Abstract
The discovery of a new lineage of helper T cells that selectively produces interleukin (IL) -17 has provided exciting new insights into immunoregulation, host defense and the pathogenesis of autoimmune diseases. Additionally, the discovery of this T cell subset has offered a fresh look at how the complexity of selective regulation of cytokine gene expression might relate to lineage commitment, terminal differentiation and immunologic memory. Information continues to accumulate on factors that regulate Th17 differentiation at a rapid pace and a few lessons have emerged. Like other lineages, Th17 cells preferentially express a transcription factor, retinoic acid-related orphan receptor (ROR)γ t, whose expression seems to be necessary for IL-17 production. In addition, signals from the T-cell receptor are a critical aspect of controlling IL-17 production and the transcription factor nuclear factor of activated T cells (NFAT) appears to be another important regulator. IL-6, IL-21 and IL-23 are all cytokines that activate the transcription factor STAT3, which has been established to be necessary for multiple aspects of the biology of Th17 cells. Similarly, TGFβ-1 is important for the differentiation of murine Th17 cells and inducible regulatory T cells (iTregs), but how it exerts its effect on IL-17 gene transcription is unknown and there are data indicating TGFβ-1 is not required for human Th17 differentiation. The extent to which Th17 cells represent terminally differentiated cells or whether they retain plasticity and can develop into another lineage such as IFNγ secreting Th1 cells is also unclear. Precisely how cytokines produced by this lineage are selectively expressed and selectively extinguished through epigenetic modifications is an area of great importance, but considerable uncertainty.
INTRODUCTION
The identification of two distinct subsets of CD4+ T helper (Th) cells by Coffman and Mossman provided enormous insight into our understanding of T cell function. This discovery led to further advances in how subsets of T cells with selective cytokine production emerge in response to different microbial pathogens, thus providing a robust model for understanding mechanisms of gene regulation and fate determination [1–3] [4]. It is therefore worth reviewing what has been learned about gene regulation in Th1 and Th2 cells as background for discussing regulation of IL-17.
Th1 cells produce the signature cytokine, IFNγ, a critical factor that enhances cellular immunity. Precisely how IFNγ production is selectively induced in Th1 cells has been the focus of much research, but is still incompletely understood. Nonetheless, the study of this important gene has provided interesting insights. Clearly, signals emanating from the T cell receptor (TCR) are of major importance and the activation and induction of transcription factors such as nuclear factor of activated T cells (NFAT) and T-box transcription factor, T-box 21 or T-box protein expressed in T cells (T-bet) are clearly key events [3]. Additionally, interleukin (IL)-12 produced by activated dendritic cells (DC) acting via the transcription factor STAT4 is particularly important in promoting Th1 differentiation.
In addition to the activation of transcription factors, we now know that epigenetic modifications of genes represent an important aspect of regulation. DNA is associated with histone proteins to form nucleosomes, the basic structure of chromatin. Active transcription generally requires chromatin modification and remodeling, which can be assayed by sensitivity to nucleases, histone tail modifications and CpG DNA methylation. The selective expression of cytokine genes in activated CD4+ T cells and their progeny represent excellent examples of the dependency upon epigenetic regulation. Recent comprehensive epigenetic analyses of Ifng locus suggests that regulatory elements are spread over a region of about 100 kilobases flanking the gene including the collective action of multiple distal regulatory elements [5, 6]. These include multiple DNase hypersensitivity sites and conserved noncoding sequences (CNS). CNS-1 resides 5 kb upsteam of the Ifng gene and is subject to Th1-specific hypersensitivity, and NFAT and T-bet bind in this region. CNS-2 is located 18 kb downstream of the Ifng gene and it undergoes Th1-specific histone hyperacetylation; this region binds T-bet and STAT5 [7, 8]. Another CNS is located upstream of the Ifng gene, 22 kb (CNS-22) from the transcriptional start site. CNS-22 binds T-bet and is associated with histone modifications typical of accessible chromatin in both T helper 1 (Th1) and Th2 cells. Deletion of this element blocked Ifng gene expression in CD4+ Th1 cells, CD8+ T cells and NK cells [9].
Epigenetic marking throughout the Ifng locus such as histone modification and DNA methylation are critical in maintaining the gene in a transcriptionally accessible/repressed state and it appears that STAT4 is essential for these modifications [10]. Overexpression of T-bet promotes epigenetic modifications; however, T-bet, is not absolutely required for remodeling of the Ifng locus [11]. Conversely, silencing of the Ifng locus in differentiating Th2 cells is dependent on GATA3 and STAT6 [5, 6, 12]. Interference with DNA methylation also results in enhanced IFNγ production, and DNA demethylation and histone acetylation are STAT-dependent [4, 13].
In contrast to Th1 cells, the hallmark cytokine secreted by Th2 cells is IL-4, which is crucial for host defense against helminths and the pathogenesis of asthma and allergy. Th2 development is initiated by TCR signaling in conjunction with IL-4 signaling with the participation of the transcription factors STAT6 and GATA3. The Th2 locus encompasses 140 kb and includes genes encoding IL-4, IL-13, Rad50, and IL-5. The Rad50 gene does not encode a cytokine, but it contains elements that regulate IL-4, IL-5 and IL-13 production (discussed below). The intergenic region between the Il4 and Il13 genes includes three DNase hypersensitivity sites. This region also includes a critical, evolutionarily conserved 400-bp noncoding sequence 1 (CNS-1), which when deleted interferes with Th2 cell differentiation, but does not interfere with mast cell IL-4 production. [14] CNS-1 is highly responsive to GATA3 in terms of chromatin remodeling and enhancer activity. Another conserved noncoding sequence, CNS-2 overlaps with a DNase hypersensitivity region present downstream of the Il4 gene. This region has enhancer activity and is bound by GATA3 and NFAT. Deletion of CNS-2 blocks IL-4, IL-5 and IL-13 production in Th2 and mast cells [15]. Another DNase I hypersensitivity site (HS1) located 1.6 kb upsteam of the Il13 locus also binds GATA3.
A locus control region (LCR) is a regulatory region with both enhancer and insulator activity and a Th2 LCR resides in the 3′ end of the Rad50 gene, which contains multiple DNase hypersensitivity sites [16]. The Th2 LCR also undergoes a number of lineage-specific epigenetic modifications including Th2-specific histone hyperacetylation and DNA demethylation.
Studies have shown that the promoters of the Th2 cytokine genes undergo intrachromosomal interactions, such that they are in close spatial proximity to form a “prepoised” core chromatin configuration even in nonlymphoid cells that do not produce these cytokines. It is thought that early cytokine production, which can occur in a STAT6-independent manner may be due to the “prepoised” structure of the Th2 locus. GATA3 may be an important factor in this organization and is known to induce chromatin remodeling. GATA3 also interacts with other transcription factors, such as members of the NFAT family, which may work in concert with GATA3 to facilitate the interaction between the Th2 LCR and the promoters of the target cytokine genes.
In T and NK cells, the LCR also interacts with the promoters of the Th2 cytokine genes, referred to as a “poised” state of intrachromasomal structure. STAT6 is likely to be required for “poised” chromatin conformation in later stages of Th2 differentiation. In this way, STAT6 contributes to the sustained tissue-specific expression of the Th2 cytokines.
Another important aspect of Th1/Th2 counterregulation is interchromosomal interactions between Th1- or Th2-specific cytokine genes [17]. In naïve T cells the promoter region of the Ifng locus (chromosome 10) and the LCR of the Th2 locus (chromosome 11) interact. Upon Th1 stimulation, Th2 genes move to a more repressed region of the nucleus while IFNγ is expressed. The gene expression under Th1 or Th2 stimulation is then regulated by intrachromosomal regulatory elements, although mechanics of this process are poorly understood.
Together, these mechanisms enable Th1 and Th2 lineage decisions to be made at a very early stage of T-helper differentiation, with respective Th1/Th2 cytokines enforcing their own expression and inhibiting alternative commitment. This is a key feature of Th1 and Th2 cells – silencing of the gene encoding signature cytokine of the opposing fate – in this respect, Th1 and Th2 cells appear to be terminally differentiated, at least in the mouse. Generally, once a CD4+ T cell has committed to one of these two lineages, conversion to the other lineage does not occur. As a result of these mechanisms, Th1 and Th2 cells develop into mature effectors with rather stable phenotypes. This is important because the helper T cell response to microorganisms is essential in shaping the immune response to eliminate these pathogens.
While the concept of Th1 vs. Th2 cells has led to enormous advances, this view of T cell differentiation is somewhat static, with stereotypical, inflexible responses. With time it has become clear that the model has a number of limitations, and that it does not reflect the true complexity of T cell responses.
New levels of complexity in fates of CD4+ T cells
Although the cytokine IL-17 was identified more than 10 years ago as a product of CD4+ T cells, it was only recently that a subset of T cells which preferentially produces IL-17 was recognized. Conversely, it was also recognized that polarized Th1 and Th2 cells do not produce IL-17 and in fact, IFNγ and IL-4 inhibit Th17 differentiation. Thus, it was proposed that IL-17-producing helper T cells or Th17 cells represent a new lineage of CD4+ T cells, which appears to be very important in the pathogenesis of autoimmune disease.
While there are data that support the notion that Th17 represents a distinct lineage, it should be noted that T cells producing both IFNγ and IL-17 have now been found in a number of circumstances in vitro and in vivo; exactly how such cells are related to Th17 or Th1 cells is unclear. In the human, these subsets (IL-17/IFNγ double producers and IL-17 single producers) can be distinguished by their chemokine receptor expression [18] (discussed below). Furthermore, Th17 cells also produce other cytokines including TNF, IL-17A, IL-17F, IL-21 and IL-22, though IL-22 can also be produced by other T cell subsets [19–22]. This has important implications for considering Th17 lineage commitment and the extent to which Th17 cells are terminally differentiated or are “works in progress”.
Another critical subset of CD4+ T cells, is regulatory T (Treg) cells. So-called natural Treg cells are CD4+ CD25+ cells found in the thymus and in the periphery that express the forkhead box protein transcription factor, Foxp3. In addition, inducible Treg cells can be generated in vitro from naïve CD4+ T cells with the appropriate cytokine stimuli. Treg cells have critical functions in preserving peripheral immunological tolerance, and it has recently been argued that Treg and Th17 cells [20, 23–26] are reciprocally regulated. In this chapter, we will discuss the various stimuli that induce Th17 and Treg differentiation focusing on transcription factors that are activated and how they might regulate the various fates of differentiating T cells.
TCR signaling and IL-17 production
The first step in the activation of any T cells relates to consequences of signals emanating from TCR occupancy; as with the differentiation of other helper T cell lineages, such signals are an important aspect of Th17 differentiation. Indeed, for memory T cells, TCR occupancy is sufficient to induce large amounts of IL-17 [27]. Similarly, NK T cells are also poised to produce large quantities of cytokines in response to TCR occupancy (2, 4–8). The TCR of this subset of T cells recognizes glycolipid antigens presented by the major histocompatibility complex class I-related protein CD1d. Upon activation by glycolipids such alpha-galactosylceramide (α-GalCer), these T cells can rapidly produce large amounts of various cytokines, such as IL-4, IFNγ, TNF-α, IL-3, and GM-CSF. Recently, a specialized subset of NK T cells, invariant NK T cells that lack the NK1.1 marker (NK1.1neg), has been found to produce large amounts of IL-17 and low levels of IFNγ and IL-4 in response to α-GalCer [28]. Another subset of T cells, gamma-delta T cells, also produces IL-17 in response to TCR occupancy [29].
TCR signaling activates a number of pathways and transcription factors. A major pathway activated by the TCR is the production of intracellular calcium and the activation of the transcription NFAT. Accordingly, the proximal promoter of the human IL17A gene contains two NFAT binding sites which appear to be important in regulation of IL-17 [30]. Co-stimulatory signaling also contributes to the regulation of IL-17 expression as CD3-induced IL-17 production is enhanced when anti-CD28 is added [30], but the pathways responsible are poorly understood. TCR signaling also induces the activation of the transcription factors NFκ B and AP-1. The role of these transcription factors in IL-17 production is not known but it would not be surprising if they played important roles.
Cytokines and the critical role of STAT3 in Th17 differentiation
In addition to TCR-dependent signals, an important aspect of CD4+ T cell differentiation is the cytokine milieu. It is now generally agreed that IL-6 is a key factor in promoting Th17 differentiation of naïve CD4+ T cells, and IL-6−/− mice have reduced, but not absent, numbers of Th17 cells [20]. Additionally, it has recently been found that IL-21 is selectively produced by Th17 cells compared to Th1 and Th2 cells. Furthermore, it is also clear that IL-21 promotes IL-17 production and inhibits IFNγ production [20–22, 31, 32]. Thus, IL-21 acts in a manner analogous to IL-4 in that the lineage-specific product promotes differentiation to the subset that produces this factor while simultaneously inhibiting other fates. Although IL-23 was initially thought to be an important inducer, it is now generally agreed that this cytokine is more important for the survival and expansion of Th17 cells [33–35]. Naïve CD4+ T cells do not express the IL-23 receptor, whereas a subset of memory T cells do express this receptor. For human Th17 differentiation, IL-23 may play a more prominent role in initial differentiation [27, 36].
IL-6, IL-21 and IL-23 all activate Janus family kinases and preferentially activate STAT3. Furthermore, in vitro Th17 differentiation is greatly impaired in STAT3-deficient T cells [37–39]. As will be discussed below, Th17 cells preferentially express the retinoic acid-related orphan receptor, RORγt; expression of this transcription factor is also impaired in T cells lacking STAT3. Conversely, retroviral overexpression of constitutively active STAT3, results in increased production of IL-17 production [38]. The Il17a/f locus has putative STAT binding sites and using chromatin immunoprecipitation (ChIP) assays, we have found that STAT3 directly binds to this promoter [40]. Thus STAT3 appears to be a direct regulator of IL-17. Whether the effect of STAT3 also includes binding to the Rorc (the gene encoding RORγ) promoter remains to be determined. Stat3 may also have additional roles in promoting Th17 differentiation. For instance, the production of IL-21 by IL-6 is also Stat3 dependent and, STAT3 appears to be a direct regulator of IL-21 production as it binds to the Il21 promoter [32]. Furthermore, IL-6, IL-21 and IL-23 can upregulate expression of the IL-23R and this too appears to be STAT3 dependent [21, 22, 27] (unpublished data). Thus, STAT3 seems to be a critical transcription factor for Th17 differentiation through at least four mechanisms: directly binding and regulating Il17a and Il17f locus and the Il21 locus, as well as regulating RORγt and IL-23R expression.
Suppressor of cytokine signaling (SOCS3) is a cytokine-inducible negative regulator of STAT3. Interestingly, deletion of SOCS3 is associated with enhanced STAT3 phosphorylation and increased IL-17 production [40, 41]. Deletion of SOCS3 is also associated with multiorgan inflammatory disease characterized as neutrophil infiltration and overexpression of factors regulated by IL-17 (G-CSF, chemokines, IL-6). These findings further support the critical functions of STAT3 in regulating IL-17.
Th17 cells and RORγt
Despite the importance of STAT3 signaling, it should be emphasized that this stimulus alone appears not to be sufficient to drive Th17 differentiation. Like Th1 and Th2 cells, Th17 cells selectively express a lineage-specific transcription factor, which is important for restricted cytokine production. For Th17 cells, this transcription factor is the retinoic acid-related orphan receptor gamma (RORγ), and is required for IL-17 production [42]. RORγ belongs to the superfamily of steroid nuclear receptors and is most closely related to a retinoic acid receptor (RAR) subfamily of transcription factors [43]. As implied by the name, the founding members of this family bind retinoic acid (RA), the active metabolite of vitamin A, but unlike other steroid receptors the ligand(s) for RORγ is unknown. RORγ is important in lymphoid organogenesis and in thymopoiesis as evidenced by the fact that RORγ −/− mice exhibit reduced numbers of double-positive and CD4+ single-positive thymocytes. RORγ −/− mice also show lack of lymph nodes and Peyer’s patches [44–46].
RORγt is a splice variant of RORγ that results from initiation by a distinct promoter and so differs from RORγ in the amino terminus of the protein encoded by the variant transcript [47]. In the normal state, RORγt is predominantly expressed in lamina propria T cells. RORγ −/− mice have reduced Th17 differentiation and are less susceptible to experimental autoimmune encephalomyelitis, indicating that this transcription factor is important with respect to autoimmunity. Conversely, overexpression of RORγt promotes IL-17 expression, and in this regard RORγt acts similarly to transcription factors such as T-bet and GATA3.
Precisely how RORγt regulates IL-17 production is not known, nor has it been defined whether RORγt directly or indirectly regulates Il17a gene transcription. One possibility is that RORγt directly binds Il17a promoter. While a potential ROR binding site is present, it has not been demonstrated to be functional. Furthermore, how RORγt is regulated is also unclear. One important means of regulating RORγt expression is through cytokines, especially IL-6 and related cytokines. IL-6 regulation of RORγt is mediated by STAT3, but it has not been established that STAT3 binds the Rorc gene. Furthermore, overexpression of active STAT3 in RORγt-deficient cells results in poor IL-17 induction, indicating STAT3 is necessary but not sufficient for IL-17 expression; conversely a constitutively expressed RORγt cannot induce IL-17 expression, suggesting that the effect of these two transcription factors act, to some extent, in parallel [22]. Another possibility is the indirect effect of RORγt for IL-17 expression as RORγt can interact with NFAT and inhibits IL-2 production (see below) [48]. It should be noted that Th17 cells also preferentially express another member of the ROR family, RORα (authors’ unpublished observations); it will be of interest to determine the extent to which RORαand RORγt have unique or redundant roles in Th17 differentiation.
As mentioned, for some retinoid receptors we know the ligand that binds and activates the receptor/transcription factor, but in the case of RORγthe ligand is unknown. This begs the obvious question as to whether a secreted product of immune cells can activate this transcription factor and in so doing regulate T cell differentiation. It is of note in this regard that dendritic cells from the gut-associated lymphoid organs tend to be immunosuppressive. Importantly, mucosal DCs also produce retinoic acid (RA) which, among other actions, enhances the expression of α4β7 and CCR9 on activated T cells, promoting gut tropism [49–52]. As discussed below, DC-derived RA negatively regulates Th17 differentiation and so one might hypothesize that other DCs or antigen-presenting cells might produce ligands for RORγt and thereby promote Th17 differentiation.
TGFβ-1 – a species-specific factor?
TGFβ-1 has pleiotropic functions in immune responses, but it is clear that this cytokine is critical for murine Th17 differentiation. That is, addition of IL-6 or IL-21 to naïve CD4+ T cells, activated by anti-CD3 and anti-CD28 is not sufficient to induce IL-17 production, even though this stimulus does appear to be sufficient to induce other cytokines like IL-22 [19, 53]. Of note is that TGFβ-1 is also a critical regulator of Treg cells [54] and the dependence of Th17 and Treg cells on TGFβ-1 is one line of evidence that supports the relatedness of these two subsets. What is less clear is how TGFβ-1 functions to promote Th17 and Treg differentiation.
TGFβ-1 inhibits both Th1/Th2 differentiation, downregulating both T-bet and GATA3, [55, 56]. TGFβ-1can also inhibits IL-2 production, and this could favor Th17 differentiation (see below) [37]. Whether TGFβ-1 has direct effects on regulating Il17 gene expression has not been reported. Although stimulation of cells with TGFβ-1 and IL-6 induces RORγt, the combination of IL-2 and TGFβ-1 upregulates Foxp3. Foxp3 expression is known to downregulate cytokines like IL-21 and IL-17, so it is particularly interesting to consider how the opposing regulation of Foxp3 and IL-17 might occur [57]. This enigma is further compounded by data indicating that Treg cells might convert to Th17 under certain circumstances [26]. Despite the critical role of TGFβ-1 in murine Th17 differentiation, this factor has been reported dispensable for human Th17 differentiation; indeed, it inhibits IL-17 production in human cells [27, 36, 58]. This is especially perplexing as the mechanistic implication is that IL-17 transcription per se may not to be highly dependent upon TGFβ-1 signaling.
The active form of TGFβ-1 initially engages a receptor comprising two subunits, TGFβRII and TGFβRI. The binding results in phosphorylation of receptor-associated SMAD2 or SMAD3, dimerization with SMAD4, which is needed for nuclear translocation, and finally binding to SMAD-binding element of the target genes. There are no data indicating whether SMADs directly bind to Il17a, Rorc or Foxp3 genes or whether they are involved in regulation of chromatin remodeling. It is of note that SMAD3 deficient mice suffer from inflammatory disease [59]. SMADs also interact with a variety of other transcription factors, including AP-1, so they could be acting in concert with factors like STAT3.
IL-1 and Th17 differentiation
Another inflammatory cytokine reported positively regulate Th17 development is IL-1. This has been documented in mouse cells and also in human cells where it synergizes with IL-6 and IL-23 [36, 58, 60]. Whereas IL-6 alone causes transient induction of RORγ, the combination of IL-1 and IL-6 results in sustained induction. Accordingly, IL-1RI−/− mice have impaired Th17 cell differentiation and reduced incidence of EAE associated with failure to induce autoantigen-specific ThIL-17 cells [61].
The IL-1 receptor has homology to Toll-like receptors, and IL-1 engagement of its receptor results in activation of complex that comprises the adapter MyD88 and IL-1 Receptor-associated kinases (IRAK). This complex recruits another adapter molecule, tumor necrosis factor receptor associated factor (TRAF)-6, which leads to the activation of other kinases including Jun kinase (Jnk) and inhibitor of kappa-B kinases (IKKs). Jnk phosphorylates the transcription factor Jun, which dimerizes with Fos to form the transcription factor AP-1. IKKs phosphorylate the inhibitor of kappa B (IkB), which degrades and releases nuclear factor kappa B to translocate to the nucleus.
IRF4 and Th17 differentiation
In addition to the activation of STAT3, TGFβ–1 signaling and induction of RORγt, other factors have recently been reported to be necessary for IL-17 induction. Recently, Brustle and colleagues have investigated the role of Interferon regulatory factor (IRF)4 in Th17 differentiation [62]. IRF4 was originally implicated as a key inducer of GATA3 expression and Th2 lineage differentiation [63–65]. Additionally, Irf4−/− and wild-type Th cells transfected with IRF4 siRNA have impaired IL-17 production despite stimulation of TGFα-1 and IL-6. Moreover, Irf4−/− mice are resistant to EAE, which is overcome by the transfer of wild-type CD4+ T cells. However, Irf4−/− mice show a degree of resistance to EAE in excess of IL-17, IL-23, RORγt or STAT3 null animals, implying that there are additional effects of IRF deficiency. IRF4 is known to interact with NFAT and this may be one mechanism through which it could regulate IL-17 production [63, 65]. It is conceivable that IRF4 might cooperate with STAT3 to induce RORγt expression. It is of note that this is a rare example of a factor required for both Th17 and Th2 development, which is in contrast with the previously recognized links with Th17 and Th1 development via IL-23 [34] and Th17 and Treg development via TGFβ-1 [23].
Regulation of IL-17 production by G-protein-coupled receptors
G-protein-coupled receptors (GPCR), or seven transmembrane receptors are the most abundant receptors in both mice and humans. Sphingosine 1-phosphate (S1P) is a biologically active lysophospholipid that binds to its receptor S1PR1, a member of the GPCR family, and has critical functions in regulating lymphocyte migration [66]. Recently, S1P was reported to have similar effects to IL-23 in terms of increasing Th17 development from splenic CD4+ T cells [67]. That is, addition of S1P during in vitro culture resulted in increased IL-17 production and reduced IFNγ and IL-4 expression. Whether S1P is a physiologically important regulator of Th17 differentiation has yet to be determined; nonetheless, the findings raise the intriguing possibility that ligands for other GPCRs might influence T cell fate determination. In this regard, it is worth considering that chemokines also bind GPCRs and might have similar actions.
S1P engagement of its receptor induces calcium mobilization and so could activate NFAT –in this way, this ligand could function similarly to TCR engagement. S1P also activates PI3’kinase. It is notable in this study that IL-23 was used as a stimulus, implying that the cells were not naïve. Whether S1P will have effects on the differentiation of naïve CD4+ T cells to Th17 cells or has its effects on memory cells needs further exploration. At the moment, the latter seems more likely.
Cytokines and the negative regulation of Th17 differentiation
Some of the earliest studies of Th17 cells documented that IFNγ and IL-4, the signature cytokines of Th1 or Th2 cells, inhibited Th17 differentiation [34, 35]. This was one of the first pieces of evidence that argued for the idea that IL-17-producing CD4+ T cells represented a separate lineage of helper T cells. This view is complicated somewhat by the realization that some IL-17-producing T cells also produce IFNγ, whereas others only produce IL-17. Nonetheless, there are data indicating that IFNγ and IL-4 clearly inhibit IL-17 production. IFNγ and IL-4 activate STAT1 and STAT6, respectively. Of note is that STAT1-deficient helper T cells have a greater propensity to produce IL-17 [34]. Additionally, after viral infection, mice lacking STAT1 develop airway disease associated increased IL-17 expression [68]. Whether these STATs can directly regulate genes involved in Th17 differentiation (Il17a, Il17f, Rorc, Il21 and Il23r) has not been examined but is clearly worthy of investigation.
In addition, IFNγ also upregulates T-bet [69] and T-bet expression is significantly reduced in Th17 cells [34, 60, 70]. T cells from T-bet-deficient mice have increased IL-17 production and, conversely, overexpression of T-bet in T cells inhibits IL-17 production in an IFNγ-independent manner [71]. Together, these data suggest that T-bet negatively regulates IL-17 production; however, the molecular mechanism is not clear. It has also been reported that T-bet binds the Il23R promoter and enhances its expression. As a result, loss of T-bet has been suggested to interfere with the expansion of IL-17-producing lymphocytes [72, 73]. Sorting out putative positive and negative regulatory effect of T-bet during Th17 development is clearly warranted.
IL-27 is an IL-12-related cytokine that consists of two subunits, p28 and EBI3. Like IL-6, its receptor is composed of gp130 but in addition has a ligand-specific subunit termed WSX-1. Despite its similarities to IL-6, IL-27 has the opposite effect in regulating IL-17 expression. Toxoplasma gondii-infected mice lacking IL-27R developed more severe CNS inflammation and enhanced Th17 differentiation [74, 75]. In contrast to IL-6, IL-27 predominantly activates STAT1 and in STAT1-deficient mice the inhibitory effect of IL-27 is abrogated.
Strongly activated by IL-12, STAT4 is well known as a critical positive regulator of Th1 differentiation and IFNγ production. As both IL-12 and IFNγ suppress IL-17 production, similar to STAT1, the expectation might be that STAT4 would negatively regulate IL-17 expression. However, two recent studies showed that in STAT4-deficient T cells, IL-17 production was decreased, indicating the positive role of STAT4 for IL-17 production [38, 76]. The decreased IL-17 production in STAT4-deficient T cells might be related to the impaired IL-23 signaling, as IL-23 also activates STAT4. It will be of interest to further examine the role of STAT4 in Th17 differentiation.
IL-2 is a well-known T cell growth factor in vitro, but deficiency of IL-2 results in severe multi-organ autoimmune disease. This is due in part to its role in promoting the differentiation of Tregs, but, in addition, IL-2 has an important role in negatively regulating Th17 differentiation in vivo and in vitro [37, 77]. STAT5a/b are two related transcription factors that are critical for IL-2 signaling. Like Il2−/− mice, STAT5-deficient mice have autoimmune disease that is associated with loss of Treg cells and expansion of Th17 cells. Thus, STAT5a/b appear to be essential for Foxp3 expression and in constraining Th17 cells. However, the mechanisms underlying the blockade of Th17 differentiation by IL-2 via STAT5 are not entirely clear. It is possible that this is mediated indirectly via Foxp3, as STAT5 is necessary for Foxp3 expression and directly binds Foxp3 promoter [78]. In this way it is possible that IL-2-mediated inhibition of IL-17 production is related to Foxp3 induction. However, STAT5 also binds to the Il17a gene, it is possible that STAT5a/b might directly repress Il17a transcription. Other possibilities are that STAT5a/b might downregulate ROR t, which in turn would downregulate IL-17 expression.
Negative regulation of Th17 cells by retinoic acid
A series of recent studies shows that RA produced by mucosal DCs reciprocally regulates Th17 and Treg differentiation [50–52, 79–81]. That is, addition of RA inhibits Th1, Th2 and Th17 differentiation. RA also downregulates expression of RORγt, and enhances the expression of Foxp3. Moreover, T cells cultured in RA and TGFβ-1 are functionally immunosuppressive. Conversely, an RAR antagonist inhibited Foxp3 expression, indicating that expression of this key transcription factor may be dependent upon endogenous production of RA and the positive effect of RARs. Thus, these new data suggest that the balance of Treg vs. Th17 differentiation might be influenced by the actions of RAR and RORγt and modulated by the production of RA and other unknown ligands. The data also suggest that the induction of Foxp3 in developing Th17 cells might curtail differentiation of this lineage. At this point, we do not know whether Foxp3 directly binds the Il17a, Rorc or Il21 promoters, but clearly these possibilities are worth considering. In this respect, it is worth noting that IL-17 and IL-21 are downregulated in Foxp3-expressing cells [57]. Currently it is not clear whether and how RORγt and Foxp3 interact to control Th17 and Treg differentiation, but this will surely be an important area to investigate in the future to determine how these lineages might be related. As discussed, TGFβ-1 produced by Treg cells can promote Th17 differentiation, and it may be that Tregs may themselves become Th17 cells [26]. Again, this speaks to a very different model from the simplistic Th1/2 dichotomy, which implies more options and more flexibility for differentiating T cells. This more complex view of T cells also provides a better conceptual framework for understanding autoimmune disease.
Ets-1 and negative regulation of Th17 development
Ets-1 is the founding member of a family of related transcription factors that recognize a conserved GGAA/T motif. Ets-1 is important for hematopoietic cell development and lymphoid development. Additionally, Ets-1 is also known to be an important positive regulator of Th1 differentiation. Ets-1 interacts with T-bet and can directly bind to Ifng promoter [82]. Recently, it has been reported that Ets-1 deficiency is associated with increased Th17 differentiation. This is associated not only with increased production of IL-17 in vivo and in vitro, but also increased production of mRNA for IL-22 and IL-23R in response to IL-6 and TGFβ-1. However, Ets-1 apparently does not bind to the Il17 promoter. Rather, it appears that Ets-1-deficient T cells make less IL-2 and have impaired responsiveness in terms of IL-2-mediated inhibition of Th17 differentiation. Thus, it appears that a major mechanism underlying the enhanced Th17 differentiation is related to inability to produce adequate amounts of IL-2 and reduced ability to respond to this cytokine. It is likely that a number of settings in which IL-2 production is impaired will be associated with increased Th17 differentiation [83].
Il17 gene regulation and epigenetic control
It should be clear that in contrast to the understanding of the Ifng gene and the Th2 locus, we are in the infancy of understanding Il17 gene regulation. Additionally, our understanding of the epigenetic control is also quite primitive. The Il17a gene is linked to the Il17f gene on chromosome 1 (human chromosome 6) in a tail-to-tail configuration. Expression of these two cytokines appears to be linked. The promoter regions of both the Il17a and Il17f genes undergo histone H3 acetylation and K4 tri-methylation in response to TGFβ-1 and IL-6, implying increased accessibility of the locus [84]. In addition, there are eight CNS in this locus, four of which reside in the intergenic region. These sites also appear to undergo preferential histone acetylation. However, aside from STAT3, which is known to bind the Il17a gene, we know essentially nothing about the details of the regulation of the Il17a/f locus. The mechanisms through which TGFβ-1 in combination with IL-6 promote accessibility to this locus are entirely unknown. Whether and how RORγt and STAT3 might contribute to the chromatin remodeling of Il17a/Il17 locus have not been assessed, but based on what we know about the regulation of the Th2 and Ifng loci it is likely that this is a strong possibility. Since there are interchromosomal interactions between the Ifng and Th2 loci, one wonders whether interactions between Il17a/f and Foxp3 loci also exist and how interchromosomal and intrachromosomal interactions might change with activation and differentiation of T cells.
Conclusions
The past several years have witnessed an explosion of information pertaining to CD4+ T cell differentiation. In addition to Th1 and Th2 cells, we know that CD4+ T cell fate includes Th17 and Treg (natural and inducible) cells. We have also learned that Th17 cells produce IL-22 and IL-21, but that production of the former is not restricted to Th17 cells. Similarly, IL-10 is a product of Treg cells, but it, too, is produced by other T cell subsets. Furthermore, although Th1 and Th17 cells are thought to be distinct subsets, cells producing both IL-17 and IFNγ are routinely identified. The question arises, what is the real relationship between all these subsets of cytokine-producing cells? To what extent are these lineages interconvertible or are they terminally differentiated? What controls plasticity and flexibility of responses? What mechanisms underlie their development and differentiation? How do these processes relate to immunologic memory? Based on what we know about Th1 and Th2 cells, it would be expected that interactions between transcription factors and epigenetic modifications, and possibly intra- and interchromosal interactions, underlie selective regulation of cytokine genes. Understanding the molecular basis of these processes will be key to understanding the fates of differentiating T cells and the possibility of using specific lineages of cells in future therapies.
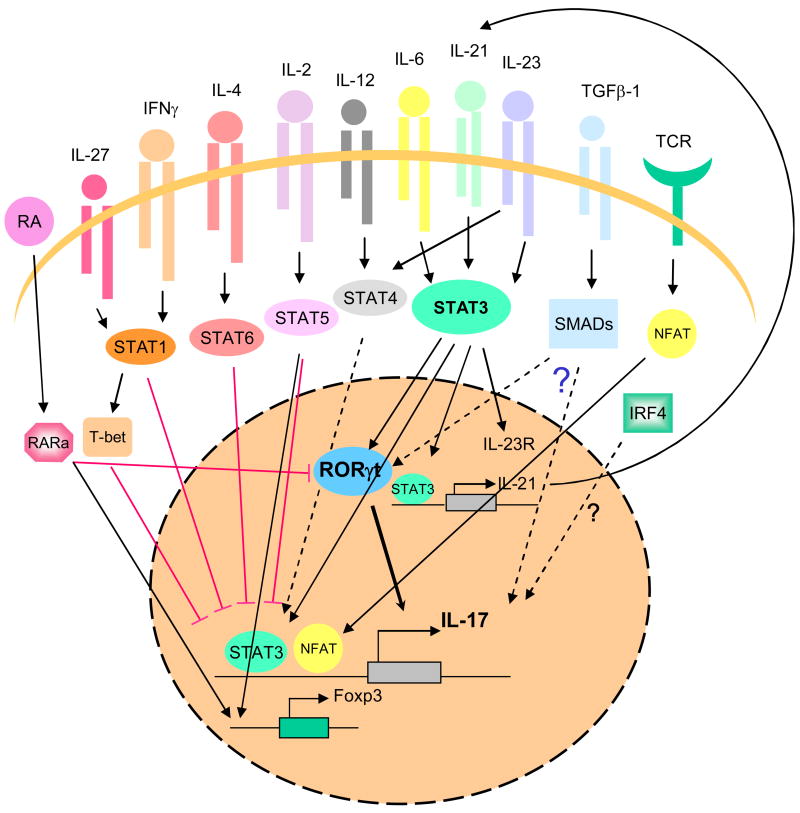
Figure 1. Signaling pathways and transcription factors that regulate Th17 differentiation.
In helper T cells, TCR stimulation activates NFAT, which likely directly regulates IL17 expression. Cytokines such IL-6, IL-21 and IL-23, activate STAT3, which also binds Il17a/f, and Il21 genes and controls Rorc and Il23r expression. TGFβ-1 signaling involves the activation of SMAD proteins. This cytokine acts in conjunction with STAT3 and Rorγt, although the mechanism through which TGFβ-1 promotes differentiation of Th17 cells is presently unknown. IRF4 is also a positive regulator of Th17 differentiation, how it acts is unclear.
Unsurprisingly, many factors inhibit Th17 differentiation. Retinoic acid, acting through its cognate receptors, downregulates IL-17 expression and upregulates Foxp3 expression. IL-2, IL-4, IFNγ and IL-27 activate STAT5, STAT6 and STAT1, respectively, also negatively regulate Il17a expression. Foxp3 also inhibits cytokine production.
References
- 1.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–57. [PubMed] [Google Scholar]
- 2.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–93. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 3.Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–44. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 4.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–79. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–42. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang S, Aune TM. Dynamic changes in histone-methylation ‘marks’ across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat Immunol. 2007;8:723–31. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- 7.Bream JH, Hodge DL, Gonsky R, Spolski R, Leonard WJ, Krebs S, et al. A distal region in the interferon-gamma gene is a site of epigenetic remodeling and transcriptional regulation by interleukin-2. J Biol Chem. 2004;279:41249–57. doi: 10.1074/jbc.M401168200. [DOI] [PubMed] [Google Scholar]
- 8.Shnyreva M, Weaver WM, Blanchette M, Taylor SL, Tompa M, Fitzpatrick DR, et al. Evolutionarily conserved sequence elements that positively regulate IFNgamma expression in T cells. Proc Natl Acad Sci U S A. 2004;101:12622–7. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, et al. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–29. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFNgamma loci accompany Th1/Th2 differentiation. J Immunol. 2002;169:647–50. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 11.Usui T, Preiss JC, Kanno Y, Yao ZJ, Bream JH, O’Shea JJ, et al. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. J Exp Med. 2006;203:755–66. doi: 10.1084/jem.20052165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spilianakis CG, Flavell RA. Epigenetic regulation of Ifng expression. Nat Immunol. 2007;8:681–3. doi: 10.1038/ni0707-681. [DOI] [PubMed] [Google Scholar]
- 13.Makar KW, Perez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB. Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nat Immunol. 2003;4:1183–90. doi: 10.1038/ni1004. [DOI] [PubMed] [Google Scholar]
- 14.Mohrs M, Blankespoor CM, Wang ZE, Loots GG, Afzal V, Hadeiba H, et al. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat Immunol. 2001;2:842–7. doi: 10.1038/ni0901-842. [DOI] [PubMed] [Google Scholar]
- 15.Solymar DC, Agarwal S, Bassing CH, Alt FW, Rao A. A 3′ enhancer in the IL-4 gene regulates cytokine production by Th2 cells and mast cells. Immunity. 2002;17:41–50. doi: 10.1016/s1074-7613(02)00334-5. [DOI] [PubMed] [Google Scholar]
- 16.Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–53. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 17.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–45. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 18.Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8:639–46. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 19.Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–9. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korn T, Bettelli E, Gao W, Awasthi A, Jager A, Strom TB, et al. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448:484–7. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–3. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 22.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007 doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 23.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–8. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 24.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–86. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shevach EM. Regulatory T cells in autoimmmunity*. Annu Rev Immunol. 2000;18:423–49. doi: 10.1146/annurev.immunol.18.1.423. [DOI] [PubMed] [Google Scholar]
- 26.Xu L, Kitani A, Fuss I, Strober W. Cutting edge: regulatory T cells induce CD4+CD25-Foxp3- T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J Immunol. 2007;178:6725–9. doi: 10.4049/jimmunol.178.11.6725. [DOI] [PubMed] [Google Scholar]
- 27.Chen Z, Tato C, Muul L, Laurence A, O’Shea JJ. Distinct Regulation of IL-17 in Human Helper T Lymphocytes. Arthritis Rheum. 2007 doi: 10.1002/art.22866. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michel ML, Keller AC, Paget C, Fujio M, Trottein F, Savage PB, et al. Identification of an IL-17-producing NK1.1neg iNKT cell population involved in airway neutrophilia. J Exp Med. 2007;204:995–1001. doi: 10.1084/jem.20061551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stark MA, Huo Y, Burcin TL, Morris MA, Olson TS, Ley K. Phagocytosis of apoptotic neutrophils regulates granulopoiesis via IL-23 and IL-17. Immunity. 2005;22:285–94. doi: 10.1016/j.immuni.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Liu XK, Lin X, Gaffen SL. Crucial role for nuclear factor of activated T cells in T cell receptor-mediated regulation of human interleukin-17. J Biol Chem. 2004;279:52762–71. doi: 10.1074/jbc.M405764200. [DOI] [PubMed] [Google Scholar]
- 31.Hoeve MA, Savage ND, de Boer T, Langenberg DM, de Waal Malefyt R, Ottenhoff TH, et al. Divergent effects of IL-12 and IL-23 on the production of IL-17 by human T cells. Eur J Immunol. 2006;36:661–70. doi: 10.1002/eji.200535239. [DOI] [PubMed] [Google Scholar]
- 32.Wei L, Laurence A, Elias K, O’Shea J. IL-21 is produced by TH17 cells and drives IL-17 production in a STAT3-dependent manner. J Biol Chem. 2007 Oct 2; doi: 10.1074/jbc.M705100200. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278:1910–4. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 34.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–32. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 35.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–41. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007 doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 37.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–81. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 38.Mathur AN, Chang HC, Zisoulis DG, Stritesky GL, Yu Q, O’Malley JT, et al. Stat3 and Stat4 Direct Development of IL-17-Secreting Th Cells. J Immunol. 2007;178:4901–7. doi: 10.4049/jimmunol.178.8.4901. [DOI] [PubMed] [Google Scholar]
- 39.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282:9358–63. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 40.Chen Z, Laurence A, Kanno Y, Pacher-Zavisin M, Zhu BM, Tato C, et al. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc Natl Acad Sci U S A. 2006;103:8137–42. doi: 10.1073/pnas.0600666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong PK, Egan PJ, Croker BA, O’Donnell K, Sims NA, Drake S, et al. SOCS-3 negatively regulates innate and adaptive immune mechanisms in acute IL-1-dependent inflammatory arthritis. J Clin Invest. 2006;116:1571–81. doi: 10.1172/JCI25660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. Cell. 2006;126:1121–33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 43.Eberl G, Littman DR. The role of the nuclear hormone receptor RORgammat in the development of lymph nodes and Peyer’s patches. Immunol Rev. 2003;195:81–90. doi: 10.1034/j.1600-065x.2003.00074.x. [DOI] [PubMed] [Google Scholar]
- 44.Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–73. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 45.Kurebayashi S, Ueda E, Sakaue M, Patel DD, Medvedev A, Zhang F, et al. Retinoid-related orphan receptor gamma (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc Natl Acad Sci U S A. 2000;97:10132–7. doi: 10.1073/pnas.97.18.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 47.He YW, Deftos ML, Ojala EW, Bevan MJ. RORgamma t, a novel isoform of an orphan receptor, negatively regulates Fas ligand expression and IL-2 production in T cells. Immunity. 1998;9:797–806. doi: 10.1016/s1074-7613(00)80645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Littman DR, Sun Z, Unutmaz D, Sunshine MJ, Petrie HT, Zou YR. Role of the nuclear hormone receptor ROR gamma in transcriptional regulation, thymocyte survival, and lymphoid organogenesis. Cold Spring Harb Symp Quant Biol. 1999;64:373–81. doi: 10.1101/sqb.1999.64.373. [DOI] [PubMed] [Google Scholar]
- 49.Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–38. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 50.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-{beta} and retinoic acid dependent mechanism. J Exp Med. 2007;204:1757–64. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–85. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benson MJ, Pino-Lagos K, Rosemblatt M, Noelle RJ. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J Exp Med. 2007;204:1765–74. doi: 10.1084/jem.20070719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung Y, Yang X, Chang SH, Ma L, Tian Q, Dong C. Expression and regulation of IL-22 in the IL-17-producing CD4+ T lymphocytes. Cell Res. 2006 doi: 10.1038/sj.cr.7310106. [DOI] [PubMed] [Google Scholar]
- 54.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26:579–91. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 55.Gorelik L, Fields PE, Flavell RA. Cutting edge: TGF-beta inhibits Th type 2 development through inhibition of GATA-3 expression. J Immunol. 2000;165:4773–7. doi: 10.4049/jimmunol.165.9.4773. [DOI] [PubMed] [Google Scholar]
- 56.Gorelik L, Constant S, Flavell RA. Mechanism of transforming growth factor beta-induced inhibition of T helper type 1 differentiation. J Exp Med. 2002;195:1499–505. doi: 10.1084/jem.20012076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–5. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 58.Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, Sallusto F. Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nat Immunol. 2007 doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- 59.Yang X, Letterio JJ, Lechleider RJ, Chen L, Hayman R, Gu H, et al. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. Embo J. 1999;18:1280–91. doi: 10.1093/emboj/18.5.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–89. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 61.Sutton C, Brereton C, Keogh B, Mills KH, Lavelle EC. A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J Exp Med. 2006;203:1685–91. doi: 10.1084/jem.20060285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007 doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 63.Hu CM, Jang SY, Fanzo JC, Pernis AB. Modulation of T cell cytokine production by interferon regulatory factor-4. J Biol Chem. 2002;277:49238–46. doi: 10.1074/jbc.M205895200. [DOI] [PubMed] [Google Scholar]
- 64.Lohoff M, Mittrucker HW, Prechtl S, Bischof S, Sommer F, Kock S, et al. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc Natl Acad Sci U S A. 2002;99:11808–12. doi: 10.1073/pnas.182425099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rengarajan J, Mowen KA, McBride KD, Smith ED, Singh H, Glimcher LH. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J Exp Med. 2002;195:1003–12. doi: 10.1084/jem.20011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosen H, Goetzl EJ. Sphingosine 1-phosphate and its receptors: an autocrine and paracrine network. Nat Rev Immunol. 2005;5:560–70. doi: 10.1038/nri1650. [DOI] [PubMed] [Google Scholar]
- 67.Liao JJ, Huang MC, Goetzl EJ. Cutting edge: alternative signaling of th17 cell development by sphingosine 1-phosphate. J Immunol. 2007;178:5425–8. doi: 10.4049/jimmunol.178.9.5425. [DOI] [PubMed] [Google Scholar]
- 68.Hashimoto K, Durbin JE, Zhou W, Collins RD, Ho SB, Kolls JK, et al. Respiratory syncytial virus infection in the absence of STAT 1 results in airway dysfunction, airway mucus, and augmented IL-17 levels. J Allergy Clin Immunol. 2005;116:550–7. doi: 10.1016/j.jaci.2005.03.051. [DOI] [PubMed] [Google Scholar]
- 69.Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, et al. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci U S A. 2001;98:15137–42. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–4. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 71.Mathur AN, Chang HC, Zisoulis DG, Kapur R, Belladonna ML, Kansas GS, et al. T-bet is a critical determinant in the instability of the IL-17-secreting T-helper phenotype. Blood. 2006;108:1595–601. doi: 10.1182/blood-2006-04-015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–8. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 73.Gocke AR, Cravens PD, Ben LH, Hussain RZ, Northrop SC, Racke MK, et al. T-bet regulates the fate of Th1 and Th17 lymphocytes in autoimmunity. J Immunol. 2007;178:1341–8. doi: 10.4049/jimmunol.178.3.1341. [DOI] [PubMed] [Google Scholar]
- 74.Stumhofer JS, Laurence A, Wilson EH, Huang E, Tato CM, Johnson LM, et al. Interleukin 27 negatively regulates the development of interleukin 17-producing T helper cells during chronic inflammation of the central nervous system. Nat Immunol. 2006;7:937–45. doi: 10.1038/ni1376. [DOI] [PubMed] [Google Scholar]
- 75.Batten M, Li J, Yi S, Kljavin NM, Danilenko DM, Lucas S, et al. Interleukin 27 limits autoimmune encephalomyelitis by suppressing the development of interleukin 17-producing T cells. Nat Immunol. 2006;7:929–36. doi: 10.1038/ni1375. [DOI] [PubMed] [Google Scholar]
- 76.Hildner KM, Schirmacher P, Atreya I, Dittmayer M, Bartsch B, Galle PR, et al. Targeting of the transcription factor STAT4 by antisense phosphorothioate oligonucleotides suppresses collagen-induced arthritis. J Immunol. 2007;178:3427–36. doi: 10.4049/jimmunol.178.6.3427. [DOI] [PubMed] [Google Scholar]
- 77.Kryczek I, Wei S, Zou L, Altuwaijri S, Szeliga W, Kolls J, et al. Cutting edge: Th17 and regulatory T cell dynamics and the regulation by IL-2 in the tumor microenvironment. J Immunol. 2007;178:6730–3. doi: 10.4049/jimmunol.178.11.6730. [DOI] [PubMed] [Google Scholar]
- 78.Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007 doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, et al. Reciprocal Th-17 and Regulatory T Cell Differentiation Mediated by Retinoic Acid. Science. 2007 doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 80.Schambach F, Schupp M, Lazar MA, Reiner SL. Activation of retinoic acid receptor-alpha favours regulatory T cell induction at the expense of IL-17-secreting T helper cell differentiation. Eur J Immunol. 2007 doi: 10.1002/eji.200737621. [DOI] [PubMed] [Google Scholar]
- 81.Elias K, Laurence A, Davidson T, Stephens GYKEMS, et al. Retinoic Acid Inhibits Th17 Polarization and Enhances FoxP3 Expression through a Stat-3/Stat-5 Independent Signaling Pathway. Blood. 2007 Oct 19; doi: 10.1182/blood-2007-06-096438. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grenningloh R, Kang BY, Ho IC. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J Exp Med. 2005;201:615–26. doi: 10.1084/jem.20041330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho I. Ets-1 is a negative regulator of Th17 differentiation. J Exp Med. 2007 doi: 10.1084/jem.20070994. submitted manuscript. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–72. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]