Abstract
The four mammalian MutL homologs (MLH1, MLH3, PMS1, and PMS2) participate in a variety of events, including postreplicative DNA repair, prevention of homeologous recombination, and crossover formation during meiosis. In this latter role, MLH1–MLH3 heterodimers predominate and are essential for prophase I progression. Previous studies demonstrated that mice lacking Mlh1 exhibit a 90% reduction in crossing over at the Psmb9 hot spot while noncrossovers, which do not result in exchange of flanking markers but arise from the same double-strand break event, are unaffected. Using a PCR-based strategy that allows for detailed analysis of crossovers and noncrossovers, we show here that Mlh3−/− exhibit a 85–94% reduction in the number of crossovers at the Psmb9 hot spot. Most of the remaining crossovers in Mlh3−/− meiocytes represent simple exchanges similar to those seen in wild-type mice, with a small fraction (6%) representing complex events that can extend far from the initiation zone. Interestingly, we detect an increase of noncrossovers in Mlh3−/− spermatocytes. These results suggest that MLH3 functions predominantly with MLH1 to promote crossovers, while noncrossover events do not require these activities. Furthermore, these results indicate that ∼10% of crossovers in the mouse are independent of MLH3, suggesting the existence of alternative crossover pathways in mammals.
MEIOTIC recombination between homologous chromosomes during prophase I can result in both crossover (CO) and noncrossover (NCO) products. Most, if not all, of these events are initiated by the same double-strand break (DSB) event induced by SPO11, a conserved protein sharing sequence similarities with the catalytic subunit of TopoVI, a type II DNA topoisomerase from archae (reviewed in Keeney and Neale 2006), but are processed through distinct and highly conserved molecular pathways, the details of which remain to be elucidated fully in mammals. What is clear from studies in a number of species is that the frequency and distribution of CO events are tightly controlled in prophase I, and that this control is essential for the accurate reductional segregation of chromosomes at the first meiotic division.
Studies in Saccharomyces cerevisiae have shown that CO and NCO events result from two distinct pathways that differ in their specific DNA intermediates and functional requirements (Allers and Lichten 2001; Hunter and Kleckner 2001). In particular, a specific group of proteins comprising the ZMM family, encoded by the yeast ZIP1, ZIP2, ZIP3, ZIP4, MSH4, MSH5, and MER3 genes, acts early in the formation of most CO but is not required for NCO formation (Borner et al. 2004). The yeast MutL homologs, Mlh1 and Mlh3, are also necessary for CO formation but act at a later step, presumably after the formation of the double-Holliday-junction intermediate (Hunter and Borts 1997; Wang et al. 1999; Argueso et al. 2004). In mammals, MSH4 and MSH5 are thought to be required at an early step of meiotic recombination, given that mutant mice are defective in synapsis and that the proteins localize along chromosome axes as early as leptonema in both males and females (Kneitz et al. 2000; Lenzi et al. 2005). These proteins have also been proposed to have a later action on CO formation on the basis of their binding activities to double Holliday junctions (Snowden et al. 2004) and the colocalization of MSH4 with MLH1 at midpachynema (Santucci-Darmanin et al. 2000).
Mammalian MLH1 and MLH3 proteins also appear to be involved and required for CO formation on the basis of localization studies and the phenotypes of Mlh1 and Mlh3 mutant mice (Kolas and Cohen 2004). At mid- to late pachynema, these proteins colocalize (Kolas et al. 2005) at the future sites of chiasmata (Marcon and Moens 2003). Both Mlh1 and Mlh3 nullizygous mice show a strong defect in chiasma formation in both sexes (Baker et al. 1996; Edelmann et al. 1996; Lipkin et al. 2002; Kan et al. 2008). However, recent analysis has revealed different properties of these two MutL homologs: MLH3 accumulates at nascent CO sites prior to MLH1 in males and can continue to do so in mice lacking a functional Mlh1 gene (Lipkin et al. 2002; Kolas and Cohen 2004). Conversely, MLH1 requires MLH3 preloading at MSH4–MSH5 sites since no MLH1 is localized to meiotic chromosomes in spermatocytes from Mlh3−/− mice (Kolas and Cohen 2004; Kolas et al. 2005). At the same time, electron-dense meiotic nodules, the sites at which recombination events are taking place, persist on meiotic chromosomes from Mlh1−/− mice but not on chromosomes from Mlh3−/− mice (Lipkin et al. 2002). Direct analysis at the DNA level has shown that MLH1 is required for most but not all CO and not required for NCO formation (Guillon et al. 2005).
To examine the role of MLH3 directly on meiotic recombination events, we examined the CO and NCO frequencies at the well-characterized Psmb9 recombination hot spot on mouse chromosome 17, using a strategy that allows for detailed analysis of CO and NCO products at this locus (Guillon and de Massy 2002; Guillon et al. 2005; Baudat and de Massy 2007b). Our analyses of recombination in male and female germ cells demonstrate that, similar to Mlh1−/− mice, Mlh3 null animals exhibit an 85–94% reduction in the number of crossings over at the Psmb9 hot spot, while the respective NCO frequencies remain unchanged. Most remaining crossovers in Mlh3−/− spermatocytes represent simple exchanges similar to those seen in wild-type mice. However, a small fraction (6%) of residual COs are complex events with exchange points far from the initiation zone and similar in profile to the residual CO events observed in Mlh1−/− spermatocytes. A significant increase of NCO frequencies was observed in spermatocytes from Mlh3−/− mice, potentially indicating a channeling of some CO intermediates into NCO products. These results suggest that MLH3 functions predominantly with MLH1 to promote crossing over and that, together, this heterodimer governs ∼90% of all CO events in mouse meiocytes.
MATERIALS AND METHODS
Generation and maintenance of hybrid mice:
Mice were housed in the Cornell University Animal Facility (Ithaca, NY) and all procedures utilizing these animals were reviewed and approved by the Cornell University Institutional Animal Care and Use Committee. Mice were maintained on standard laboratory chow under controlled conditions of light and temperature. The generation of Mlh3 mutant mice has been described previously (Lipkin et al. 2002). Mice heterozygous at the Psmb9 (wm7/b) locus and carrying one or two mutant alleles of Mlh3 (Mlh3+/+, Mlh3+/−, or Mlh3−/−) were generated by crossing the B10.A (R209) strain carrying the wm7 haplotype (named R209, B10 background) and the Mlh3+/− strain (B6 background). Progeny of Mlh3+/− mice (B6 background) crossed with Mlh3+/− wm7/wm7 (B6/B10 mixed background) mice were analyzed, providing Mlh3+/−, Mlh3+/+, and Mlh3−/− littermates. Testes were dissected from mice at 11, 13, 15, 17, and 19 days postpartum (pp), while ovaries were taken from mice at 1 day pp. Genomic DNA preparations were used for subsequent PCR analysis of CO and NCO products.
Detection and mapping of COs and NCOs:
COs were detected in two ways: (1) direct selection with two rounds of allele-specific PCR with primers specific for B6 on one side and for R209 on the other side of the hot spot (Guillon and de Massy 2002) and (2) by a protocol allowing the detection of NCO in parallel to CO. In this protocol, B6-R209 (BR) COs were detected using in the first round of amplification (PCRI) a B6 allele-specific forward primer and a universal reverse primer. In the secondary amplification reactions (PCRII) a B6 allele-specific forward primer was used in combination with an R209 allele-specific reverse primer. Using the same PCRI products as substrates, an alternative secondary amplification is performed to detect NCO products with a gene conversion at the BsrFI site. This is achieved using an R209 BsrFI-specific forward primer with a B6 allele-specific reverse primer. Although the BsrFI site is located near the center of the hot spot, given the short sizes of gene conversion tracts among NCOs, we estimate that those covering BsrFI represent less than half of the total (F. Baudat and B. de Massy, personal communication). For direct selection of COs, PCRI can be performed on pools of testis DNA containing up to 10,000 amplifiable molecules, whereas for the parallel detection of COs and NCOs, PCRI is performed on pools of testis DNA containing up to 400 amplifiable molecules. Both methods gave similar values of CO frequencies, which were therefore determined by averaging independent experiments using either protocol. For mapping analysis, the CO molecules were obtained from direct selection, using at the second PCR forward and reverse primers located at positions 402 and 4315, respectively, therefore leading to the amplification of a 4-kb product (see Figure 3). Details of the procedures and primer sequences are as described in Guillon et al. (2005). A small-scale experiment on 24 pools at all time points in wild type and Mlh3−/− indicated that frequency of RB crossover molecules was similar to that of BR molecules as expected (data not shown). No significant difference of CO or NCO frequencies could be detected between +/+ and +/− mice. COs were first mapped by RFLP at 10 sites in the amplified region. Those showing unexpected patterns were sequenced over the whole 4-kb region. NCOs were tested for the presence of StyI and SphI sites and were sequenced upstream of the BsrFI site. Those showing single-site conversions at BsrFI were sequenced downstream of this site. Additional CO events were selected and analyzed from Mlh1−/− mice for mapping analysis.
Figure 3.—
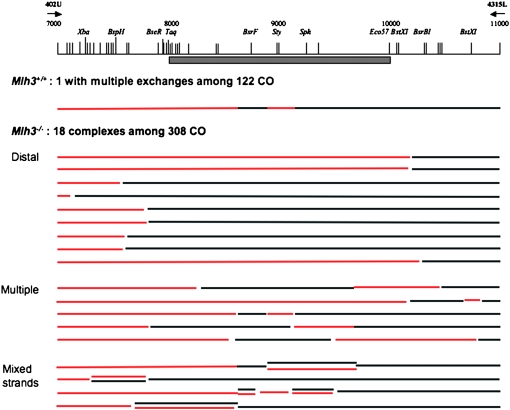
Mapping of exchange points among complex CO products. One hundred twenty-two COs and 308 COs were mapped in wt and Mlh3−/− (respectively) from genomic DNA of day 17 and day 19 pp testes. They were obtained as 4-kb molecules amplified with primers at positions 402 and 4315. The hot spot, identified by a shaded bar, is located between the TaqI and Eco57I sites and centered in the BsrFI–StyI region. Red and black lines represent B6 and R209 haplotypes, respectively. Those molecules with unusual positions of exchange points, i.e., more than one exchange point or an exchange point outside the TaqI–Eco57I interval, were sequenced and divided into three categories: molecules with a distal exchange point, multiple exchanges, and mixed strands.
RESULTS
A specific reduction of CO frequencies both in spermatogenesis and in oogenesis in Mlh3−/− mice:
Using a strategy and protocols similar to those developed for the analysis of recombination events in Mlh1−/− mice (Guillon et al. 2005), we have measured the frequency of COs and NCOs at the Psmb9 recombination hot spot. The general strategy is as follows: mice heterozygous at Psmb9 (wm7/b) and homozygous mutant at Mlh3 (Mlh3−/−) were generated along with Mlh3+/− or Mlh3+/+ controls. Analyses in males were performed on genomic DNA extracted from testes of mice from 11 to 19 days old, before the massive induction of apoptosis occurring at metaphase I (Lipkin et al. 2002). Analyses in females were performed on ovaries from newborn mice. CO and NCO molecules were detected by allele-specific PCR on small pools of genomic DNA. Two protocols were used, one allowing the detection of CO products only and one allowing the parallel detection of COs and NCOs. The allele-specific PCR strategy we have used allows the recovery of all CO molecules in the 4-kbp interval tested but only a fraction of NCOs, those with gene conversion tracts covering the central BsrFI site (see materials and methods).
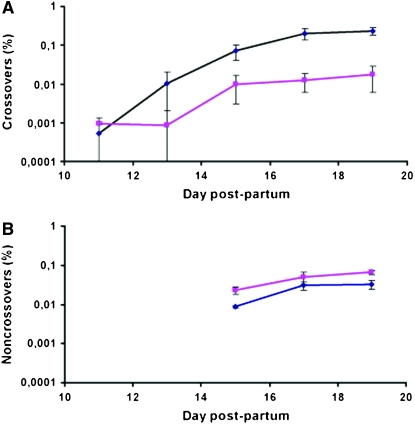
During meiosis in males, the frequency of CO events was found to be 85–94% lower in spermatocytes from Mlh3−/− males compared to wild type at all time points during the first wave of spermatogenesis where a significant number of events could be recovered, i.e., at 13, 15, 17, and 19 days. This reduction in CO content was statistically significant at 15, 17, and 19 days, using a one-tailed t-test (P-values of 0.0455, 0.0004, and 0.0004 at 15, 17, and 19 days, respectively). The kinetics of CO formation were similar in wild-type and Mlh3−/− spermatocytes, with a plateau reached at 17 days when cells progressing through the first wave of meiosis reach the end of the pachytene stage (Figure 1A). The analysis of events in female meiosis also indicated a 94% reduction in CO frequencies (one tailed t-test, P < 0.0001; Table 1).
Figure 1.—
CO and NCO frequencies in Mlh3−/− mice. (A) CO frequencies in wild-type (wt) (+/+ or +/−) and Mlh3−/− mice during the first wave of spermatogenesis. Values correspond to twice the frequency of BR recombinant molecules and were the average from two measurements at 11 and 13 days and from five to eight measurements at 15, 17, and 19 days. Frequencies are given ± standard error (SE) (blue diamonds, wt; pink squares, Mlh3−/−). (B) NCO frequencies in wt (+/+ or +/−) and Mlh3−/− mice during the first wave of spermatogenesis. Values correspond to the frequencies of NCOs with a gene conversion selected at BsrFI on the B6 parental genome and were the average of two, four, and three measurements at 15, 17, and 19 days, respectively. Each frequency measurement was obtained from allele-specific PCR on 96 pools. Frequencies are given ± SE (blue diamonds, wt; pink squares, Mlh3−/−).
TABLE 1.
CO and NCO frequencies in ovaries from wt (+/+ and +/−) and Mlh3−/− mice
| CO frequencies % | Standard error (%) | No. of events | |
|---|---|---|---|
| Mlh3−/− | 0.0087 | 0.0103 | 8 |
| Mlh3+/+ and Mlh3+/− | 0.140 | 0.044 | 108 |
| Ratio wt/mutant | 16 | ||
| NCO frequencies % | Standard error (%) | No. of events | |
| Mlh3−/− | 0.051 | 0.035 | 35 |
| Mlh3+/+ and Mlh3+/− | 0.046 | 0.012 | 45 |
| Ratio wt/mutant | 0.9 |
CO frequencies reported are two times the frequencies of BR recombinant molecules measured. The NCO frequencies are NCOs selected at BsrFI on the B6 chromosome. The numbers of recombinant events detected are provided.
In striking contrast, no reduction in NCO frequencies in Mlh3−/− mice was observed either in spermatocytes or in oocytes (Figure 1B and Table 1). In fact, we detected an increase of 67–171% in NCO rates in Mlh3−/− spermatocytes between days 15 and 19 postpartum. These differences between wild-type and Mlh3−/− mice are statistically significant (one-tailed t-test, P-values of 0.0291, 0.0386, and 0.0036 at 15, 17, and 19 days, respectively). The small difference between wild-type and Mlh3−/− mice observed in oocytes was not significant, however (P = 0.43963).
Properties of residual crossovers in the absence of MLH3:
We have mapped the positions of exchange points among 122 and 308 CO products in wild-type and Mlh3−/− mice, respectively. In wild-type mice, all COs but one were simple exchange products with an exchange point in the interval between TaqI and Eco57I. The distribution of exchange points was similar to that in previous analyses (Guillon and de Massy 2002; Guillon et al. 2005).
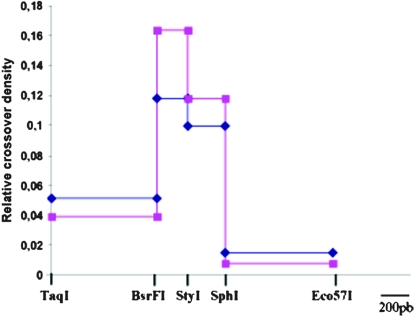
In Mlh3−/− males, most CO products (94%, 290/308) have a structure similar to wild type, with one exchange point within the TaqI and Eco57I interval and with the highest density in the BsrFI–StyI interval. The distribution is globally similar to that observed in wild type, as in previous analyses, but with a higher density in the BsrFI–StyI interval, suggesting that the conversion tracts associated with these CO events are shorter (Figure 2). The distributions of wild-type and Mlh3−/− COs are indeed statistically different (chi square, P = 0.028). However, the remaining fraction of CO molecules (6%, 18/308) in Mlh3−/−males displays unusual structures, and these are not observed in wild-type control males. Among these unusual recombinant molecules, we could identify three types of molecules: COs with one single exchange point located distal to the hot spot, COs with multiple exchange points (with mosaic structure), and COs containing mixed-strand information (Figure 3). These CO products with mixed strands could be either molecules containing heteroduplex DNA or a mixture of a CO with a simple exchange and a CO with a mosaic structure. This second possibility is statistically compatible with the number of CO events per pool in these experiments (supplemental Table 1). In any case, these observations indicate a modification of the processing of recombination intermediates in Mlh3−/− males. The mapping of these products suggests that recombination intermediates extend farther away from the initiation zone than those generated in wild-type meiocytes and/or that the repair of heteroduplex structures is altered in the absence of MLH3 protein. Although a qualitatively similar observation was made in Mlh1−/− males (Guillon et al. 2005), we noted that the relative proportion of COs with a single distal exchange point was higher in the absence of MLH3 as compared to that observed by Guillon et al. in the absence of MLH1. To more accurately compare the two mutant strains, we repeated these CO analyses from Mlh1−/− spermatocytes using identical conditions to, and in parallel with, our studies in Mlh3−/− spermatocytes. Although the relative proportion of COs with a distal exchange point was higher in Mlh3−/− spermatocytes compared to Mlh1−/− spermatocytes (50–55% vs. 17–30%), the difference is not statistically significant (supplemental Figure 1 and supplemental Table 1). It appears therefore that the absence of MLH1 or MLH3 has similar consequences for residual CO processing.
Figure 2.—
Mapping of exchange points among simple CO products in Mlh3+/+ and Mlh3−/− mice. CO products were recovered from genomic DNA from the testes of 17- and 19-day pp mice. COs were obtained by amplification of a 4-kb interval by primers located at positions 402 and 4315 (see Figure 3). Only those with a single exchange in the TaqI–Eco57I interval are integrated in this graph. The relative crossover density is the proportion of COs in each interval (relative to all COs between TaqI and Eco57I) divided by its length. Intervals are defined by RFLP sites TaqI, BsrFI, StyI, SphI, and Eco57I (blue diamonds, wt; pink squares, Mlh3−/−).
No detectable change in NCO maps in Mlh3−/− mice:
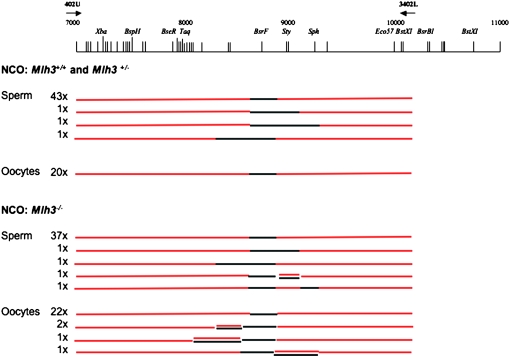
The NCO products were selected at the BsrFI site where flanking markers are at 329 and 214 bp to the left and right, respectively. In both wild-type and Mlh3−/− spermatocytes most of the NCO products selected at BsrFI have a conversion tract including only the BsrFI marker (95 and 88%, respectively). Some rare products were observed to coconvert a marker flanking BsrFI. In addition, in spermatocytes and oocytes from Mlh3−/− mice, some rare molecules were found to have mosaic structures or mixed strands (Figure 4). However, no significant difference in the frequencies of these different products could be detected between wild-type and Mlh3−/− male or female mice.
Figure 4.—
Mapping of NCO products in Mlh3+/+ and Mlh3−/− mice. Both left and right sides of independent NCO events with a conversion at BsrFI were selected by allele-specific PCR and sequenced. Most events are converted at the BsrFI marker only. DNA was extracted from either newborn ovaries or testes of day 17 and day 19 pp wild-type and Mlh3−/− mice.
DISCUSSION
The ability to detect recombinant molecules directly by allele-specific PCR has allowed us to analyze, at the DNA level, the effect of MLH3 deletion on meiotic recombination in mice, in terms of both frequency and mapping of CO and NCO events at the Psmb9 hot spot. Our results show a differential effect between COs and NCOs with a strong reduction of CO frequency (85–94%) whereas NCO frequency is not affected or slightly increased. The analysis of COs detected in the absence of MLH3 reveals a fraction of events that differ in the location of exchange points compared to wild-type meiocytes. This suggests that MLH3 plays a role in the processing of recombination intermediates.
The role of MLH3 and MLH1 in CO formation:
The reductions of CO frequencies during the first wave of spermatogenesis and during oogenesis of Mlh3−/− mice, demonstrated herein, reveal an essential role for MLH3 for CO formation. This finding is consistent with the reduction of chiasmata in Mlh3 −/− males (Lipkin et al. 2002) and implies that the effect on chiasma is not due to a defect in chiasma stabilization but results directly from a defect in CO formation. The quantitative effect on CO formation is identical to that previously described for Mlh1−/− mice (Guillon et al. 2005), a result consistent with the similar defect on chiasma formation also described in these two mutants. This suggests that both proteins act in the same pathway with respect to CO formation, which is supported by immunological studies that have shown the colocalization of MLH1 and MLH3 at pachynema (Kolas et al. 2005) and at sites of chiasmata (Marcon and Moens 2003). At the same time, however, subtle differences in MLH1 and MLH3 focus dynamics indicate that the loss of either gene product might not necessarily have the same effect on CO frequency in Mlh1−/− and Mlh3−/− animals. First, in males MLH3 foci form earlier (early pachynema) and possibly in greater initial numbers as compared to MLH1; second, meiotic nodules observed by electron microscopy are still detected in Mlh1−/− males but are absent in Mlh3−/− males (Lipkin et al. 2002); and third, the presence of MLH1 foci depends on preloading of MLH3, whereas a significant number of MLH3 foci are still detected in Mlh1−/− (Kolas et al. 2005). These differences in cytological observations compared to the similar defect in CO formation in Mlh3−/− and Mlh1−/− males suggest that the persistent CO events observed at the Psmb9 locus in Mlh1−/− males are not due to residual MLH3 activity and that neither MLH1 nor MLH3 can function independently of each other in the context of meiotic recombination events.
The localization of MLH1 and MLH3 to meiotic nodules is independent of the somatic repair MutS homolog heterodimers, MutSα (MSH2/MSH6) or MutSβ (MSH2/MSH3) and instead operates through interactions with the meiosis-specific MutS complex, MutSγ, that consists of MSH4 and MSH5. This suggestion is supported by the interaction between human MSH4 and MLH3 (Santucci-Darmanin et al. 2002) and by the interaction and colocalization of MSH4 and MLH1 in mice (Santucci-Darmanin et al. 2000) as well as in humans (Oliver-Bonet et al. 2005). According to in vitro analysis of mismatch repair activity, MutL proteins are thought to be adaptors able to transduce signals, possibly involving protein conformational changes. MLH1/MLH3 might serve to modulate the accumulation of MSH4/MSH5 sliding clamps by stabilizing these structures and/or by facilitating the sliding of MSH4/MSH5 along the chromosomes, thereby facilitating the loading of additional factors at the Holliday-junction structure and biasing these sites toward a CO fate. If recombination intermediates are not converted into COs in Mlh1−/− and Mlh3−/− meiocytes, at least two outcomes can be envisioned: either they are left unresolved or they are processed into alternative products. We suggest that a fraction of these intermediates are actually resolved into NCOs with the dissolution activity of BLM-TOP3α (Wu and Hickson 2003). The increase of NCO frequency detected in Mlh3−/− spermatocytes is consistent with this hypothesis. At the quantitative level, we cannot determine what fraction of COs is channeled into NCOs, as this would require knowledge of the gene conversion tract length and distribution among the NCO molecules. In addition, although the effect of unresolved intermediates on meiotic prophase progression is unknown, the analysis of RAD51C hypomorph mutant mice has led to the suggestion that unresolved intermediates would induce chromosome breakage (Kuznetsov et al. 2007), a phenotype not observed in either Mlh1−/− or Mlh3−/− mice.
A residual activity of CO formation in the absence of MLH3:
Our results also clearly show that a residual level of crossing over, ∼10% of wild type, occurs in Mlh3−/− mice. If this subset of events also occurs in wild-type meiocytes, one implication is that MLH1/MLH3 foci would not be expected to mark all sites of COs. We note, in fact, that a small difference in terms of genetic map length is observed when estimated from MLH1 foci or from genetic mapping (Baudat and de Massy 2007a). Despite this, however, it is clear that the major pathway for CO formation in mouse is MLH3 (and MLH1) dependent. In S. cerevisae, it has been established that most of the Mlh1-independent COs result from the Mus81/Mms4 pathway (Hoffmann et al. 2003; Argueso et al. 2004; Hollingsworth and Brill 2004), while in Schizosaccharomyces pombe, all COs require Mus81/Mms4 (Smith et al. 2003). In the case of the latter organism, single, not double, Holliday junctions have been shown to be intermediates of meiotic recombination (Cromie et al. 2006), and these are readily cleaved by the Mus81/Mms4 proteins in vitro (Boddy et al. 2001; Osman et al. 2003). Whether an analogous, “alternate” pathways exists in mice remains to be seen. The normal fertility of MUS81 mutant mice (Hollingsworth and Brill 2004; Dendouga et al. 2005) is compatible with the idea that any MUS81-driven recombination events would concern only a small fraction of CO events and/or would be elicited only in the absence of appropriate MLH1/MLH3 activity.
The mapping of the remaining COs that occur in Mlh3−/− meiocytes also provides additional information about COs generated by the MutLγ-independent pathway. Two categories of CO products are detected: the majority (94%) have a single exchange point with a distribution similar to that seen in meiocytes from wild-type animals, and a smaller population (6%) have atypical exchange points (discussed below). Interestingly, the majority class shows a slight difference in terms of distribution from that of wild type, with a narrower clustering of exchange points around the predicted region of initiation, suggesting that the mean length of the associated conversion tract is smaller. A similar effect was observed in Mlh1 −/− animals (Guillon et al. 2005). This suggests a difference between the two pathways (MutLγ dependent or independent), either in the formation or in the processing of intermediates. Whether the resolution of this MutLγ-independent class of COs is also independent of MutSγ remains to be seen. What is clear, however, is that the absence of mismatches among the COs recovered in the majority class indicates that the heteroduplex DNA generated during the formation of these COs have been repaired in the absence of MLH3. Given that a similar conclusion was reached from the analysis of Mlh1 −/− male mice, if a MutL activity is involved in mismatch repair, it is more likely to involve PMS2 assuming it can act with either MLH1 or some other partner. In vitro biochemical data indicate that PMS2 and MLH3 do not form a complex with each other, ruling out a role for MLH3 in heteroduplex repair, even in Mlh1−/− animals.
Exchanges at distal locations in the absence of MLH3:
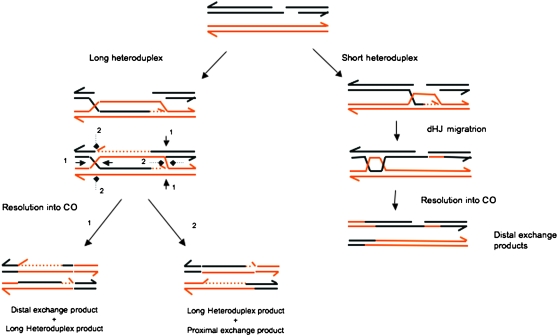
The detailed mapping of CO products from Mlh3−/− males reveals a small fraction (6%) of products with unusual properties, not seen in wild-type meiocytes, but also detected in the absence of MLH1. Fifteen of 18 of these CO events exhibit an exchange point located >1 kb away from the region of initiation. Among these distal events, 9 have a single exchange point. Such products could be derived from a DSB repair event exemplified in Figure 5, either resulting from intermediates with a long heteroduplex on one side of the DSB or through migration of the double Holliday junction. Interestingly, long heteroduplex intermediates lead to a distal exchange product without (or with short) heteroduplex only for one type of resolution (orientation 1 in Figure 5). The short heteroduplex formed could be either undetectable or repaired by a short patch repair pathway. Resolution in orientation 2 generates a product with a long heteroduplex and a reciprocal product with exchange points close to initiation. The detection of an increased number of events extending to more distal positions has been observed in pms1 and msh2 mutants of S. cerevisiae (Alani et al. 1994). Such observations might indicate either that the heteroduplex extends to distal positions even in a MutL wild-type context or that the absence of mismatch repair leads to an increase in heteroduplex length. Furthermore, the detection of molecules with mosaic structures suggests that heteroduplex DNA has been repaired in a noncontinuous way, possibly by an alternative repair pathway, such as the short patch repair pathways described in msh2 and pms2 mutant strains of S. cerevisiae and S. pombe (Fleck et al. 1999; Coic et al. 2000; Hoffmann et al. 2005) and in msh6 mutants of Drosophila melanogaster (Radford et al. 2007b).
Figure 5.—
Alternative ways to generate distal exchange points on CO products. Two alternative ways are considered for the formation of COs with a distal exchange point without long heteroduplexes. One implies the formation of a long asymmetric heteroduplex and the other the migration of a double Holliday junction. In the first alternative, products differ depending on the directionality of resolution. Only resolution 1 gives rise to a molecule with a distal exchange and without a long heteroduplex. In the second alternative, migration of a double Holliday junction and resolution can lead to a CO with a distal exchange point and with small stretches of heteroduplex DNA.
In conclusion, our work provides precise information on the activity of MLH3 and its major partner, MLH1. The activity of MLH3 on crossover control does not seem to be redundant to that of PMS2 or PMS1 in mice, and this meiosis-specific property of MLH3 seems to be conserved among species. In Arabidopsis thaliana Mlh3 mutants show a defect in chiasma formation with 30% residual activity (Franklin et al. 2006). In some species, such as Caenorhabditis elegans, D. melanogaster, and S. pombe, Mlh3 is absent and it is postulated that either Pms2 substitutes for the CO function of Mlh3 in these species or crossover control does not require the MutLγ complex. This hypothesis could be consistent in D. melanogaster and S. pombe, where the nature of recombination intermediates might differ from those occurring in mice as suggested by the requirement for COs on Mus81 in S. pombe (Smith et al. 2003) and on mei9 in D. melanogaster (Radford et al. 2007a). The crossover control activity of this MutLγ complex is thought to require the MutSγ complex (MSH4 and MSH5) on the basis of immunolocalization and interaction analysis. Functional analyses in both S. cerevisiae and C. elegans have shown that Msh4 was indeed required for crossover formation. In mice, however, testing directly the effect of MSH4 or MSH5 on crossing over is not currently feasible given that these mutants have earlier defects in the process of recombination, disrupting the progression of meiotic prophase probably before DSBs can eventually be repaired. In light of the current results, together with previous studies demonstrating the importance of MLH1 in the majority of CO events in mice, we hypothesize that MSH4–MSH5 heterodimers function at the same cohort of CO events, suggesting that the residual 5–10% of COs that are not dependent on mismatch repair function are instead processed through an alternative pathway that is analogous, functionally if not compositionally, to the yeast Mus81 pathway.
Acknowledgments
We thank J. Kim Holloway, Cornell University, for invaluable discussion and for reading this manuscript. This work was supported by grants from the Centre National de la Recherche Scientifique, Association pour la Recherche contre le Cancer (4626 and 3723) and Agence National de la Recherche (ANR-06-BLAN-0160-01) to B.d.M., by research funding from the National Institute of Child Health and Human Development to P.E.C. (HD41012), and by start-up funding from Cornell University College of Veterinary Medicine, also to P.E.C.
References
- Alani, E., R. A. Reenan and R. D. Kolodner, 1994. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics 137 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers, T., and M. Lichten, 2001. Intermediates of yeast meiotic recombination contain heteroduplex dna. Mol. Cell 8 225–231. [DOI] [PubMed] [Google Scholar]
- Argueso, J. L., J. Wanat, Z. Gemici and E. Alani, 2004. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 168 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, S. M., A. W. Plug, T. A. Prolla, C. E. Bronner, A. C. Harris et al., 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13 336–342. [DOI] [PubMed] [Google Scholar]
- Baudat, F., and B. de Massy, 2007. a Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 15 565–577. [DOI] [PubMed] [Google Scholar]
- Baudat, F., and B. de Massy, 2007. b Cis- and trans-acting elements regulate the mouse Psmb9 meiotic recombination hotspot. PLoS Genet. 3 e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, M. N., P. H. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates et al., 2001. Mus81-eme1 are essential components of a Holliday junction resolvase. Cell 107 537–548. [DOI] [PubMed] [Google Scholar]
- Borner, G. V., N. Kleckner and N. Hunter, 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117 29–45. [DOI] [PubMed] [Google Scholar]
- Coic, E., L. Gluck and F. Fabre, 2000. Evidence for short-patch mismatch repair in Saccharomyces cerevisiae. EMBO J. 19 3408–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie, G. A., R. W. Hyppa, A. F. Taylor, K. Zakharyevich, N. Hunter et al., 2006. Single Holliday junctions are intermediates of meiotic recombination. Cell 127 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendouga, N., H. Gao, D. Moechars, M. Janicot, J. Vialard et al., 2005. Disruption of murine Mus81 increases genomic instability and DNA damage sensitivity but does not promote tumorigenesis. Mol. Cell. Biol. 25 7569–7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann, W., P. E. Cohen, M. Kane, K. Lau, B. Morrow et al., 1996. Meiotic pachytene arrest in MLH1-deficient mice. Cell 85 1125–1134. [DOI] [PubMed] [Google Scholar]
- Fleck, O., E. Lehmann, P. Schar and J. Kohli, 1999. Involvement of nucleotide-excision repair in msh2 pms1-independent mismatch repair. Nat. Genet. 21 314–317. [DOI] [PubMed] [Google Scholar]
- Franklin, F. C., J. D. Higgins, E. Sanchez-Moran, S. J. Armstrong, K. E. Osman et al., 2006. Control of meiotic recombination in Arabidopsis: role of the MutL and MutS homologues. Biochem. Soc. Trans. 34 542–544. [DOI] [PubMed] [Google Scholar]
- Guillon, H., and B. de Massy, 2002. An initiation site for meiotic crossing-over and gene conversion in the mouse. Nat. Genet. 32 296–299. [DOI] [PubMed] [Google Scholar]
- Guillon, H., F. Baudat, C. Grey, R. M. Liskay and B. de Massy, 2005. Crossover and noncrossover pathways in mouse meiosis. Mol. Cell 20 563–573. [DOI] [PubMed] [Google Scholar]
- Hoffmann, E. R., P. V. Shcherbakova, T. A. Kunkel and R. H. Borts, 2003. MLH1 mutations differentially affect meiotic functions in Saccharomyces cerevisiae. Genetics 163 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, E. R., E. Eriksson, B. J. Herbert and R. H. Borts, 2005. MLH1 and MSH2 promote the symmetry of double-strand break repair events at the HIS4 hotspot in Saccharomyces cerevisiae. Genetics 169 1291–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, N. M., and S. J. Brill, 2004. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 18 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, N., and R. H. Borts, 1997. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 11 1573–1582. [DOI] [PubMed] [Google Scholar]
- Hunter, N., and N. Kleckner, 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106 59–70. [DOI] [PubMed] [Google Scholar]
- Kan, R., S. Fei, N. K. Kolas, E. Avdievich, B. Kneitz et al., 2008. Comparative analysis of meiotic progression in female mice bearing mutations in genes of the DNA mismatch repair pathway. Biol. Reprod. 78 462–471. [DOI] [PubMed] [Google Scholar]
- Keeney, S., and M. J. Neale, 2006. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem. Soc. Trans. 34 523–525. [DOI] [PubMed] [Google Scholar]
- Kneitz, B., P. E. Cohen, E. Avdievich, L. Zhu, M. F. Kane et al., 2000. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14 1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Kolas, N. K., and P. E. Cohen, 2004. Novel and diverse functions of the DNA mismatch repair family in mammalian meiosis and recombination. Cytogenet. Genome Res. 107 216–231. [DOI] [PubMed] [Google Scholar]
- Kolas, N. K., A. Svetlanov, M. L. Lenzi, F. P. Macaluso, S. M. Lipkin et al., 2005. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J. Cell Biol. 171 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov, S., M. Pellegrini, K. Shuda, O. Fernandez-Capetillo, Y. Liu et al., 2007. RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J. Cell Biol. 176 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzi, M. L., J. Smith, T. Snowden, M. Kim, R. Fishel et al., 2005. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis i in human oocytes. Am. J. Hum. Genet. 76 112–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkin, S. M., P. B. Moens, V. Wang, M. Lenzi, D. Shanmugarajah et al., 2002. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 31 385–390. [DOI] [PubMed] [Google Scholar]
- Marcon, E., and P. Moens, 2003. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics 165 2283–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Bonet, M., P. J. Turek, F. Sun, E. Ko and R. H. Martin, 2005. Temporal progression of recombination in human males. Mol. Hum. Reprod. 11 517–522. [DOI] [PubMed] [Google Scholar]
- Osman, F., J. Dixon, C. L. Doe and M. C. Whitby, 2003. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12 761–774. [DOI] [PubMed] [Google Scholar]
- Radford, S. J., S. McMahan, H. L. Blanton and J. Sekelsky, 2007. a Heteroduplex DNA in meiotic recombination in Drosophila mei-9 mutants. Genetics 176 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford, S. J., M. M. Sabourin, S. McMahan and J. Sekelsky, 2007. b Meiotic recombination in Drosophila Msh6 mutants yields discontinuous gene conversion tracts. Genetics 176 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santucci-Darmanin, S., D. Walpita, F. Lespinasse, C. Desnuelle, T. Ashley et al., 2000. MSH4 acts in conjunction with MLH1 during mammalian meiosis. FASEB J. 14 1539–1547. [DOI] [PubMed] [Google Scholar]
- Santucci-Darmanin, S., S. Neyton, F. Lespinasse, A. Saunieres, P. Gaudray et al., 2002. The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Hum. Mol. Genet. 11 1697–1706. [DOI] [PubMed] [Google Scholar]
- Smith, G. R., M. N. Boddy, P. Shanahan and P. Russell, 2003. Fission yeast mus81.eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics 165 2289–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden, T., S. Acharya, C. Butz, M. Berardini and R. Fishel, 2004. hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15 437–451. [DOI] [PubMed] [Google Scholar]
- Wang, T. F., N. Kleckner and N. Hunter, 1999. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. USA 96 13914–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L., and I. D. Hickson, 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426 870–874. [DOI] [PubMed] [Google Scholar]