres represent the positions of the spin labels “reported” by the individual domains, determined from C48-spin labeling data (Fig. 3e). These structures were obtained from those in panel (a) by moving the two proteins away from each other along the Z-axis and rotating the complex around this axis by ~30°. (d) Same structures as in (c) (rotated around the Z axis) with the spin labels reconstructed from C12-SL experiments (Fig. 3f). The coloring of the SL-representing spheres in (c) and (d) is the same as in Fig. 3, i.e. the SL positions derived from measurements on SL-Ub and UQ1-UBA are colored blue and light blue, respectively. The pairs of residues that give intermolecular NOEs are shown in ball-and-stick in panels (c, d) and colored green (Q49 - L584) and orange (G47 - N561). Throughout this figure, Ub is colored blue and UQ1-UBA is light blue.
It is instructive to discuss here the effect of the experimental restraints on the calculated structures of the complex. Due to their orientational degeneracy, dipolar couplings alone cannot establish unambiguously the orientation of one protein with respect to the other in a complex 42; 43. In general, eight symmetry-related conformations of the Ub/UQ1-UBA complex are possible from solely RDC data (not shown). However, only two of them (schematically shown as conformations I and II in Fig. 4c,d) agree with the binding interface mapped by the CSP measurements (Fig. 2). The relative orientations of UQ1-UBA and Ub differ between the two conformations by a 180° rotation of one of these proteins (e.g. UQ1-UBA) about the principal axis (Z) of the alignment tensor. The initial docking calculation that included only ambiguous intermolecular constraints from the CSP mapping yielded the conformation-I-like structures as the most energetically favorable, although the convergence was poor. With the RDC data included, both conformations were among the calculated structures, although conformation II had a somewhat lower overall energy. However, as evident from Fig. 4c,d, this structure disagrees with the NOEs and spin labeling data; in this figure, panels (c) and (d) display the results obtained for the SL attached to C48 and C12 in Ub, respectively. Only after the unambiguous, NOE- and PRE-derived intermolecular distance restraints were included in the calculation, did the final lowest-energy ensemble contain exclusively type-I structures. Note that the PRE restraints derived independently from spin labeling at two Ub positions (12 and 48) located on the opposite sides of the hydrophobic patch and separated by ~20 Å agree with each other and with the observed intermolecular NOEs. The final structure of the complex is in excellent agreement with the RDC data for both proteins, with the overall correlation coefficient of 0.99 and the quality R-factor of 0.10 (Supplementary Fig. 1).
Independent validation of the derived structures of UQ1-UBA and the UQ1-UBA/monoUb complex using 15N relaxation measurements
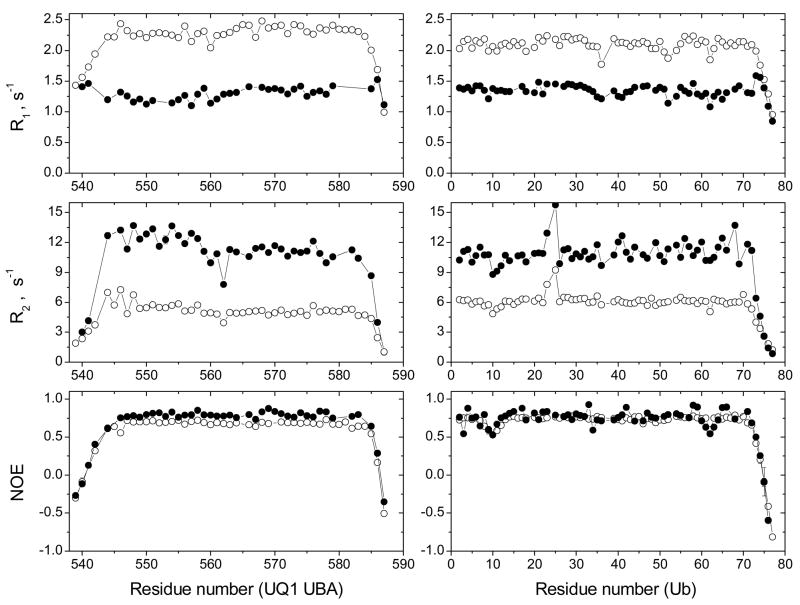
15N relaxation measurements in UQ1-UBA provided an independent verification of the calculated UQ1-UBA structure. The 15N R1, R2, and heteronuclear NOE in UQ1-UBA all show a decrease at both N and C termini (Fig. 5), indicating high backbone flexibility in these regions of the construct, consistent with the greater structural variability at the termini in the calculated structural ensemble (Fig. 1). It is worth mentioning here that the NOEs at the termini are below 0.5 (Fig. 5), and the squared order parameters are S2 < 0.5 (data not shown). In contrast, the backbone order parameters in the rest of the UQ1-UBA construct (S2=0.84±0.04 for residues 546–584 and 0.86±0.04 in the elements of secondary structure) are at the level typical for a well-folded protein, thus confirming that the structure of UQ1-UBA is well defined. Analysis using ROTDIF 44 program showed that the UQ1-UBA structure obtained here is in good agreement with the observed orientational dependence of the 15N relaxation rates. The overall rotational correlation time (Table 3) obtained from this analysis (3.30±0.13 ns) agrees well with the expected tumbling time of UQ1-UBA (3.56 ns predicted using HYDRONMR 45), thus confirming that UQ1-UBA behaves as a monomer in solution under these conditions.
Figure 5.
NMR characterization of the changes in the overall tumbling and backbone dynamics of UQ1-UBA and monoUb upon complex formation. Shown are the 15N relaxation rates (R1, R2) and hetero-NOE for both binding partners, UQ1-UBA (left panels) and Ub (right panels), in the free (open circles) and bound (solid circles) states.
Table 3.
Characteristics of the overall rotational diffusion derived from 15N relaxation data for Ub and UQ1-UBA, free in solution and in the complex, using ROTDIF analysis 44.
| Dxxa | Dyya | Dzza | αb | βb | τcc | |
|---|---|---|---|---|---|---|
| UBA (free) | 4.41 (0.10) | 4.41 (0.10) | 6.33 (0.51) | 157 (13) | 84 (10) | 3.30 (0.13) |
| Ub (free) | 3.49 (0.11) | 3.49 (0.11) | 4.47 (0.32) | 106 (5) | 151 (17) | 4.37 (0.17) |
| UBA (bound) | 1.55 (0.04) | 1.55 (0.04) | 2.54 (0.15) | 10 (8) | 91 (7) | 8.86 (0.29) |
| Ub (bound) | 1.64 (0.06) | 1.64 (0.06) | 2.44 (0.14) | 1 (5) | 83 (9) | 8.73 (0.36) |
| UBA/Ub complex | 1.57 (0.03) | 1.57 (0.03) | 2.53 (0.08) | 8 (4) | 86 (4) | 8.80 (0.20) |
The numbers in the parentheses represent estimated standard deviations for the corresponding parameters.
Principal values of the overall rotational diffusion tensor, in 107 s−1. The data presented here were obtained assuming an axially symmetric diffusion tensor. Detailed analysis (Supplementary Material) indicates that this model provides a significantly better fit than the isotropic tumbling model, whereas a fully anisotropic diffusion model was not statistically warranted in all these cases.
Euler angles (in degrees) describing the orientation of the principal axes frame of the overall rotational diffusion tensor with respect to the coordinate frame of a given protein.
The overall rotational correlation time, in 10−9 s.
The formation of the UQ1-UBA/monoUb complex is independently confirmed by the overall decrease in 15N R1 concomitant with an increase in 15N R2 (Fig. 5), indicating an increase in the overall tumbling time (hence the size) of the protein. These data allowed us to determine the stoichiometry of the complex. Thus, both UQ1-UBA and Ub in the bound state sense practically the same value of the 15N relaxation rate: R1(bound Ub) = R1(bound UQ1-UBA) = 1.3 s−1 and R2(bound Ub) = R2(bound UQ1-UBA) = 13 s−1. Since the measurements for the target protein were carried out when the titrant protein was in excess, we can conclude that the stoichiometry of the UQ1-UBA/Ub complex is 1:1. This qualitative conclusion is further supported by quantitative analysis of the relaxation data. Indeed, the overall tumbling time derived from ROTDIF analysis of the 15N relaxation data for UQ1-UBA and Ub in the complex was 8.86±0.29 ns and 8.73±0.36 ns, respectively; and a similar value (8.80±0.20 ns) was obtained when analyzing these data for both domains together (Table 3). The 2-fold increase in the tumbling time compared to the isolated Ub (4.4 ns) and UQ1-UBA (3.3 ns) is consistent with the expected size of a 1:1 complex. In fact, the overall tumbling time predicted based on the structure of the complex ranges from 9.7 ns if all residues are included to 8.5 ns when the 9 flexible unstructured N-terminal UBA residues are clipped. The 1:1 stoichiometry of the UQ1-UBA/Ub complex also agrees with the interface mapping data (Fig. 2) that show CSPs only on one side of each molecule.
Interestingly, in Ub-bound UQ1-UBA, the 15N R2/R1 ratio (which is sensitive to NH-bond’s orientation relative to the global rotational diffusion tensor 44) is systematically higher in helix α1 (11.2±0.7) than in helices α2 and α3 (8.4±0.4 and 7.9±0.4, respectively). This observation supports the calculated structure of the UQ1-UBA complex (Figs. 4a, 8a), in which helix α1 is oriented in the direction of the largest dimension of the complex (which determines the axis of the fastest tumbling of the molecule, thus the highest R2/R1 value 46), while the other two helices are almost perpendicular to this axis (hence lower R2/R1 values). Furthermore, there is an excellent agreement between the overall rotational diffusion tensors of the complex “reported” by the individual domains (Table 3) and the diffusion tensor derived using the calculated structure of the complex (i.e. analyzing both domains together). Because the 15N relaxation data were not included in the structure calculation, this agreement serves as an independent validation of the derived structure of the UQ1-UBA/Ub complex.
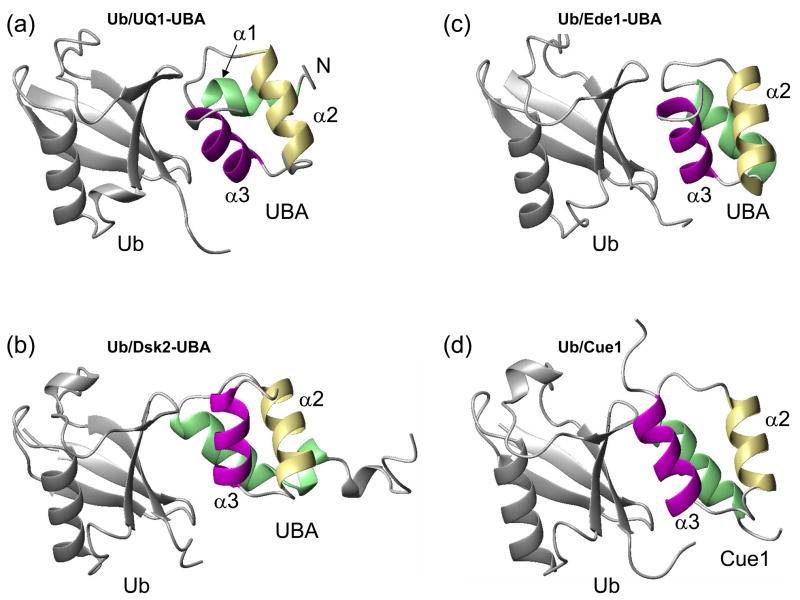
Figure 8.
Comparison of the structure of (a) UQ1-UBA/monoUb complex derived here with the published structures of monoUb complexes with (b) Dsk2-UBA, (c) Ede1-UBA, and (d) Cue2-CUE1 domains (PDB codes 1WR1, 2G3Q, and 1OTR, respectively). Helices in UBAs are colored green (α1), khaki (α2), and magenta (α3) to guide the eye.
It is worth pointing out that the termini of UQ1-UBA remain flexible in the bound state (Fig. 5), thus indicating that the additional N-terminal residues present in the construct used in this study are not involved in and hence do not affect Ub binding. This is also supported by a separate study (not shown) in which a shorter UBA construct missing the five N-terminal residues (QNPEV) induced very similar CSPs in Ub.
UQ1-UBA undergoes subtle structural changes upon binding to ubiquitin
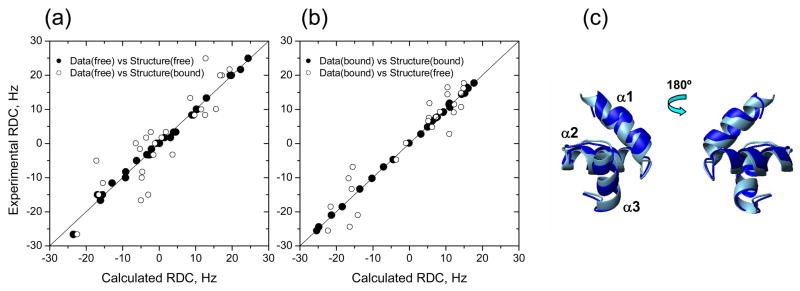
As mentioned above, the similarity of the 1H-15N NMR spectra of the free and Ub-bound UQ1-UBA suggests that the overall fold and the local electronic environment in the backbone are not significantly perturbed upon binding. Indeed, when the structures of the free and Ub-bound UQ1-UBA are superimposed (Fig. 6), it is difficult to discern their difference, but the backbone RMSD (0.92 Å for the secondary structure) is greater than the corresponding RMSDs within the NMR ensembles of the free (0.28 Å) and bound (0.32 Å) UQ1-UBA. A detailed analysis showed that Ub binding caused a slight reorientation of the UQ1-UBA helices, such that the inter-helical angles increased from 57° to 65° (α1/α2) and from 124° to 135° (α2/α3), while the α2/α3 angle remained practically unchanged (115° versus 118°).
Figure 6.
Comparison of the experimental and back-calculated RDCs derived from the UQ1-UBA structures indicates structural changes in UQ1-UBA upon Ub binding. The RDC data for the (a) free and (b) Ub-bound UQ1-UBA were fit to UQ1-UBA structures in the free and Ub-bound states. The solid line corresponds to absolute agreement. (c) Superimposition of the UQ1-UBA structures in the free (light blue) and Ub-bound (blue) states.
In order to verify that the differences between the two structures are real and not an artifact of the docking procedure (recall that no intra-molecular NOEs were included in the HADDOCK calculation), we used RDCs which are highly sensitive to bond orientations in a protein and thus provide the means to monitor subtle structural changes resulting from binding. For this purpose, we compared the experimental RDC values with those back-calculated for a given structure using program ALTENS 47. The analysis (Fig. 6, Supplementary Table 4) showed that although the 1H-15N RDCs measured for the complex are in excellent agreement with the structure of bound UQ1-UBA (the correlation coefficient r=0.999) they fit rather poorly (r=0.946) the structure of free UQ1-UBA. The disagreement is also evident from a significant increase in the quality R-factor 39, from 0.02 to 0.23. (It is worth emphasizing that this disagreement between the experimental RDC data measured in the bound state and the structure of free UBA is independent of the results of docking.) It is then not surprising that the RDCs measured in free UQ1-UBA, when compared to the structures of the free and Ub-bound UQ1-UBA, also show a significant decrease in the correlation coefficient (from 0.997 down to 0.88), accompanied by an increase in the R-factor from 0.05 to 0.33. All these results indicate a change in the UQ1-UBA structure upon binding to Ub. This conclusion is independently supported by our 15N relaxation data: the residuals of fit from ROTDIF analysis of the data measured in the bound state increased by ~3-fold when atom coordinates of bound UQ1-UBA were replaced with those for the free protein.
In contrast, Ub’s structure is much less affected by the complex formation (Supplementary Table 4). The RDCs for bound Ub fit well to both the free Ub structure and Ub in the complex, with the correlation coefficients of 0.994 and 0.997, respectively, and very low R-factors (0.06 and 0.04). The observed differences between Ub and UBA reflect greater structural rigidity/stability of the former, likely due to a much bigger number of hydrophobic contacts stabilizing its core (compared to UBA) and the network of hydrogen bonds rigidifying the β-sheet in Ub.
Exploring UQ1-UBA interactions with di-ubiquitin chains
We then used CSP mapping to determine and compare the UQ1-UBA-binding surface on both Ubs in K48- and K63-linked Ub2 chains. The primary goals of these studies were to understand the structural basis of the absence of chain-linkage selectivity in the case of UQ1-UBA domain 21 and to examine its binding preference toward the distal or the proximal Ub in each of these two chains. Our CSP mapping data (Fig. 7) indicate that, regardless of the linkage, UQ1-UBA binds directly to the hydrophobic patch on both Ubs in Ub2. The CSP patterns are similar between the two ubiquitins in the chain as well as between the two chains and show the largest perturbation in K48, followed by Q49 (Fig. 7). Combined with slow exchange in the K48 signal, this suggests strong UQ1-UBA binding to these sites in both chains. Moreover, there is a strong similarity between the UBA-induced CSPs in each Ub unit in these chains and those in monoUb (Fig. 2a). Also the pattern of the CSPs in UQ1-UBA upon binding to K48-Ub2 (Supplementary Fig. 2) is very similar to that in the complex with monoUb (Fig. 2b). These data suggest that intermolecular contacts between UQ1-UBA and the individual Ubs in these chains are practically the same as with monoUb.
Figure 7.
Chemical shift perturbation mapping of Ub2’s surface involved in UQ1-UBA binding. Shown are CSPs as a function of residue number in (a) distal and (b) proximal domains in K48-Ub2, (c) distal and (d) proximal domains in K63-Ub2, and (e, f) ribbon representations of the corresponding Ub2s colored by the strength of the CSPs. Note that the Δδ values for K48 (>1 ppm) and Q49 (~0.7 ppm) exceed the vertical scale used here. The location of G76 (distal Ub) and the side chain (shown in red stick) of K48 or K63 (proximal Ub) that form the isopeptide bond linking the two Ubs in Ub2 is indicated in (e) and (f).
The stoichiometry of the Ub2/UQ1-UBA binding was determined using longitudinal 15N relaxation time (T1) measurements. As illustrated elsewhere 30, the 15N T1 is a sensitive indicator of the rate of overall rotational diffusion of a protein and can be directly related to the protein’s size (hence molecular weight). For proteins with molecular weight above ~6 kDa (at these experimental conditions), i.e. comparable in size to or bigger than monomeric UQ1-UBA or Ub, the 15N T1 increases with the overall size of the molecule. A comparison of the 15N T1 values for various proteins studied here is shown in Table 4. The average 15N T1 for both K48-Ub2 and K63-Ub2 in the UQ1-UBA-bound state is consistent with the size of the complex expected for a 2:1 stoichiometry (UBA:Ub2). In addition, the titration curves from the Ub2/UQ1-UBA binding assays fit the best to a two-binding-sites model while those for monoUb/UQ1-UBA titration fit the best to the one-binding-site model.
Table 4.
Longitudinal 15N relaxation time data measured in various constructs studied here and the corresponding values of the molecular weight (MW).
| Sample | T1a, ms | MW range b (in kDa), estimated from 15N T1 | MW c (in kDa) expected | ||
|---|---|---|---|---|---|
| UQ1-UBA (free) | 432±12 | 5.0 – 6.8 | 5.7 | ||
| Ub (free) | 471±10 | 8.7 – 9.7 | 8.8 | ||
| K48-Ub2 (free) | 695 ± 32 d | 16.3 – 18.3 | 17.3 | ||
| K48-Ub2 (bound) | 1201 ± 71 (Proximal Ub) | 1037 ± 45 (Distal Ub) | 30.4 – 34.6 | 26.4 – 29.0 | 28.6 (Ub2+2xUBA) |
| K63-Ub2 (bound) | 1173 ± 61 (Proximal Ub) | 1026 ± 76 (Distal Ub) | 30.0 – 33.5 | 25.1 – 29.7 | 28.6 (Ub2+2xUBA) |
The T1 values reported here (mean ± standard deviation) are for secondary structure residues only.
The molecular weight range corresponds to a ± one standard deviation interval around the mean 15N T1.
The molecular weights reported here reflect 15N labeling.
From ref. 30.
The affinity of UQ1-UBA binding to Ub2 was assessed by fitting the CSPs in the individual residues measured in the course of the titration (Supplementary Fig. 3) assuming a 2:1 stoichiometry. The resulting microscopic Kd values (Table 2) indicate strong binding affinity of UQ1-UBA for both K63- and K48-linked Ub2.
The effect of the Ub-Ub linkage on di-ubiquitin/UQ1-UBA interaction
The effect of Ub2 linkage on Ub2/UQ1-UBA interaction was examined by comparing the CSPs and the Kd values for Ub units in Ub2 with those for monoUb. Although all Ub domains (as well as monoUb) show very similar CSP patterns indicating that the same regions are perturbed, detailed quantitative comparison revealed domain-specific differences reflecting differential effect of UQ1-UBA binding on the local structure of Ub (Supplementary Fig. 4).
In K63-Ub2, the strong similarity of the CSP profiles in both Ubs (Fig. 7, see also Supplementary Fig. 4) and their resemblance to the CSPs in monoUb agree with the extended conformation of this chain 48, in which the two Ubs behave as two separate entities upon UQ1-UBA binding (see also Fig. 9). The site-specific CSPs in K48-Ub2 show somewhat greater differences from those in monoUb and K63-Ub2 (Supplementary Fig. 4). This likely reflects the fact that UQ1-UBA binding to K48-Ub2 is associated with a transition of this chain from the predominantly closed conformation (with the hydrophobic Ub/Ub interface 47) to an open form with a UBA bound to each Ub domain. Note in this regard that the CSPs in both Ubs in K48-Ub2 are greater than the corresponding chemical shift differences between the open and closed forms of K48-Ub2 47, thus further supporting the 2:1 stoichiometry model in which each Ub unit in this chain binds a UQ1-UBA domain. The CSPs observed in K48-Ub2 also reflect the involvement of K48 (proximal Ub) and the C-terminus of the distal Ub in the isopeptide linkage, which could interfere with the UBA binding negatively (via steric effect) or positively, via additional Ub/UBA interactions. In fact, the somewhat higher affinity for K48-Ub2 and its proximal domain indicates that the isopeptide linkage has a positive effect on UBA binding.
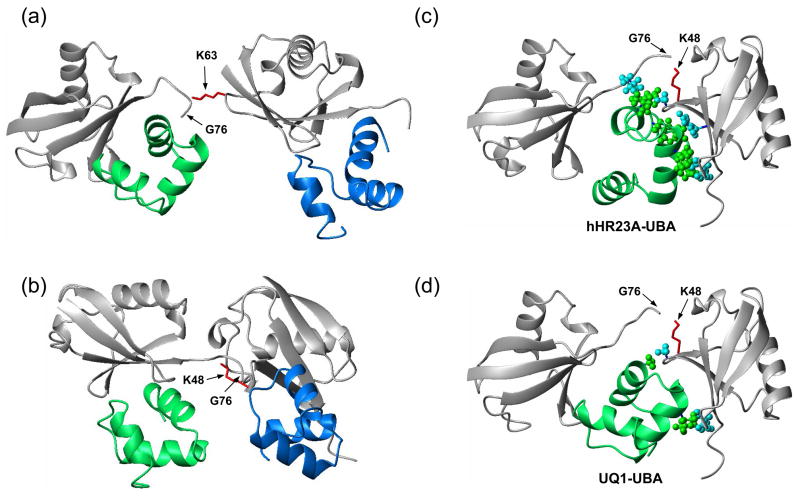
Figure 9.
Structural models show how UQ1-UBA can bind (a) K63- and (b) K48-linked Ub2 chains and help rationalize (c,d) the differences in linkage selectivity between UQ1-UBA and hHR23A-UBA2. The structures in (a, b) were obtained by superimposition of the UQ1-UBA/Ub complex onto the distal and proximal Ubs of each chain, i.e. assuming that UQ1-UBA interacts with each Ub unit in the same way as with monoUb. The latter is justified by the fact that the CSPs in each Ub in these chains upon UQ1-UBA binding are almost identical to those in monoUb (cf Figs 2a and 7). The K63-Ub2 structure is from ref. 48, the K48-Ub2 structure shown in (b) corresponds to a fully open conformation of the chain reported in refs. 55; 56. Comparison of the intermolecular contacts stabilizing the hHR23A-UBA2/K48-Ub2 complex 30 (panel (c)) with those in a hypothetical model of a similar complex for UQ1-UBA (panel (d)) shows that the interactions that favor the formation of a 1:1 complex in the former are missing in the latter. The structure model in panel (d) was obtained by replacing the distal-Ub/UBA2 pair in (c) with the Ub/UQ1-UBA structure determined in this study (Fig. 8a); this replacement is justified by the fact that the CSPs observed both in the distal Ub and in UQ1-UBA are essentially the same as in the monoUb/UQ1-UBA complex. In both structures (c,d), the “canonical” Ub-binding side (loop 1 and helix α3) of the corresponding UBA domain is in contact with the hydrophobic patch on the distal Ub, and only the side chains of residues forming contacts between the UBA and the proximal Ub or the Ub-Ub linker are shown in ball-and-stick, colored green (UBA) and cyan (Ub). In all these Ub2 structures (grey) the distal Ub is on the left, the proximal Ub is on the right. The location of G76 (distal Ub) and the side chain (shown in red stick) of K48 or K63 (proximal Ub) that form the isopeptide bond linking the two Ubs in Ub2 is indicated. The UBAs bound to the distal and proximal Ubs are colored green and blue, respectively.
Discussion
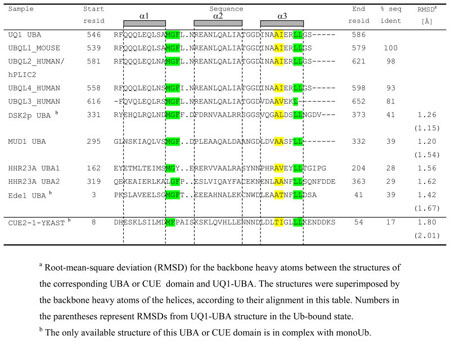
Sequence comparison indicates high conservation of residues F547 through G585 in the ubiquilin family (Table 5), suggesting the functional importance of the UBA domain. However, the sequence similarity to other UBAs, including that of Dsk2 (the yeast orthologue of UQ1-UBA), is rather low, and the structural similarity was not obvious until the UQ1-UBA structure has been determined in this study.
Table 5.
Sequence alignment of various UBA domains and a CUE domain. The sequence alignment of the non-ubiquilin UBAs is based on the superimposition of their structures (all three helices) with that of UQ1 UBA.
|
Our data show that UQ1-UBA forms a compact three-helix bundle structure typical for UBA domains. Other known structures include the UBA domains of hHR23A 28, Mud1 49, Dsk2 38, and Ede1 50. Despite the low sequence similarity among these UBAs, they all share some common sequence patterns (Table 5) including the MGF motif (loop 1) and a di-leucine motif at the end of α3, both of which reportedly are involved in Ub binding. The latter is in agreement with our CSP mapping data (Fig. 2) and the UQ1-UBA/Ub complex structure (Fig. 3). Despite the overall fold similarity with the other UBAs, UQ1-UBA has noticeable structural differences. Thus, the α1/α3 inter-helical angle in UQ1-UBA is 57° (65° in the bound state), which is close to that in Dsk2-UBA (52 °) but greater than in hHR23A’s UBA1 and UBA2 (44° and 27°, respectively) and Ede1-UBA (34°), and significantly different from the Cue2-CUE1 domain, where the two helices are almost parallel (19°) to each other (Fig. 8). The backbone heavy atoms of all 3 helices in UQ1-UBA superimpose with the corresponding atoms of the other UBAs with the RMSD values ranging from 1.2 Å (Mud1-UBA) to ~1.6 Å (hHR23A UBAs) to 1.8 Å (Cue2-CUE1) (Table 5). As in other UBAs, F559 makes close contacts with several residues, L554, L580, L583, and therefore is important to the formation/stabilization of the hydrophobic core of UQ1-UBA 28; 38.
Analysis of the structure of UQ1-UBA/Ub complex shows a close contact between the MGF motif of UBA and residues 46–48 of Ub and between helix α3 of UQ1-UBA and β-strands of Ub, particularly β4 and β5, which is similar to Ub complexes with Dsk2-UBA, Ede1-UBA, and CUE domains (Fig. 8). In contrast, helix α1 of UQ1-UBA points away from the hydrophobic surface of Ub and is not involved in any interaction with Ub. Unlike Dsk2-UBA and CUE domains, α3 of UQ1-UBA is packed across the β-sheet and oriented almost perpendicularly to the direction of the β-strands of Ub, such that L584 of UQ1-UBA makes hydrophobic contacts with side chains of I44, Q49, and V70 while E581 is posed to form a salt bridge with R72 and/or R74 of Ub. The combination of these latter interactions is unique to UQ1-UBA and could be the cause of its enhanced affinity for Ub. In fact, our CSP data show that Q49-E51 of Ub are significantly perturbed in both monoUb and Ub2 upon titration with UQ1-UBA, which, has not been reported in the case of CUE2-1/Ub, Dsk2-UBA/Ub, and Ede1-UBA/Ub studies.
Interestingly, despite the low sequence homology between the UBAs of UQ1 and Dsk2 (Table 5) their structures in the Ub-bound state superimpose relatively well (RMSD =1.15 Å). There is certain similarity in the contacts between these UBAs and Ub, which involve the MGF-motif’s interaction with the β3/β4 loop in Ub and helix α3 contacting I44 of Ub. However, the actual orientation of the two UBAs on Ub surface differs by ~48°, such that the secondary structure elements of the UQ1-UBA/Ub and Dsk2-UBA/Ub complexes superimpose rather poorly, with the backbone RMSD of 3.2 Å. Interestingly, a detailed analysis shows that the structure and inter-residue contacts in UQ1-UBA/Ub complex resemble those in the Ede1-UBA/Ub complex (Fig. 8), and both complexes superimpose rather well, with the backbone RMSD of 2.0 Å.
The Kd of ~20 μM (Table 2) measured here agrees with the previous study 21 that concluded that UQ1-UBA domain is one of the tightest Ub binders. This Kd value is also similar to 14.8 μM reported for Dsk2-UBA 38. Both UQ1-UBA and Dsk2-UBA bind to Ub tighter than most of the known UBDs, including Ede1-UBA (89 μM), CUE2-1 (150 μM), hHR23A-UBA2 (400–500 μM), hHR23B-UBAs (310–360 μM) 51, and Mud1-UBA (390 μM).
UQ1-UBA showed a somewhat higher affinity for Ub2 over monoUb, regardless of the linkage (Table 2). These results generally agree with the surface plasmon resonance data 21 in which UQ1-UBA binds to Ub4 stronger (Kd=1.2 μM for K48-Ub4 and 0.5 μM for K63-Ub4) than to monoUb (Kd=27 μM). However, the difference in the microscopic Kd values in Table 2 for monoUb and Ub2s is not dramatic. Combined with the similarity between the perturbation maps for monoUb and Ub2s (Figs. 2 and 7), this suggests that UQ1-UBA does not strongly discriminate between monoUb and a Ub unit in the context of Ub2. Given that Ub monomer appears to be the unit of recognition of UQ1-UBA in polyUb, the higher affinity of UQ1-UBA for Ub2 and longer chains could be caused by an increase in the effective local concentration of Ub in the chains (i.e. when a UBA dissociates from Ub, it has a higher probability to bind to a Ub unit in the same chain compared to binding to a Ub in another chain). The Ub-Ub linkage could also play a role here, particularly in the case of K48-linked chains, because of the involvement of K48 and the neighboring residues in the Ub/UQ1-UBA interaction. In this regard, it is instructive to compare UQ1-UBA binding to Ub2 with that of hHR23A-UBA2. As mentioned above, UQ1-UBA shares certain structural similarity with hHR23A-UBA2 (Table 5). However, our binding studies revealed a striking difference in their interaction with Ub. HHR23A-UBA2 binds monoUb weakly (Kd ~ 400–500 μM 48; 52), but the presence of the extended hydrophobic pocket in K48-Ub2 allows it to bind UBA2 in a sandwich-like manner, which greatly enhances its binding affinity (Kd ~ 8 μM) 30. In contrast, UQ1-UBA binds very tightly already to monoUb. Moreover, the perturbed surface on UQ1-UBA upon binding to K48-Ub2 is almost identical to that involved in the interaction with monoUb, with no significant CSPs on the backside (α2) of the UBA (Supplementary Fig. 2). Also the CSPs measured in both Ubs in K48-Ub2 are very similar to those observed in monoUb upon UQ1-UBA binding. This indicates a different mode of binding (compared to hHR32A-UBA2) and suggests that the hydrophobic pocket in K48-Ub2 that allowed hHR23A-UBA2 to simultaneously interact with both Ubs, may have less or no significant impact on enhancing the UQ1-UBA binding. In K48-Ub2, the proximal Ub appears to be the primary binding site for hHR23A-UBA 30 whereas UQ1-UBA shows almost no preference toward the proximal Ub in both K48- and K63-Ub2s.
In terms of binding stoichiometry, different UBAs may interact with polyUb chains differently. Thus, whereas two hHR23A-UBA2 domains bind K63-Ub2, a single UBA2 binds to K48-Ub2 at the UBA2:Ub2 molar ratios < 2 30; 48, which is due to the hydrophobic binding pocket formed in the K48-Ub2. Note that a second UBA2 domain can bind to the chain when UBA2 is in significant excess 30. A 1:1 stoichiometry also predominates in the high affinity binding of the UBL-UBA1 construct from hHR23A to K48-Ubn chains of n=2–6 20; it is possible however that other complexes with different stoichiometry can also exist albeit with lower affinity. The same stoichiometry has also been reported for the K48-Ub2/Mud1-UBA complex, based on analytical sedimentation equilibrium measurements 49. It should be pointed out here that all these UBA domains (and hHR23A-UBA1 in the presence of UBL) belong to the class of UBAs that show binding preference toward K48-linked chains. Our data show that, in contrast to these UBAs, UQ1-UBA binds both K48-Ub2 and K63-Ub2 chains in a 2:1 stoichiometry. This is perhaps not surprising, given that UQ1-UBA shows little linkage selectivity in binding to Ub2 (this study) and Ub4 21. In fact, UQ1-UBA represents a class of UBA domains that bind monoUb strongly and do not discriminate between polyUb chains of different linkages 21.
In order to gain structural insights into possible reasons for these differences, we used the structure of the K48-Ub2/hHR23A-UBA2 complex 30 to examine whether UQ1-UBA could also form interactions favoring a similar complex with Ub2 (Fig. 9). In the Ub2/UBA2 complex, the “canonical” Ub-binding side of the UBA2 (loop 1 and helix α3) faces the distal Ub. This sandwich-like complex is stabilized by multiple contacts between helix α3 and residues A46, G47 of the proximal Ub as well as the Ub-Ub linker (L73 of the distal, K48 of the proximal Ub) and also by the hydrophobic interactions between helix α2 (the backside of UBA2) and the proximal Ub (H68, L8). These interactions are responsible for the tight binding of UBA2 to Ub2 and longer chains despite the low affinity of UBA2 for monoUb. A replacement of the distal-Ub/UBA2 pair in this complex with the Ub/UQ1-UBA structure obtained in this study shows that due to different orientation of UQ1-UBA and due to the presence of different amino acid residues at the corresponding positions in this UBA, most of the stabilizing contacts, including those with the linker region, are absent (Fig. 9c,d). This suggests that the extended hydrophobic pocket in K48-Ub2 contributes little (if any) to the strength of UQ1-UBAs binding to this chain. Thus, a UQ1-UBA bound to the distal Ub is expected to interact only weakly (if at all) with the proximal Ub. This would then allow a second UQ1-UBA to bind the proximal Ub, thus resulting in a 2:1 stoichiometry of the complex. A similar prediction can be made for UQ1-UBA already bound to the proximal Ub. Because structurally K48-Ub4 is a dimer of Ub2’s 53, these conclusions are expected to be relevant to Ub4 and longer chains.
Structure-based models in Fig. 9a,b show how UQ1-UBA can bind to K63- and K48-linked Ub2 chains. While two UBAs can readily bind to K63-Ub2 (which is in extended conformation 48), the closed conformation of K48-Ub2, predominant at physiological conditions 47, must open to allow UBA access to the hydrophobic patches on both Ub units 54. Furthermore, the (partially open) conformation of K48-Ub2 observed in its complex with hHR23A is not sufficient for this purpose (Fig. 9c,d), and full opening of this chain is required in order to accommodate two UBAs. (Note in this regard that K48-Ub2 undergoes fast transitions between the closed and open states 55; 56.) No direct contact between the UQ1-UBAs and the Ub-Ub linker is evident in these structural models, in agreement with the abovementioned lack of linkage selectivity. Atomic-resolution structures of these complexes are expected to shed light on the differences between UQ1-UBA and hHR23A-UBA2 binding, and, more generally, on the structural basis for the variability in linkage selectivity between UBA domains. It is worth mentioning that due to high sequence similarity among the UBA domains in the ubiquilin/PLIC family, the results obtained here are relevant to other UBAs in this family, including hPLIC2.
It should be pointed out that the structures shown in Fig. 9a,b might not relate to the physiological mode of polyUb binding for those proteins containing only one UBA domain. These structural models, however, are likely to be relevant to proteins with more than one UBA (or other UBDs). Note also that, the mono- and polyUb binding properties of the isolated UQ1-UBA domain examined here might differ from those of the full-length UQ1, as it is possible that the rest of the protein can affect the binding affinity and/or stoichiometry of the interaction. Although in the UQ1-UBA:monoUb complex (Fig. 4a), as well as in the models of UQ1-UBA:Ub2 complexes (Fig. 9a,b), the N-terminus of UQ1-UBA (where the rest of UQ1 is attached) points away from Ub and Ub2, thus suggesting that the rest of the protein might not affect the binding sterically, the presence of specific interactions cannot be excluded. In fact, the possibility of regulation of UBA domains by their environment has been suggested previously 21 based on the difference in polyUb binding properties of the hHR23A-UBA1 domain, in the isolated form and in the context of the UBL-UBA1 construct of the same protein.
UBL-UBA-containing proteins Rad23 and Dsk2 have been identified as Ub receptors for the proteasome in budding yeast. Their homologues, including hHR23A and UQ1/hPLIC1, respectively, were predicted to play a similar role in human cells 11. According to a recently proposed model 27, hHR23A helps build the final contact between polyUb-protein conjugates and the 26S-proteasome by interacting simultaneously with proteasome, via UBL domain, and with ubiquitinated proteins, via UBA domain. Whether Dsk2 family proteins play the same role as a shuttling factor for polyubiquitinated proteins to the proteasome is still unclear. In contrast to Rad23 proteins, UBA domains of the Dsk2 family appear to have little preference toward a particular linkage in polyUb chain. The results presented here for UQ1-UBA suggest that this could be due to a combination of (i) strong UBA binding to the individual Ub units in the chain and (ii) the lack of intermolecular contacts that would allow the UBA domain to interact simultaneously with two or more Ub units in the chain. The difference between the two UBA domains in their interaction with mono- and polyUb could be a reflection of their specific and diverse function at the physiological conditions which remains to be determined.
Materials and Methods
Protein constructs
The 52 amino acid UQ1-UBA construct used in this study contains full-length UBA domain of human UQ1, residues 546–586 (Swiss-Prot entry Q9UMX0). In addition, this construct has a ten-residue N-terminal extension (GSPEFQNPEV, numbered 536–545 for consistency), of which GSPEF is from the fusion junction and the rest are part of the natural UQ1 sequence beyond the UBA domain, and one Serine residue as the C-terminal extension, numbered 587.
Recombinant ubiquitins Ub (D77), Ub(K48R), Ub (K48C), Ub(T12C) and segmentally isotope-labeled Ub2s linked via K48 (K48-Ub2) or K63 (K63-Ub2) were prepared as described 47; 48. Monomeric Ub (D77) as well as Ub2 constructs, unlabeled or 15N labeled on the distal (Ub2-D) or proximal (Ub2-P) Ub units (with respect to the free C-terminus), were used for NMR titration assays.
Expression and purification of UQ1-UBA
Bacterial expression plasmid encoding GST-fusion of the UBA domain of UQ1 was introduced into the BL21 (pJY2) cells. Bacteria were grown at 37 °C in LB medium, induced with 1 mM IPTG at OD600 ~ 0.6, and incubated for another 5 hours. The harvested cells were lysed with 0.4 mg/mL lysozyme in 20 mM Tris buffer at pH 7.6, containing 1mM PMSF, 50 μM TLCK, 2.5 μg/mL leupeptin, 2.5 μg/mL soybean trypsin inhibitor, and 0.02% Triton X-100. GST-UQ1-UBA fusion proteins were purified from the cell lysate using 20 mL column (Bio-rad Econo-Pac) packed with 10 mL glutathione (GSH) beads. After the binding of GST-UQ1-UBA, the column was washed thoroughly with PBS buffer before incubation with 200 units of thrombin at room temperature for 12 hours. Released UQ1-UBA was then eluted with PBS and further purified by Hitrap benzamidine FF column (1 mL) to remove thrombin. 15N-labeled UQ1-UBA was expressed in Escherichia coli cells grown in M9 minimal media with 15NH4Cl as the sole source of nitrogen.
NMR experiments
Most of the NMR data were acquired at 23 °C on a Bruker Avance 600 MHz spectrometer. The NMR samples were exchanged into a buffer containing 20 mM sodium phosphate (pH6.8), 7% D2O, and 0.02% (w/v) NaN3.
NMR signal assignment and NOE data collection
The backbone and side chain chemical shift assignments of UQ1-UBA were obtained from a combination of phase-sensitive homonuclear 2D TOCSY, 2D NOESY, and 3D 15N-edited TOCSY-HSQC experiments. The HMQC-J experiment 57 was conducted to verify the secondary structure assignment of UQ1-UBA. The inter-proton distance restraints for structure calculation of the free UBA domain were derived from homonuclear 2D NOESY spectra. 15N-filtered 2D NOESY was used to obtain NOEs between the side chains of 15N-labeled UQ1-UBA and the backbone amide protons in perdeuterated Ub. All NMR spectra were processed using XWINNMR (Bruker) and analyzed using XEASY/CARA 58; 59 and in-house software.
Torsion angles were calculated using program TALOS 60. Hydrogen bonds in the helical elements were determined based on a combination of dihedral angle values and characteristic NOE patterns, including HN-HN NOEs between the adjacent residues, as well as between residues i and i+3.
Residual dipolar coupling measurements
The 1H-15N residual dipolar couplings (RDCs) were measured in liquid crystalline medium composed of a mixture of n-alkyl-poly(ethylene glycol) (C12E5) and n-hexanol as described 61. The RDCs were obtained from the difference in the 1H–15N couplings measured in the oriented (23 °C) and in the isotropic phase (33 °C) using a pseudo-3D variant of the IPAP 1H-15N HSQC experiment. The alignment tensor was calculated using PALES 62 and an in-house program ALTENS 47.
RDCs for the UQ1-UBA/monoUb complex were obtained from two separate experiments, with one of the binding partners 15N-labeled (at 0.6 mM concentration) at a time and mixed with a 1.3 molar equivalent of the unlabeled protein to ensure that most of the labeled protein is in the bound state. Both samples were prepared at the same time using one alignment medium stock to assure the same degree of alignment in both experiments (verified separately by the split of the 2H signal). The RDCs for Ubs in the K48-Ub2/UQ1-UBA complex were measured in a similar way, using in this case two separate experiments, for Ub2-D or Ub2-P constructs (~0.6 mM) in the presence of a ~3 molar equivalent of unlabeled UQ1-UBA.
The comparison of the experimental RDCs with a protein structure was performed as follows. Fist, the alignment tensor for a particular protein structure was obtained directly from the experimental RDCs using program ALTENS 47 based on the singular-value decomposition approach 63. Then this alignment tensor was used to back-calculate the expected RDC values for each NH group in the protein.
15N relaxation measurements and analysis of protein dynamics
The 15N relaxation measurements were performed as described in ref 46, and include rates of 15N longitudinal (R1) and transverse (R2) relaxation and the 15N-1H cross-relaxation rate measured via steady-state 15N{1H} NOE. The experiments were carried out on UQ1-UBA alone as well as on both UQ1-UBA and Ub in the complex; the experimental data were analyzed as detailed elsewhere 46. Briefly, the 15N relaxation rates and heteronuclear NOEs were analyzed using ROTDIF 44 and assuming various overall rotational diffusion models (Supplementary Material), and the resulting diffusion tensor was used as input into computer program DYNAMICS 46; 64 for extracting the “model-free” parameters characterizing backbone dynamics.
NMR Titration studies of Ub:UQ1-UBA binding
The interaction surfaces on Ub and UQ1-UBA were mapped using chemical shift perturbation (CSP) approach. A series of 1H-15N HSQC spectra were recorded while titrating the 15N -labeled protein (Ub or UQ1-UBA) with the unlabeled ligand (UQ1-UBA or Ub, respectively) until the ligand-to-protein molar ratio reached >2:1. In general, the assay was carried out starting with a 0.7–1.0 mM sample of 15N-labeled protein and adding increasing amounts of a concentrated stock solution of the ligand. A change in a peak position in the spectra in the course of titration indicates perturbation in the electronic environment of the corresponding nucleus as a result (direct or indirect) of the binding interaction. These shifts were quantified using combined amide CSPs calculated as Δδ= [(ΔδH)2+(ΔδN/5)2]1/2, where ΔδH and ΔδN are the observed chemical shift changes for 1H and 15N, respectively. The binding affinity between UQ1-UBA and Ub was quantified by fitting the observed CSPs (Δδ) as a function of the protein and ligand concentrations to various stoichiometry models as detailed elsewhere 48.
Similar titration experiments were performed to map the binding surface on UQ1-UBA and both Ub units in Ub2 for the UBA:Ub2 interactions. Both K48- and K63-linked Ub2 chains were studied, in order to examine whether the linkage-specific conformational differences between the two chains have any effect on the UQ1-UBA binding. The titrations started at 0.75 mM of the 15N-labeled protein (Ub2-D, or Ub2-P) and continued until the UBA:Ub2 molar ratio of >2.8. A similar assay for 15N-UBA as the observed protein (0.7 mM starting concentration) and K48-Ub2 as the ligand continued until [Ub2]/[UBA]=2.
Site-directed Spin Labeling
A paramagnetic spin label (SL), (1-oxy-2,2,5,5-tetramethyl-3-pyrroline-3-methyl) methanesulfonate, was covalently attached to Ub through the side chain of a cysteine as described 65. Two Ub residues, K48 and T12, were mutated to a cysteine for this purpose in two separate experiments, in order to allow unambiguous determination of the interprotein orientation in the Ub/UQ1UBA complex. The paramagnetic relaxation enhancement (PRE) effect was measured as a ratio of signal intensities in the 1H-15N HSQC spectra recorded with SL in the oxidized (paramagnetic) and reduced state as detailed elsewhere 65. To measure the PREs in the Ub:UQ1-UBA complex, the latter was prepared by mixing SL-Ub with 15N-UQ1-UBA at a 1.3:1 molar ratio. 15N-Ub-SL was used to measure PREs in Ub, in order to verify the attachment of the SL and determine its position on Ub. The relative position of the unpaired electron of SL with respect to the 15N-labeled species (Ub or UBA) was determined from the global fit (Fig. 3) of the observed PREs using in-house program SLfit 55. The PRE-derived intermolecular distance constraints for the HADDOCK calculation were obtained by measuring the distances between selected amide protons and the reconstructed position of the SL (Supplementary Table 3). These were included in the docking as distance restraints between a given NH and the Cα atom of the residue (T12 or K48 in Ub) at the position where the SL was attached. As the main purpose of the PRE-derived restraints was to select the proper intermolecular orientation (see above), they were set as rather loose restraints, with a 5 Å tolerance.
Structure calculations
Structure calculation of the UQ1-UBA was performed iteratively using program ARIA, version 2.0 66 combined with CNS 67. Experimental constraints used in the calculation included NOE-derived distance restraints, torsion angles, hydrogen bonds, and long-distance orientational constraints from the RDCs (see Supplementary Material).
The structure of the UQ1-UBA/Ub complex was determined using protein-protein docking program HADDOCK41. The calculation included ambiguous interaction restraints (AIRs) derived from the CSPs (Supplementary Table 2), unambiguous distance restraints from intermolecular NOEs and PREs (Supplementary Table 3), and orientational constraints from the RDCs measured for both proteins in the bound state. The starting atomic coordinates for Ub were from the solution structure of free Ub (PDB code 1D3Z); the coordinates for UQ1-UBA were from the NMR structure determined in this study as described above. The AIRs for the docking were defined by the following criteria: residues with chemical shift change greater than 0.07 ppm and with relative side chain accessibility above 50% were defined as “active”; residues neighboring active residues and having relative solvent accessibility about 50% were defined as “passive”. Flexible segments were defined as stretches of active and passive residues plus one sequential neighbor. Various strategies of incorporating RDCs into protein docking are discussed in ref 68. A combined VEAN-SANI 68 approach was used here, in which the intervector projection angle restraints were used in iteration 0 to drive the docking and then direct RDC restraints were used in iteration 1 and in the final explicit solvent refinement.
Supplementary Material
Acknowledgments
The original UQ1-UBA expression plasmid was a kind gift from M. Monteiro. We thank Ranjani Varadan for initial help with the measurements and for useful discussions. This work was supported by NIH grant GM065334 to D.F. S.R. was supported by NIH grant GM60372 to Cecile Pickart and presently by a research grant from Deutsche Forschungsgemeinschaft, Bonn. Atom coordinates of the UQ1-UBA domain and the UQ1-UBA/Ub complex have been deposited in the Protein Data Bank, the accession numbers are 2JY5 and 2JY6, respectively.
Footnotes
Dedication. This paper is dedicated to the memory of Cecile M. Pickart, our wonderful colleague and mentor, who initiated and inspired this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Di Fiore PP, Polo S, Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat Rev Mol Cell Biol. 2003;4:491–7. doi: 10.1038/nrm1124. [DOI] [PubMed] [Google Scholar]
- 3.Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5:461–6. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 4.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–72. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 5.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–61. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 8.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–6. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 10.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 11.Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–9. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 12.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–21. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 13.Harper JW, Schulman BA. Structural complexity in ubiquitin recognition. Cell. 2006;124:1133–6. doi: 10.1016/j.cell.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–72. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc Natl Acad Sci U S A. 1996;93:861–6. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beal RE, Toscano-Cantaffa D, Young P, Rechsteiner M, Pickart CM. The hydrophobic effect contributes to polyubiquitin chain recognition. Biochemistry. 1998;37:2925–34. doi: 10.1021/bi972514p. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann K, Bucher P. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- 18.Madura K. The ubiquitin-associated (UBA) domain: on the path from prudence to prurience. Cell Cycle. 2002;1:235–44. [PubMed] [Google Scholar]
- 19.Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 20.Raasi S, Orlov I, Fleming KG, Pickart CM. Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J Mol Biol. 2004;341:1367–1379. doi: 10.1016/j.jmb.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 21.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 22.Prakash S, Prakash L. Nucleotide excision repair in yeast. Mutat Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 23.Medicherla B, Kostova Z, Schaefer A, Wolf DH. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004;5:692–7. doi: 10.1038/sj.embor.7400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 25.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Rao H, Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J Biol Chem. 2002;277:11691–5. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- 27.Kang Y, Chen X, Lary JW, Cole JL, Walters KJ. Defining how ubiquitin receptors hHR23a and S5a bind polyubiquitin. J Mol Biol. 2007;369:168–76. doi: 10.1016/j.jmb.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dieckmann T, Withers-Ward ES, Jarosinski MA, Liu CF, Chen IS, Feigon J. Structure of a human DNA repair protein UBA domain that interacts with HIV-1 Vpr. Nat Struct Biol. 1998;5:1042–1047. doi: 10.1038/4220. [DOI] [PubMed] [Google Scholar]
- 29.Withers-Ward ES, Mueller TD, Chen IS, Feigon J. Biochemical and structural analysis of the interaction between the UBA(2) domain of the DNA repair protein HHR23A and HIV-1 Vpr. Biochemistry. 2000;39:14103–14112. doi: 10.1021/bi0017071. [DOI] [PubMed] [Google Scholar]
- 30.Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 31.Mah AL, Perry G, Smith MA, Monteiro MJ. Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J Cell Biol. 2000;151:847–862. doi: 10.1083/jcb.151.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massey LK, Mah AL, Ford DL, Miller J, Liang J, Doong H, Monteiro MJ. Overexpression of ubiquilin decreases ubiquitination and degradation of presenilin proteins. J Alzheimers Dis. 2004;6:79–92. doi: 10.3233/jad-2004-6109. [DOI] [PubMed] [Google Scholar]
- 33.Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 34.Kleijnen MF, Alarcon RM, Howley PM. The ubiquitin-associated domain of hPLIC-2 interacts with the proteasome. Mol Biol Cell. 2003;14:3868–3875. doi: 10.1091/mbc.E02-11-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng P, Scott CW, Cho NH, Nakamura H, Chung YH, Monteiro MJ, Jung JU. Kaposi’s sarcoma-associated herpesvirus K7 protein targets a ubiquitin-like/ubiquitin-associated domain-containing protein to promote protein degradation. Mol Cell Biol. 2004;24:3938–48. doi: 10.1128/MCB.24.9.3938-3948.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao L, Tu H, Shi ST, Lee KJ, Asanaka M, Hwang SB, Lai MM. Interaction with a ubiquitin-like protein enhances the ubiquitination and degradation of hepatitis C virus RNA-dependent RNA polymerase. J Virol. 2003;77:4149–4159. doi: 10.1128/JVI.77.7.4149-4159.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glockzin S, Ogi FX, Hengstermann A, Scheffner M, Blattner C. Involvement of the DNA repair protein hHR23 in p53 degradation. Mol Cell Biol. 2003;23:8960–9. doi: 10.1128/MCB.23.24.8960-8969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohno A, Jee J, Fujiwara K, Tenno T, Goda N, Tochio H, Kobayashi H, Hiroaki H, Shirakawa M. Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition. Structure. 2005;13:521–532. doi: 10.1016/j.str.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 39.Clore GM, Garrett DS. R-factor, free R, and complete cross-validation for dipolar coupling refinement of NMR structures. J Am Chem Soc. 1999;121:9008–12. [Google Scholar]
- 40.Zuiderweg ER. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 41.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 42.Bax A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci. 2003;12:1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fushman D, Varadan R, Assfalg M, Walker O. Determining domain orientation in macromolecules by using spin-relaxation and residual dipolar coupling measurements. Progress in Nuclear Magnetic Resonance Spectroscopy. 2004;44:189–214. [Google Scholar]
- 44.Walker O, Varadan R, Fushman D. Efficient and accurate determination of the overall rotational diffusion tensor of a molecule from (15)N relaxation data using computer program ROTDIF. J Magn Reson. 2004;168:336–345. doi: 10.1016/j.jmr.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 45.Garcia de la Torre J, Huertas ML, Carrasco B. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J Magn Reson. 2000;B147:138–46. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
- 46.Hall JB, Fushman D. Characterization of the overall and local dynamics of a protein with intermediate rotational anisotropy: Differentiating between conformational exchange and anisotropic diffusion in the B3 domain of protein G. J Biomol NMR. 2003;27:261–275. doi: 10.1023/a:1025467918856. [DOI] [PubMed] [Google Scholar]
- 47.Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 48.Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 49.Trempe JF, Brown NR, Lowe ED, Gordon C, Campbell ID, Noble ME, Endicott JA. Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain. EMBO J. 2005;24:3178–3189. doi: 10.1038/sj.emboj.7600797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson KA, Hicke L, Radhakrishnan I. Structural basis for monoubiquitin recognition by the Ede1 UBA domain. J Mol Biol. 2006;358:713–24. doi: 10.1016/j.jmb.2006.02.059. [DOI] [PubMed] [Google Scholar]
- 51.Ryu KS, Lee KJ, Bae SH, Kim BK, Kim KA, Choi BS. Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a. J Biol Chem. 2003;278:36621–36627. doi: 10.1074/jbc.M304628200. [DOI] [PubMed] [Google Scholar]
- 52.Mueller TD, Kamionka M, Feigon J. Specificity of the interaction between ubiquitin-associated domains and ubiquitin. J Biol Chem. 2004;279:11926–11936. doi: 10.1074/jbc.M312865200. [DOI] [PubMed] [Google Scholar]
- 53.Eddins MJ, Varadan R, Fushman D, Pickart CM, Wolberger C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J Mol Biol. 2007;367:204–11. doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]
- 54.Dickinson BC, Varadan R, Fushman D. Effects of cyclization on conformational dynamics and binding properties of Lys48-linked di-ubiquitin. Protein Sci. 2007;16:369–78. doi: 10.1110/ps.062508007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryabov Y, Fushman D. Interdomain mobility in di-ubiquitin revealed by NMR. Proteins. 2006;63:787–96. doi: 10.1002/prot.20917. [DOI] [PubMed] [Google Scholar]
- 56.Ryabov YE, Fushman D. A model of interdomain mobility in a multidomain protein. J Am Chem Soc. 2007;129:3315–27. doi: 10.1021/ja067667r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kay LE, Bax A. New methods for the measurement of NH-CalphaH coupling constants in 15N-labeled proteins. J Magn Reson. 1990;86:110–126. [Google Scholar]
- 58.Bartels C, Xia T, Guntert P, Billeter M, Wuthrich K. The program XEASY for computer-supported NMR spectral analysis. J Biomol NMR. 1995;5:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 59.Keller R. The Computer Aided Resonance Assignment Tutorial. CANTINA Verlag; 2004. [Google Scholar]
- 60.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 61.Ruckert M, Otting G. Alignment of Biological Macromolecules in Novel Nonionic Liquid Crystalline Media for NMR Experiments. J Am Chem Soc. 2000;122:7793–7797. [Google Scholar]
- 62.Zweckstetter M, Bax A. Prediction of sterically induced alignment in a dilute liquid crystalline phase: Aid to protein structure determination by NMR. Journal of the American Chemical Society. 2000;122:3791–3792. [Google Scholar]
- 63.Losonczi JA, Andrec M, Fischer MW, Prestegard JH. Order matrix analysis of residual dipolar couplings using singular value decomposition. J Magn Reson. 1999;138:334–342. doi: 10.1006/jmre.1999.1754. [DOI] [PubMed] [Google Scholar]
- 64.Fushman D, Cahill S, Cowburn D. The main-chain dynamics of the dynamin pleckstrin homology (PH) domain in solution: analysis of 15N relaxation with monomer/dimer equilibration. J Mol Biol. 1997;266:173–94. doi: 10.1006/jmbi.1996.0771. [DOI] [PubMed] [Google Scholar]
- 65.Varadan R, Assfalg M, Fushman D. Using NMR spectroscopy to monitor ubiquitin chain conformation and interactions with ubiquitin-binding domains. In: Deshaies RJ, editor. Ubiquitin and Protein Degradation, Methods in Enzymology, Vol.399 part B. part B. Vol. 399. 2005. pp. 177–192. [DOI] [PubMed] [Google Scholar]
- 66.Linge JP, Nilges M. Influence of non-bonded parameters on the quality of NMR structures: a new force field for NMR structure calculation. J Biomol NMR. 1999;13:51–9. doi: 10.1023/a:1008365802830. [DOI] [PubMed] [Google Scholar]
- 67.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 68.van Dijk AD, Fushman D, Bonvin AM. Various strategies of using residual dipolar couplings in NMR-driven protein docking: application to Lys48-linked di-ubiquitin and validation against 15N-relaxation data. Proteins. 2005;60:367–81. doi: 10.1002/prot.20476. [DOI] [PubMed] [Google Scholar]
- 69.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–5. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.