Abstract
Advances in computer power, methodology, and empirical force fields now allow routine “stable” nanosecond-length molecular dynamics simulations of DNA in water. The accurate representation of environmental influences on structure remains a major, unresolved issue. In contrast to simulations of A-DNA in water (where an A-DNA to B-DNA transition is observed) and in pure ethanol (where disruption of the structure is observed), A-DNA in ≈85% ethanol solution remains in a canonical A-DNA geometry as expected. The stabilization of A-DNA by ethanol is likely due to disruption of the spine of hydration in the minor groove and the presence of ion-mediated interhelical bonds and extensive hydration across the major groove.
The polymorphic structure and conformational flexibility of DNA play an important role in a variety of biological processes. Sequence specific flexibility and deformability are important in transcriptional regulation; specific examples include transcription factor-induced bending of DNA (1), unwinding of the DNA helix by zinc finger proteins (2), and the transportation of one duplex through another by type II DNA topoisomerases (3). In addition to structural deformations induced by ligand or protein binding, conformational transitions in DNA among the various canonical forms are also biologically relevant. A particularly intriguing example, which increases resistance to UV radiation damage in Gram-positive bacteria, is the induction of an A-form geometry in an otherwise B-DNA duplex by α/β-type small acid-soluble spore proteins (4). Local transitions to Z-DNA in alternating purine–pyrimidine segments may also help promote and regulate DNA condensation (5). These conformational transitions occur because the conformation of DNA is strongly dependent upon the environment. Variations in ionic strength and identity, sequence, water activity, and ligand/protein binding all can modulate DNA structure.¶ An example is the B-DNA to A-DNA transition observed in 76%, 80%, or 84% ethanol (vol/vol) solutions of DNA fibers in the presence of Na+, K+, or Cs+, respectively (9). In contrast, the more strongly hydrated counterions (Li+ and Mg2+) inhibit the transition to A-DNA; instead, B-DNA to C-DNA (7) or B-DNA to P-DNA (8) transitions are observed. Although a number of theories have been advanced regarding the molecular mechanisms underlying the stabilization of A-DNA in mixed water and ethanol solutions, no one has solved a structure of an “isolated” A-DNA in an ethanolic solution. In this report, we demonstrate that molecular dynamics simulations can provide this molecular level description. In contrast to pure water, where an A-DNA to B-DNA transition is observed on a roughly 500 ps time scale (16), simulations of d[CCAACGTTGG]2 in pure ethanol or mixed water ethanol show a profoundly different behavior. Simulations starting from an A-DNA structure in pure ethanol move away from the canonical geometry [∼4.0 Å all atom root-mean-square deviation (RMSd) by ∼2 ns] and local distortions in the helicoidal parameters are evident. In a mixed water and ethanol solution, A-DNA remains in a A-like geometry for greater than 3 ns of molecular dynamics.
Molecular dynamics simulations on nucleic acids have a relatively rich history starting with the first simulations of Levitt (17) and Karplus et al. (18). The early simulations [see reviews by Beveridge et al. (19, 20)] were generally limited to short time scales (<200 ps), typically displayed anomalous structure such as base pair fraying, and demonstrated the need for including some representation of solvent and a reasonable treatment of the highly charged phosphate backbone. Advances in computer power, empirical force field representations, and the development of more reasonable means to handle the long-ranged electrostatic interactions now allow routine “stable” nanosecond length unrestrained molecular dynamics simulations of nucleic acids in water with explicit counterions [for a review see Louise-May et al. (21)]. From these studies, it has become clear that a well balanced force field is necessary to properly represent the expected structural preferences and dynamics. However, the precise balance in the force field has not been fully put to the test. Although our previous simulations demonstrate that B-DNA is favored over A-DNA in water (6) as is expected, B-DNA could be artificially stable under all conditions. Therefore, simulations were run under conditions that are expected to stabilize A-DNA using the same d[CCAACGTTGG]2 sequence, the same force field (22), and an equivalent simulation protocol‖ applying the particle mesh Ewald (PME) method (25) implemented into amber4.1 (26). The results suggest that the current DNA simulation protocols have reached a sufficient level of realism that they can reasonably represent the environmental effect on DNA structure.
When A-DNA is placed in pure ethanol (with or without counterions)**, the structure moves away from a canonical A geometry, with all atom RMSd values rising monotonically and approaching ≈4 Å after ≈2 ns of simulations. The structures visited during the dynamics are characterized by local distortions in the helicoidal parameters. Similar behavior was observed in a 1.5 ns simulation of B-DNA in pure ethanol. These distortions in the structure are not surprising since no water is present in these simulations to solvate the DNA; even under extremely dehydrating conditions, A-DNA still has some tightly associated water (15). Moreover, in solutions of greater than 90% ethanol, A-DNA to P-DNA transitions are typically encountered (8, 9). P-DNA is a highly aggregated form of DNA that appears to have lost all of its base stacking. In these simulations, because we are only simulating one structure in a periodic box, the nucleic acid cannot aggregate. However, the simulations in pure ethanol suggest that we are seeing some secondary structural collapse in the DNA [as is evident by low twist (≈18°), high roll (≈23°), and large rise (≈4.8 Å) at some base pair steps], which is consistent with the experiment. However, the simulations have not been run long enough to definitively support this claim. Despite this, these results are presented because the behavior observed in pure ethanol is clearly different than is observed in water or in a mixed water and ethanol solution.
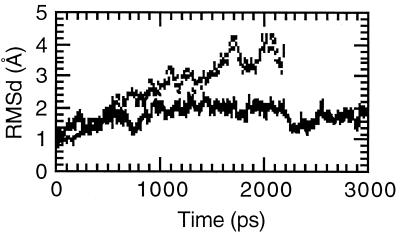
In solutions with greater than ≈80% ethanol it is expected that A-DNA should be stable. To see if our models could reproduce this behavior, simulations were run in mixed water and ethanol solutions. To avoid a very long equilibration protocol to hydrate the DNA when the nucleic acid is placed into a pre-equilibrated box of water and ethanol, “hydrated” DNA was placed into a box of pure ethanol. In other words, a snapshot from a previous trajectory of A-DNA was taken‡‡ and stripped of all the waters except the 500 waters closest to any DNA or counterion atom. This leads to ≈20 waters per nucleotide and ≈6 waters per counterion, which is equivalent to the hydration expected for B-DNA (13, 15). A 3 ns molecular dynamics simulation was performed on A-DNA in this mixed ethanol/water solution††. Fig. 1 presents the RMSd to A-DNA over the course of the simulations in pure ethanol and mixed water and ethanol solutions. The structure in the mixed water and ethanol solution stays within ≈2.0 Å of canonical A-DNA. The helicoidal parameters presented in Table 1 confirm that the structure is canonical A and moreover even closer to canonical A than the A-RNA structure calculated previously for this sequence (23). The structure does not display the intrastrand stacking at the central CpG step seen in corresponding A-RNA simulations (23) and in a number of A-DNA crystal structures (29, 30). The differences from canonical A-DNA are an average pucker value that is higher than expected, which indicates significant sugar repuckering to C2′-endo and an axial rise that is slightly higher than expected; this leads to a structure that is perhaps closer to A′-DNA (31).
Figure 1.
RMSd over the course of the A-DNA simulation. The RMSd of all DNA atoms from canonical A-DNA is shown as a function of time for the simulation of A-DNA in mixed ethanol/water solution (solid line) and A-DNA in pure ethanol (broken line).
Table 1.
Selected helicoidal and backbone angle values
| Canonical A | A-DNA | A-DNAV2=0.3 | A-RNA | B (in water) | |
|---|---|---|---|---|---|
| Twist, ° | 32.7 | 32.6 | 31.0 | 30.9 | 30.9 |
| Rise, Å | 2.56 | 2.92 | 2.95 | 2.70 | 3.26 |
| Inclination, ° | 19.1 | 15.8 | 10.9 | 15.0 | 4.9 |
| x-disp., Å | −5.43 | −4.0 | −3.4 | −5.19 | −2.96 |
| RMS to A, Å | 0.00 | 1.66 | 1.68 | 2.09 | 3.85 |
| Pucker, ° | 13.1 | 80.1 | 72.7 | 22.6 | 122.8 |
| χ, ° | 205.8 | 222.6 | 220.7 | 201.7 | 234.2 |
| δ, ° | 84.3 | 102.8 | 96.7 | 79.3 | 116.6 |
Values of selected average helicoidal parameters, backbone angles, and sugar pucker values are presented for a canonical A-DNA model structure (canonical-A), the simulation of A-DNA in mixed ethanol/water solution (A-DNA, 2,000–3,000 ps), the simulation of B-DNA with the modified V2 term on the O–C–C–O torsions (from 1.0 to 0.3 kcal/mol), which underwent a B-DNA to A-DNA transition (A-DNAV2=0.3, 2,000–3,000 ps), A-RNA in pure water (A-RNA, 1,030–2,030 ps) (23), and B-DNA in pure water (B-DNA, 400–1,400 ps) (16). All of the helicoidal values were calculated using the Dials and Windows interface (27) to Curves (28) from average structures over 1 ns portions of the trajectories (except in the case of the canonical A-DNA structure). The average structures were created by RMS fitting all DNA atoms at 1 ps intervals from the trajectories and coordinate averaging. Helicoidal values reported are averages over all base pairs or base pair steps where appropriate. The sugar pucker, χ and δ angles are 1 ps averages over 1 ns portions of the dynamics, averaged over all nucleotides (except in the case of the canonical A model structure). It should be noted, as discussed in our previous work (23), that the means of the calculated values from each snapshot at 1 ps intervals over the same range are notably different in the rise, x-displacement (x-disp.), and base pair inclination from those reported in this table and are more B-like. The means of the values calculated from each individual snapshot at 1 ps intervals over the same range for the rise, x-displacement, and base pair inclination are 3.15 Å, −2.67 Å, and 6.3°, respectively, for the A-DNA trajectory and 3.48 Å, −1.27 Å, and −6.2°, respectively, for the B-DNA trajectory (16).
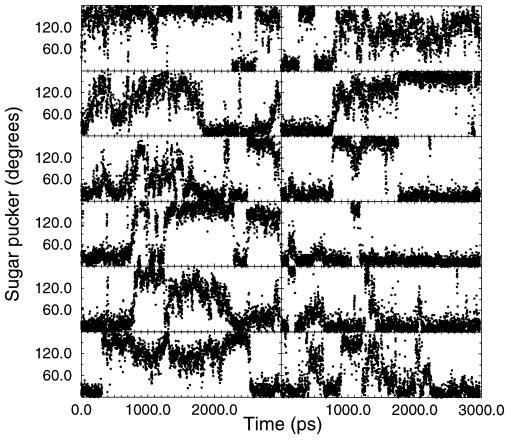
The observation of a stable A-DNA structure, despite this higher average pucker and repuckering between C3′-endo and C2′-endo, suggests that it is not a difference in the relative stability of C3′-endo sugar puckers in ethanolic solution compared with pure water that leads to the stabilization of A-DNA. Fig. 2 presents the time course of the central six sugar puckers. Sugar repuckering occurs at every nucleotide with the terminal residues displaying more C2′-endo puckering on average. The time course is shown to demonstrate that there is not a general trend toward C2′-endo puckers at later parts of the simulation, or in other words, we are not seeing a slow transition to B-DNA. Interestingly, the terminal groups, particularly the guanines, spend a significant time with C3′-endo puckers. This suggests that the structure is in the process of transitioning to a B-DNA structure; however, the water and counterions in the major groove appear to inhibit this process in the central core of the duplex and stabilize A-DNA.
Figure 2.
Time course of the sugar puckers. The time course of the sugar pucker pseudorotation phase as a function of time from the simulation of A-DNA in mixed water/ethanol is displayed for the central six residues. From top to bottom: on the left the sequence is AACGTT and on the right is the corresponding paired base, or TTGCAA.
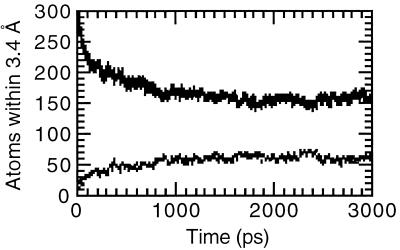
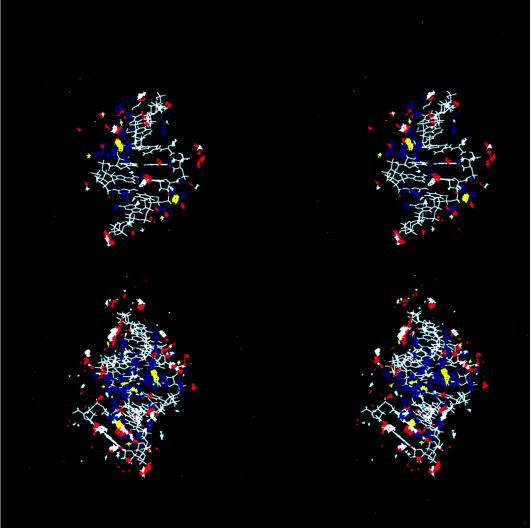
A-DNA is generally less hydrated than B-DNA; this is part of the reason why a B-DNA to A-DNA transition is observed in mixed water and ethanol solutions. Fig. 3 shows the number of water and ethanol oxygen atoms within 3.4 Å of any DNA atom over the course of the simulation in the mixed solvent. As the simulation proceeds, water is clearly being “pulled” away from the DNA. By ≈2 ns, ≈15 waters and ≈6.0 ethanols per base pair are associated with the DNA. To better visualize the water and counterion association, contours of the water oxygen, ethanol oxygen, and carbon (C5), and Na+ counterion density over the final nanosecond of the trajectory are displayed as a stereoview in Fig. 4. Despite fewer waters per base pair, the DNA is still extensively hydrated. Water and counterion density is extensive in the major groove (Fig. 4 Lower) with a large body of closely associated water bridging the two strands in a bend across the major groove. The extensive hydration and counterion association likely overcomes the interstrand phosphate repulsion and helps stabilize A-DNA. In addition to the extensive hydration transverse to the major groove, spines of hydration are also apparent as is extensive hydration on the major groove side of the backbone. The hydration in the major groove is more extensive than was seen in corresponding simulations of A-RNA in water (23) and the hydration of the backbone is greater than was seen in simulations of B-DNA in water (16). The minor groove is also extensively hydrated; however, the density appears lower than is seen in the simulations of A-RNA in water. The terminal base pair steps at both ends of the helix shows extensive ethanol association in the minor groove (Fig. 4 Lower). Ethanol is also apparent in the central part of the minor groove and appears to partially disrupt the spine of hydration in the minor groove. The minor groove side of the backbone also displays significant ethanol oxygen association.
Figure 3.
Closely associated oxygen atoms. Graph represents the number of water oxygens (solid line) and ethanol oxygens (broken line) within 3.4 Å of any DNA atom over the course of the A-DNA simulation in mixed water/ethanol solution.
Figure 4.
Hydration and counterion association in grooves of A-DNA. Shown is a stereoview into the minor (Upper) and major (Lower) grooves of the average structure from the final nanosecond of the A-DNA simulation in mixed water/ethanol. Contoured average water oxygen (blue), ethanol oxygen (red), ethanol methyl carbon C5 (white), and Na+ counterion (yellow) density is also displayed. These data are generated (25) by RMS fitting all atoms of the DNA to the first frame, at 1 ps intervals. Then a grid is constructed around the DNA (50 Å3), and for each frame in the trajectory, if the center of the atom of interest is within a particular grid element (each 0.5 Å3), a counter is updated. This grid is then contoured using the midasplus (32) density delegate (written by Christian Schafmeister, University of California, San Francisco). The water and counterion density is contoured at 15.0 hits per grid element (or roughly 3.6 times bulk water density) and the ethanol atoms are contoured at 8.0 hits per grid element (or roughly 4.8 times bulk ethanol density). To simplify the view in Upper and avoid obscuring the view of the hydration into the minor groove, the rear clipping plane is set so that the major groove density in the background is not as apparent; this clips some of the DNA structure in the background and the terminal base pairs (which are visible in the lower figure).
Less apparent hydration in the minor groove and extensive hydration and ion association in the major groove is consistent with the experimental observation of ions in the major groove of A-DNA. A-DNA is presumably stabilized not only by ions and hydration in the major groove, but likely by absence of stabilizing counterion association and, consistent with the “groove binding model” (6, 12), by hydration in the minor groove.
To see if a spontaneous B-DNA to A-DNA transition would be observed, simulations were also run starting with B-DNA under the same environmental conditions (≈85% ethanol). In these the structure remains in a B-DNA structure for more than 3 ns. Thus, we cannot claim that the lowest free energy in our model with 85% ethanol is A-DNA, but only that the ethanol environment clearly stabilizes A-DNA compared with simulations in pure water. However, a subtle modification in one torsion parameter, reduction of the V2 term in the O–C–C–O torsions from 1.0 to 0.30 kcal/mol to better stabilize the C3′-endo sugar pucker, allows a spontaneous B-DNA to A-DNA transition in ≈85% ethanol at ≈1.5 ns (see the A-DNAV2=0.3 averages in Table 1). The transition to A-DNA is rapid and characterized by a drop in the RMSd to cannonical A-DNA from ≈4 Å at 1,350 ps to less than 2.5 Å by 1,500 ps. At 1,350 ps, although approximately six of the nucleotides display predominantly C3′-endo sugar puckers, the structure is still closer to canonical B-DNA. By 1,500 ps, repuckering of eight additional nucleotides from C2′-endo to C3′-endo facilitates the transition to A-DNA. It should be noted that in pure water (with the modified torsion parameters) the B-DNA average structure obtained (after an A-DNA to B-DNA transition) is less cleanly B-DNA (with average rise of 3.15 Å, x-displacement of −3.34 Å, and inclination of 5.82° in the average structure over 1,655–1,755 ps) than is observed with the original model.
These results suggest that molecular dynamics simulations, with a state-of-the-art force field and proper treatment of the long-ranged electrostatics, can reasonably represent the effect of the environment on DNA structure. How quantitatively accurate these results are can be further tested with high resolution NMR studies in these solvent conditions. However, the simulations demonstrate the stabilization of B-DNA and A-RNA in water, unstable A-DNA and B-DNA in pure ethanol, and stabilization of A-DNA in mixed water ethanol solution. Consistent with the experiment, these studies suggest that the relative stabilization of, and transition between, A- and B-form geometries involves subtle differences in the specific hydration and counterion association with the nucleic acid. Whereas B-DNA is stabilized by extensive hydration in the minor groove and by hydrated counterions in the minor groove anchoring the two strands (Na+ < K+ < Cs+), A-DNA is stabilized by major groove hydration and ion association, and also by ion-mediated interhelical bonds across the major groove and between duplexes (Na+ > K+ > Cs+) (9). In conclusion, we have shown that state-of-the-art simulations, which reproduce the experiment well for B-DNA in water (16), show the expected environmental dependence in pure ethanol and mixed water and ethanol solutions.
Acknowledgments
We would like to acknowledge the Pittsburgh Supercomputing Center (MCA93S017P) and Cray Research for use of the Cray T3D and T3E, the University of California, San Francisco, Computer Graphics Laboratory (RR-1081), Jim Vincent and Ken Merz for the original Message Passing Interface version of amber, and Tom Darden for helpful discussions. P.A.K. is grateful for research support from the National Institutes of Health (NIH) (Grant CA-25644). T.E.C. would like to acknowledge research support from NIH Biotechnology Training Grant GM08388 and as a UCSF Chancellor’s Graduate Research Fellow.
ABBREVIATION
- RMSd
root-mean-square deviation
Footnotes
Examples of environmentally induced changes in secondary structure include increasing ethanol concentrations leading to B-DNA to A-DNA (6), C-DNA (7), or P-DNA (8) transitions depending on the nature of the counterion (9); binding of neomycin or spermine to induce a B-DNA to A-DNA transition (10) or binding of distamycin A or netropsin to induce an A-DNA or Z-DNA to B-DNA transition (11); or binding of Co(NH3)63+ to induce a B-DNA to A-DNA or Z-DNA transition (10). Common to each of these means to transition among the various structural forms seems to be both a subtle modulation of the hydration of DNA and the ionic association with the negatively charged phosphate backbone and grooves. Models such as the groove binding model (6, 12) explain the stabilization of B-DNA by ions interacting directly with base atoms in the minor groove (Cs+ > K+ > Na+) or indirectly via water bridges (Li+). B-DNA is also stabilized by extensive solvation of the grooves and backbone (13), such as the “spine of hydration” in the minor groove (14). A-DNA, on the other hand, is significantly less hydrated (13, 15) and is stabilized by aggregation and interhelical contacts (9) and also by ions interacting primarily in the major groove [such as Co(NH3)63+] (10) and with the phosphates. There is a profound dependence on the nature of the ionic species; for example, the square planer Pt(NH3)42+ ion and strongly hydrated Mg2+ and Li+ ions do not induce a B-DNA to A-DNA transition (10).
All simulations were performed applying the particle mesh Ewald method within amber4.1 with constant pressure and temperature (300 K) with Berendsen temperature coupling, SHAKE on hydrogens, a 2 fs time step, and a 9 Å cutoff applied to the Lennard–Jones interactions. Differences from our previous studies (16, 23) include the use of an all-atom ethanol model and slightly longer equilibration periods when the DNA is held fixed (100 ps in pure ethanol and ≈300 ps in the water/ethanol with the ions and water held fixed as well) to better equilibrate the ethanol prior to production simulations. An all-atom flexible ethanol model was developed using the standard nonbonded and intramolecular parameters from the Cornell et al. (22) force field and charges generated from a restrained electrostatic potential fit (24) to a 6–31 G* wave function of the minimum geometry of ethanol. The charges and atom types for the CH3CH2OH molecule are as follows: CT, −0.0990; HC, 0.0345; CT, 0.3318; H1, −0.0294; OH, −0.6718; and HO, 0.4143. These charges correspond to a total dipole moment of 1.77 Debye, which is about 5% higher than the gas-phase dipole moment for ethanol of 1.68 Debye. Molecular dynamics simulations of a periodic box of 213 ethanol molecules under constant pressure conditions yield, with this parameter set, a density of 0.781 g/cm3 and a heat of vaporization of 9.989 kcal/mol; these compare well with the experimental values of 0.789 g/cm3 (error −1.0%) and 9.67 kcal/mol (error 3.2%), respectively. When PME is applied, the density (0.800 g/cm3) and head of vaporization (10.21 kcal/mol) increase slightly.
The simulation of A-DNA in pure ethanol with Na+ counterions had 929 ethanol molecules and was placed in a box of ≈58 Å by ≈46 Å by ≈46 Å and represented 9,011 atoms. The simulation without counterions had 941 ethanol molecules in a box of roughly the same size and represented 9,101 atoms; the net charge of −18.0 on the system was neutralized by smearing it over all atoms or by subtracting −18.0/9101 from the charge on each atom. The latter simulation was run, since in the simulation with counterions, the nonhydrated counterions never moved away from the phosphate atoms, which could have lead to the distortions in the structure. However, when the counterions were removed, the simulation moved away from canonical A-DNA more rapidly. A 1.5 ns simulation of the average B-DNA structure from previous simulations (16) was also run in a box (≈59 Å by 47 Å by 47 Å) of 1,162 ethanol molecules and 18 Na+ counterions.
The A-DNA “snapshot” was generated and equilibrated from a canonical A-form geometry as described previously (16), except that that the pucker was forced to remain C3′-endo by the addition of a flatwell restraint on the C1′–C2′–C3′–C4′ torsion to keep the angle between 30° and 40°, as discussed in our previous work (23); this prevents the structure from undergoing an A-DNA to B-DNA transition. The snapshot represents the structure after 20 ps of production dynamics; at this point, the ions are still in rather close proximity to the phosphate groups of the nucleic acid.
The simulation contained 500 waters and 1,240 ethanol molecules in addition to the DNA atoms and 16 Na+ counterions. Based on the experimental densities, this is ≈85.5% ethanol by volume. The equilibration protocol involved performing ≈300 ps of dynamics with the DNA, Na+ ions, and water held fixed to allow the ethanol to equilibrate. The box size was ≈60 Å by 50 Å by 50 Å and the PME charge grid was 64 Å by 54 Å by 54 Å. Otherwise, the simulation conditions were equivalent to our previous work (23).
References
- 1.Ansari A Z, Bradner J E, O’Halloran T V. Nature (London) 1995;374:371–375. doi: 10.1038/374370a0. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y, Berg J M. Biochemistry. 1996;35:3845–3848. doi: 10.1021/bi952384p. [DOI] [PubMed] [Google Scholar]
- 3.Berger J M, Wang J C. Curr Opin Struct Biol. 1996;6:84–90. doi: 10.1016/s0959-440x(96)80099-6. [DOI] [PubMed] [Google Scholar]
- 4.Mohr S C, Sokolov N V H A, He C, Setlow P. Proc Natl Acad Sci USA. 1991;88:77–81. doi: 10.1073/pnas.88.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levin-Zaidman S, Reich Z, Wachtel E J, Minsky A. Biochemistry. 1996;35:2985–2991. doi: 10.1021/bi9525482. [DOI] [PubMed] [Google Scholar]
- 6.Ivanov V I, Minchenkova L E, Schyolkina A K, Poletayev A I. Biopolymers. 1973;12:89–110. doi: 10.1002/bip.1973.360120109. [DOI] [PubMed] [Google Scholar]
- 7.Zehfus M H, Johnson W C J. Biopolymers. 1984;23:1269–1281. doi: 10.1002/bip.360230711. [DOI] [PubMed] [Google Scholar]
- 8.Bokma J T, Johnson W C J, Blok J. Biopolymers. 1987;26:893–909. doi: 10.1002/bip.360260609. [DOI] [PubMed] [Google Scholar]
- 9.Piskur J, Rupprecht A. FEBS Lett. 1995;375:174–178. doi: 10.1016/0014-5793(95)01206-t. [DOI] [PubMed] [Google Scholar]
- 10.Robinson H, Wang A H-J. Nucleic Acids Res. 1996;4:676–682. doi: 10.1093/nar/24.4.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burckhardt G, Walter A, Zimmer C. J Biomol Struct Dyn. 1996;4:671–676. doi: 10.1080/07391102.1996.10508879. [DOI] [PubMed] [Google Scholar]
- 12.Bartenev V N, Golovamov E I, Kapitonova K A, Mokulskii M A, Volkova L I, Skuratovskii I Y. J Mol Biol. 1983;169:217–234. doi: 10.1016/s0022-2836(83)80181-8. [DOI] [PubMed] [Google Scholar]
- 13.Wolf B, Hanlon S. Biochemistry. 1975;14:1661–1670. doi: 10.1021/bi00679a018. [DOI] [PubMed] [Google Scholar]
- 14.Drew H R, Dickerson R E. J Mol Biol. 1981;3:535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- 15.Falk M, Hartman K A, Lord R C. J Am Chem Soc. 1963;85:397–391. [Google Scholar]
- 16.Cheatham T E, III, Kollman P A. J Mol Biol. 1996;259:434–444. doi: 10.1006/jmbi.1996.0330. [DOI] [PubMed] [Google Scholar]
- 17.Levitt M. Cold Spring Harbor Symp Quant Biol. 1983;47:251–262. doi: 10.1101/sqb.1983.047.01.030. [DOI] [PubMed] [Google Scholar]
- 18.Tidor B, Irikura K K, Brooks B R, Karplus M. J Biomol Struct Dyn. 1983;1:231–252. doi: 10.1080/07391102.1983.10507437. [DOI] [PubMed] [Google Scholar]
- 19.Beveridge D L, Swaminathan S, Ravishanker G, Withka J M, Srinivasan J, Prevost C, Louise-May S, Langley D R, DiCapua F, Bolton P H. In: Water and Biological Molecules. Westhof E, editor. New York: Macmillan; 1993. pp. 165–225. [Google Scholar]
- 20.Beveridge D L, Ravishanker G. Curr Opin Struct Biol. 1994;4:246–255. [Google Scholar]
- 21.Louise-May S, Auffinger P, Westhof E. Curr Opin Struct Biol. 1996;6:289–298. doi: 10.1016/s0959-440x(96)80046-7. [DOI] [PubMed] [Google Scholar]
- 22.Cornell W D, Cieplak P, Bayly C I, Gould I R, Merz K M, Jr, Ferguson D M, Spellmeyer D C, Fox T, Caldwell J W, Kollman P A. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 23.Cheatham T E, III, Kollman P A. J Am Chem Soc. 1997;119:4805–4825. [Google Scholar]
- 24.Bayly C I, Cieplak P, Cornell W D, Kollman P A. J Phys Chem. 1993;40:10269–10280. [Google Scholar]
- 25.Essmann U, Perera L, Berkowitz M L, Darden T, Lee H, Pedersen L G. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 26.Pearlman D A, Case D A, Caldwell J W, Ross W R, Cheatham T E, III, DeBolt S, Ferguson D M, Seibel G, Kollman P A. Comp Phys Commun. 1995;91:1–41. [Google Scholar]
- 27.Ravishanker G, Swaminathan S, Beveridge D L, Lavery R, Sklenar H. J Biomol Struct Dyn. 1989;6:669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- 28.Lavery R, Sklenar H. J Biomol Struct Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 29.Ramakrishnan B, Sundaralingam M. J Biomol Struct Dyn. 1993;11:11–26. doi: 10.1080/07391102.1993.10508706. [DOI] [PubMed] [Google Scholar]
- 30.Eisenstein M, Shakked Z. J Mol Biol. 1995;248:662–678. doi: 10.1006/jmbi.1995.0250. [DOI] [PubMed] [Google Scholar]
- 31.Saenger W. In: Principles of Nucleic Acid Structure. Cantor C E, editor. New York: Springer; 1984. [Google Scholar]
- 32.Ferrin T E, Huang C C, Jarvis L E, Langridge R. J Mol Graphics. 1988;6:13–27. [Google Scholar]