Abstract
From chestnut rose, a promising fruit crop of the Rosa genus, powdery mildew disease-resistant and susceptible genotypes and their F1 progeny were used to isolate nucleotide-binding-site (NBS)-encoding genes using 19 degenerate primer pairs and an additional cloning method called overlapping extension amplification. A total of 126 genes were harvested; of these, 38 were from a resistant parent, 37 from a susceptible parent, and 51 from F1 progeny. A phylogenetic tree was constructed, which revealed that NBS sequences from parents and F1 progeny tend to form a mixture and are well distributed among the branches of the tree. Mapping of these NBS genes suggested that their organization in the genome is a “tandem duplicated cluster” and, to a lesser extent, a “heterogeneous cluster.” Intraspecific polymorphisms and interspecific divergence were detected by Southern blotting with NBS-encoding genes as probes. Sequencing on the nucleotide level revealed even more intraspecific variation: for the R4 gene, 9.81% of the nucleotides are polymorphic. Amino acid sites under positive selection were detected in the NBS region. Some NBS-encoding genes were meiotically unstable, which may due to recombination and deletion events. Moreover, a transposon-like element was isolated in the flanking region of NBS genes, implying a possible role for transposon in the evolutionary history of resistance genes.
PLANTS utilize two distinct defense systems to recognize and respond to pathogen challenges (Chisholm et al. 2006; DeYoung and Innes 2006; Jones and Dangl 2006). The first line of defense against pathogens is basal defense, which uses pattern recognition receptors to recognize pathogen-associated molecular patterns, the common features of pathogens such as bacterial flagellin. The second line of defense acts strongly to specifically recognize pathogen effectors, using resistance (R) genes. Many R genes have now been cloned from a wide variety of plant species (Hammond-Kosack and Jones 1997; Dangl and Jones 2001), of which the largest class encodes a nucleotide-binding site (NBS) and a series of carboxy-terminal leucine-rich repeats (LRRs). The NBS domain contains several strictly ordered motifs—i.e., P-loop, kinase-2, kinase-3a, and Gly--Leu--pro--Leu (GLPL) motifs—that are highly conserved across plant species; they therefore have been used extensively to identify and classify NBS-encoding genes. By using PCR amplification with degenerate primers targeted to these conserved motifs, a large number of NBS sequences with homology to R genes, so-called R-gene homologs (RGHs), have been isolated from various plant species (Kanazin et al. 1996; Yu et al. 1996; Collins et al. 1998; Leister et al. 1998; Shen et al. 1998; Zhu et al. 2002; Ashfield et al. 2003). RGHs are abundant in plant genomes. Approximately 160, for example, are present in the Arabidopsis genome (Meyers et al. 2003). Genetic analysis has shown that RGHs tend to occur in clusters and often map to major resistance genes or quantitative trait loci (QTL; Kanazin et al. 1996; Collins et al. 1998; Young 2000; Donald et al. 2002; Ashfield et al. 2003; Calenge et al. 2005; Welter et al. 2007).
The large number of RGH sequences provides a facile system for studying the evolution of this gene family across plant taxa (Grube et al. 2000; Pan et al. 2000b; Bai et al. 2002; Meyers et al. 2003; see review in McHale et al. 2006). After analysis of >400 NBS-encoding homologs of 26 genera, Meyers et al. (1999) demonstrated that in genomes these RGHs frequently reside in “mega-clusters” consisting of smaller clusters with several members each, and they phylogenetically fall into two distinct groups: Toll and interleukin-1 receptor (TIR) and non-TIR subfamilies. Cannon et al. (2002) investigated an even larger number of sequences—800 RGHs from 30 genera—and suggested that evolutionary histories are different for the TIR and non-TIR subfamilies. With the increasing information in databases and the growing power of computational biology, researchers have shifted their focus to the comparative genetics of RGHs on/within the family level. Comparative analysis of tomato and Arabidopsis revealed remarkably rapid evolution of RGHs during radiation of plant families (Pan et al. 2000a). In a more in-depth study, Plocik et al. (2004) comparatively analyzed the RGHs within a specific family (Asteraceae) and indicated that gene duplication and loss events occur and change the composition of these gene subfamilies over time. Comparative analyses continued to be further applied in RGHs within a specific species, most of which concentrated on wild and cultivated plants such as apple (Lee et al. 2003), strawberry (Martinez Zamora et al. 2004), and potato (Kuang et al. 2005). However, relatively few studies have comparatively analyzed RGH patterns between resistant and susceptible lines within a fruit tree species, and the behaviors of these RGHs during meiosis have been ignored.
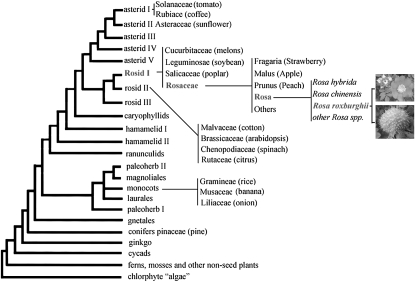
Chestnut rose, a rare fruit crop in southwest China, belongs to the Rosaceae family and Rosa genus (Figure 1). It has recently been labeled as one of the three promising new fruit crops in China (Wen and Deng 2005) due to its fruits having a high content of vitamin C (2000–3000 mg/100 g fresh weight) and displaying high levels of superoxide dismutase activity; it therefore is believed to have senescence-retarding and cancer-preventing effects (Ma et al. 1997). Unfortunately, chestnut rose crops are suffering from powdery mildew disease caused by the fungi Sphaerotheca pannosa. An indigenous cultivar from Guizhou province of China, Guinong no. 6, however, is highly resistant to powdery mildew. Within this species the other cultivar Guinong no. 5 is highly susceptible to powdery midew (supplemental Figure 1). Classical genetic analyses have showed that Guinong no. 6 and Guinong no. 5 are very similar in many evolutionary characteristics, such as growth habit and tree and leaf shape, and that they share a nearly identical genetic background. The relatively close phylogenetic relationship between Guinong no. 6 and Guinong no. 5 makes comparative analysis of R-gene candidates a particularly attractive approach to understanding how the resistance specificity was generated.
Figure 1.—
Phylogenetic position of chestnut rose in higher plant taxa. The flower and fruit of the chestnut rose are shown. Modified from Figure 1 of Ku et al. (2000).
In this study, we aim to clone RGHs from resistant and susceptible genotypes of chestnut rose (Rosa roxburghii) and their F1 progeny for comparative analysis of polymorphisms of the RGHs to identify genetic parameters relevant to the resistance specificity properties. Genetic mapping and evolutionary analysis provide us with insights into the genomic organization and evolutionary pattern of RGHs in the chestnut rose genome and allow us to detect amino acid sites under positive selection in the NBS-coding region. A transposon-like element was isolated in the flanking region of the NBS domain. In addition, some RGH meiotic stability was investigated.
MATERIALS AND METHODS
Plant materials:
R. roxburghii cv. Guinong no. 6 (a powdery mildew disease-resistant genotype) and R. roxburghii cv. Guinong no. 5 (susceptible genotype) and an F1 population of 109 plants derived from an intraspecific cross between them were used for molecular cloning, genetic mapping, and powdery mildew disease phenotyping. Disease phenotypes were evaluated as previously reported (Xu et al. 2005).
Isolation of NBS-encoding genes from resistant genotype, susceptible genotype, and their F1 progeny:
For resistant and susceptible parents, genomic DNA were extracted and used for PCR amplification. For F1 progeny, bulk DNA was constructed for molecular cloning. RGHs were broadly obtained as much as possible by employing two cloning methods: (1) PCR amplification with 19 degenerate primer pairs targeted to P-loop and GLPL motifs and (2) an overlapping extension approach, which used a target strategy to capture TIR-type RGHs from the genome (Xu et al. 2005). PCR products were cloned into pMD18-T vector (Takara Bio). Recombinant plasmid DNA was extracted by alkaline lysis (Sambrook et al. 1989). Each clone was reamplified with M13 universal primers and then subjected to restriction analysis using three restriction endonucleases (TaqI, HaeIII, and HinfI). On the basis of the restriction patterns, representative clones of each type were used for sequencing in both directions on an ABI 3730 sequencer.
For the purpose of comparative analysis, several sets of previously published RGHs from other plant species were used. Sixteen Arabidopsis RGHs were retrieved from At-Rgenes (http://niblrrs.ucdavis.edu/data_links.php) in March 2006. Fifteen NBS sequences were selected from cereal genomes described by Pan et al. (2000b) and 10 sequences from a coniferous species (Pinus monticola) were used to determine whether the chestnut rose NBS gene subfamily had evolved before or after the monocot/dicot and angiosperm/gymnosperm separation. The other sequences were from Medicago truncatula (Zhu et al. 2002), Cicer arietinum, Pennisetum glaucum, Cajanus cajan, Sorghum bicolor (Meyers et al. 1999), and unpublished data from Poncirus trifoliate in our laboratory.
Isolation of flanking sequences of NBS-encoding genes:
To isolate flanking sequences of NBS-encoding genes, a method similar to GenomeWalker Universal kit (CLONTECH) was used except that thermal asymmetric interlaced PCR (TAIL–PCR) was introduced to eliminate artificial PCR products. An adaptor was designed according to Calenge et al. (2005). The adapter had a short and a long arm, leaving the 5′ part of the adapter single stranded and the 3′ part containing a HpaII endonuclease restriction site that sticks to a digested DNA fragment. Three micrograms of genomic DNA was digested with 50 units of HpaII endonuclease enzymes for 8–16 hr and deactivated at 65° for 10 min. Then the HpaII adapter was ligated to the ends of the digested DNA fragments by adding 40 units of T4 ligase overnight at 16°. The ligated products were used as templates for PCR amplification with an adapter primer and a specific primer in the NBS region. The adapter primer was identical to the single-stranded 5′ part of the HpaII adaptor, and extension of the 3′-end of the short arm in the PCR reaction was effectively blocked by an amino linker. To increase the specificity of PCR products, TAIL–PCR was used (supplemental Table 3).
Gene-specific PCR amplification, Southern blotting, and genetic mapping:
Forty-two gene-specific primers were designed with primer3 software (http://www-genome.wi.mit.edu/cgi-bm/primer/primer3-www.cgi). The PCR cycling conditions were as follows: 94° for 3 min followed by 32 cycles of 94° for 30 sec, an annealing temperature (supplemental Table 2) for 1 min, and 72° for 1 min. PCR products were separated by electrophoresis on 1.5% agarose gel. Genomic DNA (10 μg) of both parents and a DNA bulk constituted from F1 progeny were digested with six restriction endonucleases (i.e., EcoRI, HindIII, BamHI, EcoRV, DraI, and TaqI). Southern hybridization was performed according to Xu et al. (2005).
Molecular markers were scored in the 109 F1 individuals according to Ritter et al. (1990). Linkage analysis and QTL detection were performed using Map Manager QTXb20 (Meer et al. 2004), with the linkage criterion being set at P = 0.05.
Sequence analyses:
Phylogenetic and evolutionary analyses were carried out according to Xu et al. (2007). Sequences were aligned using CLUSTALX (Thompson et al. 1997) and manually edited in GENEDOC (http://www.psc.edu/biomed/genedoc/). All the sequence statistics were output from GENEDOC, and Duncan's multiple range tests were performed by SPSS11.0 soft (SPSS, Chicago). Neighbor-joining trees using Kimura's two-parameter model and maximum parsimony phylogenetic trees were constructed, and bootstrap numbers were calculated by heuristic search in PAUP* 4.0 (Sinauer Associates, Sunderland, MA). The trees were visualized using the program TREEVIEW (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Evidence for positive selection was evaluated by using a maximum-likelihood method (Yang 1997), which allows searches for individual amino acid sites under selection. Site-specific models M7 and M8 in program CODEML from PAML soft were run to detect positive selection at individual sites. Model M7 is a special case of model M8 that assumes no selection, whereas model M8 allows for positively selected sites (Yang et al. 2000). Likelihood ratio tests (LRTs) were used to determine which model fit the observed data. The significance of the LRTs was evaluated by comparing it to a χ2 statistic. When the LRTs were significant, the Bayesian approach was used to calculate posterior probabilities of each positively selected site.
Three-dimensional structures for RGHs were constructed by homology modeling using MODELLER software 9v2 (Marti-Renom et al. 2000). The X-ray diffraction structural data from APAF-1 (PDB code 1z6t) were used as template to model the NBS-encoding genes. The sequence alignments of APAF-1 and NBS genes were obtained from PHYRE, a threading service software. Amino acids identified as possible targets of positive selection were mapped onto the three-dimensional structure using VMD software (Humphrey et al. 1996; Tiffin 2004).
RESULTS
NBS-encoding genes in resistant parent, susceptible parent, and F1 progeny:
Fragments of predicted sizes (500–700 bp) were cloned and sequenced from the resistant line (Guinong no. 6), the susceptible line (Guinong no. 5), and their F1 progeny. Of the 150 clones, 126 showed significant similarity (E-values < 10−5) to the known R genes or published RGHs as revealed by BlastX analysis in GenBank. For the remaining 24 sequences, 15 encompassed repetitive elements, while the remaining 9 sequences gave no or weak blast hits in the database (data not shown).
Conceptual translations of the above 126 sequences (38 from resistant parents, 37 from susceptible parents, and 51 from F1 progeny) revealed the presence of premature stop codons in 4 clones from the resistant line, 7 clones from the susceptible line, and 19 clones from F1 progeny. Interestingly, the Bk174 pseudogene carried six direct repeat oligos of 18 nucleotides (CAGCCTTCCTTTAGCCCT). The remaining 96 sequences (34 from resistant parents, 30 from susceptible parents, and 32 from F1 progeny) were identified as RGHs by the presence of a continuing open reading frame (ORF) and by characteristic motifs such as GVGKTT (P-loop), LLVLDDVW/D (kinase-2), and GLPL motif (Meyers et al. 1999; Pan et al. 2000b). Moreover, differences were observed in these motifs among genotypes; Sequence logos were made from the P-loop, kinase-2, and GLPL motifs to illustrate the differences among resistant parent, susceptible parent, and F1 progeny (supplemental Figure 2). Sequence diversity was analyzed on nucleotide and amino acid levels (supplemental Table 1). Within genotypes for resistant parents, sequence nucleotide identity averaged 52%; for susceptible parents, it was 57%. Unexpectedly, for the intergenotype between resistant and susceptible, average nucleotide identity was 54%, higher than that within the resistant genotype (supplemental Figure 1).
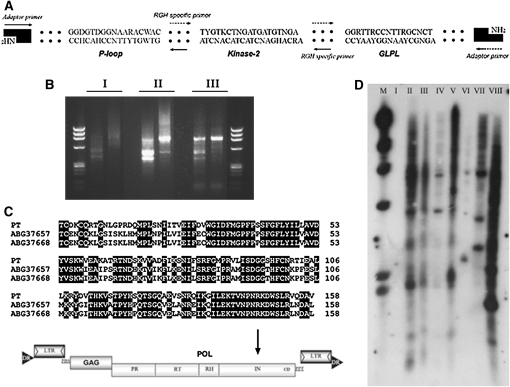
A transposon-like element is proximal to the NBS region:
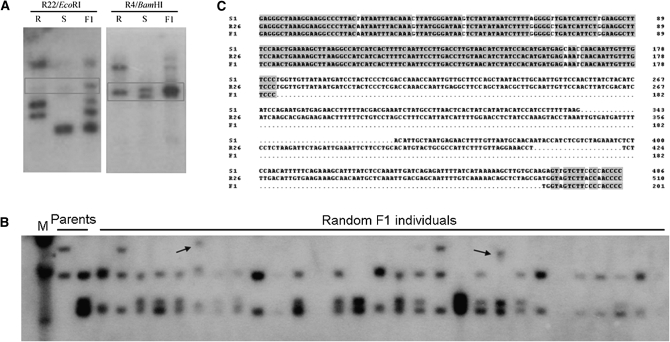
After the HpaII adaptor was ligated to digested DNA fragments, ligated products were used as templates to isolate genes proximal to the NBS-encoding region. Thirteen NBS-specific primers in combination with the adaptor primer were tested by PCR amplification. One primer combination produced a specific product of ∼500 bp after three rounds of PCR. Sequencing revealed that this fragment contains the adapter sequence at one end and an NBS primer sequence at the other end, indicating that this procedure efficiently excludes adapter–adapter fragments. The sequence was further characterized by sequence similarity searches in the GenBank databases using BLASTX program. The sequence was highly homologous to a published retrotransposon from Populus trichocarpa, with 66% identity. Southern analysis with this sequence as probe produced a classical pattern of transposon; i.e., a large number of copies and patterns look like smears, further confirming the existence of a transposon gene in the proximal region of NBS-encoding genes (Figure 2).
Figure 2.—
A transposon-like element was isolated close to the NBS domain. (A) The strategy for isolating flanking sequences, for which an adaptor was designed. Arrows show the location of the PCR primers. (B) PCR products of three rounds (I, II, and III) of TAIL–PCR. (C) The sequence was highly homologous to a transposon. (D) RFLP pattern confirmed the gene as a transposon and polymorphism was detected among crops in Rosaceae. I, negative control; II, resistant parent; III, susceptible parent; IV, R. roxburghii cv. Guinong no.1; V, R. sterilis; VI, Prunus persica; VII, Pyrus communis; VIII, Malus baccata.
Phylogenetical analysis of the NBS genes from resistant, susceptible parents and the F1 generation:
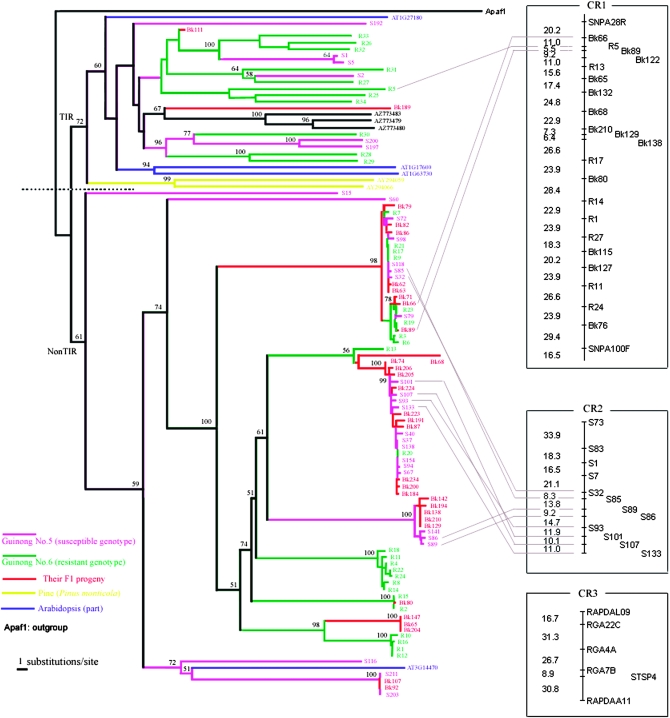
The 96 sequences from chestnut rose and 87 previously published sequences and mammalian apoptosis-related protein Apaf1 (outgroup) were used in phylogenetic analysis. Trees were created by the neighbor-joining method and the reliability of the trees was established by conducting 1000 bootstrap resampling steps (Figure 3). The overall impression of this tree is that NBS genes from resistant/susceptible genotypes and F1 progeny are well distributed among branches of this tree. This suggests that these genes probably arose from common ancestors that existed before the variety split within species. Two major branches designated as TIR and non-TIR subfamilies were obtained and well supported by high bootstrap value. The two branches were further confirmed by the presence or absence of characteristic subfamily-specific motifs; for example, RNBS-A-nonTIR (FDLxKxWVSVSDDF) and RNBS-A-TIR (LQxQLLSxxL) motifs are mutually exclusive in the two groups, consistent with previous results from Meyers et al. (1999) and Pan et al. (2000b).
Figure 3.—
Phylogenetic analysis and genetic mapping of chestnut rose NBS genes. The tree was constructed by the neighbor-joining method with human APAF1 as outgroup by PAUP* 4.0 software. Different colors denote different sources. Bootstrap values (1000 replicates) with only values >50% are shown on the branches. (Right) Three clusters were obtained by MAPMAKER and Map Manager QTXb20 software. The phylogenetic position of some RGH genes is connected to their locations in the genetic map by lines, illustrating two forms of cluster in the chestnut rose genome: the “tandem duplicated cluster,” genes that occupy the same phylogenetic lineage within a cluster, and, conversely, the “heterogeneous cluster.”
Five NBS genes from susceptible lines, 11 from resistant lines, 2 from F1 progeny, 3 from Arabidopsis, and 2 from Pinus monticola constitute the TIR branch. This TIR branch showed rather distantly clustered nodes, reflecting relative longevity of diversification within species. Some branches contained sequences from M. truncatula, implying that the ancient ancestor of chestnut rose and legumes contained multiple TIR sequences that have since diverged. The phylogenies of the TIR group showed that some chestnut rose TIR-type NBS sequences are phylogenetically closed to Pinus TIR-type sequences, indicating the antiquity of these sequences and that the origin of this gene family may predate the evolutionary split of the plant lineages leading to angiosperms and gymnosperms.
The non-TIR branch contains 62 members, with 19 from susceptible lines, 18 from resistant lines, 22 from F1 progeny, 1 from Arabidopsis, and 2 from Oryza sativa. Long branch lengths and closely clustered nodes were observed in this branch, indicating ancient divergence into separate lineages followed by more recent duplication and diversification.
The “describetrees” command was used in the PAUP software to output branch-tree-length statistics. The resistant line has a total length of 2001 units for 34 sequences, or 58.8/sequence; the susceptible line has a total length of 1590 units for 30 sequences, or 53.0/sequence; and the F1 progeny have a total length of 2018 units for 32 sequences, or 63.5/sequence. The multiple-comparisons LSD test reveals that tree distances for resistant and susceptible lines and F1 progeny are significantly different, with the difference of 3.2 ± 0.7 unit between resistant/susceptible genotypes.
Genetic mapping reveals three clusters of NBS genes in the chestnut rose genome:
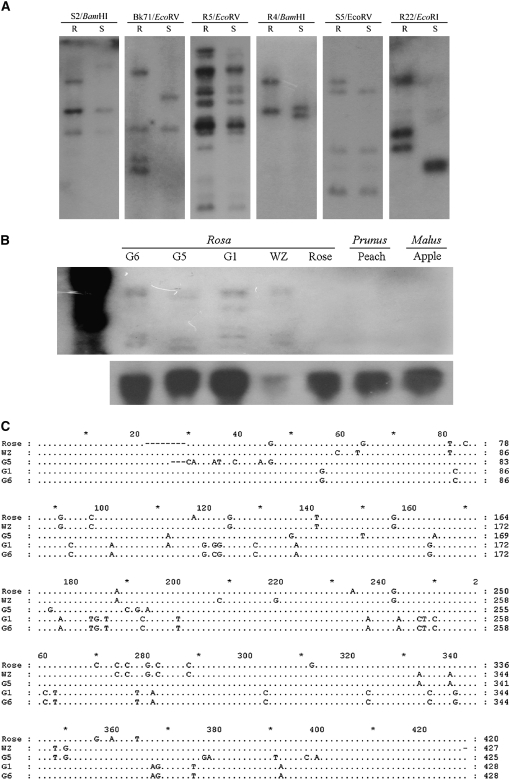
A sequence-tagged site (STS) marker was used to map the NBS genes because it can be easily applied by designing gene-specific primers. Forty-two gene-specific primer pairs (12 from the resistant genotype, 17 from the susceptible genotype, and 13 from the F1 generation genotype) were designed and used for scoring 109 F1 progeny (supplemental Table 2). Conventional RFLP was also employed to map the genes as previously described (Xu et al. 2005). Eleven representative clones in combination with five restriction endonucleases were used to detect polymorphisms between resistant/susceptible lines. Six clones yielded distinct polymorphisms (Figure 4) and were then used to survey Southern blots of the F1 progeny.
Figure 4.—
Rapid evolution of NBS-encoding genes in chestnut rose. (A) Intraspecific variation of copy number and size between resistant (R) and susceptible (S) parents. (B) Interspecific polymorphisms in Rosaceae family. (Top) A rapidly evolving gene. (Bottom) A rather old gene, which shows a uniform band among different species in Rosaceae. (C) Nucleotide polymorphism detected among different species. G6, G5, and G1 denote different cultivars in R. roxburghii; WZ, R. sterilis; and rose, R. chinensis.
Three clusters of NBS genes were obtained (Figure 3). The largest one, designated CR1, includes 23 STS markers developed from the resistant genotype and F1 progeny. NBS genes from the susceptible genotype form a single cluster CR2 composed of 12 loci. The third cluster CR3, with 6 loci, contains a major QTL that located between RGA22C marker and RAPDAL09 marker for powdery mildew resistance. Notably, among the 109 F1 progeny, 26 individuals tended to be resistant, and 25 of them were genotyped with the RGA22C marker; while 33 individuals were susceptible, all of them were absent from this marker. Therefore, RGA22C could be used as a molecular marker for efficient selection of resistant progeny (Xu et al. 2005).
When comparing the mapping data with phylogenic clades, we observed that RGHs from the same phylogenetic clade tend to cluster together. S101, S93, S133, and S107 were closely related in the same clade and sequence similarity between them was >90%; they clustered closely with a genetic distance of ∼10 cM in cluster CR2. The same phenomenon could be seen in cluster CR1. This phenomenon suggests that the tandem duplication followed by divergence occurred recently. This kind of cluster was termed the “tandem duplicated cluster” according to Leister (2004). However, sometimes NBS genes from distinguished clades clustered together. For example, in cluster CR1, the R5 gene, phylogenetically belonging to the TIR group, was flanked by Bk89 and Bk66 genes, which are a non-TIR type gene. This kind of cluster was termed the “heterogeneous cluster,” as suggested by Leister (2004).
Rapid evolution of NBS-encoding genes in chestnut rose:
The RFLP patterns of NBS-encoding genes were quite different between resistant and susceptible genotypes (Figure 4). Fewer numbers of hybridizing fragments were detected for the five probes R22, S5, R5, Bk71, and S2 in the susceptible genotype in comparison to the resistant genotype. For the R4 probe, the RFLP pattern is different between the two genotypes although both have two copy numbers. Further sequencing on the nucleotide level revealed a considerable sequence polymorphism between the two genotypes (the intraspecific variation; Figure 4). Within the NBS-encoding region 42 polymorphic sites were observed; of these, 15 result in silent substitutions, and 27 involve nonconservative amino acid changes. Approximately 9.81% of the total 428 nucleotides are polymorphic for this gene, which is higher than that observed in the Arabidopsis RPS2 gene (Caicedo et al. 1999). The intraspecific copy number and sequence nucleotide variation strongly suggest the rapid evolution and rearrangement of NBS-encoding genes in the chestnut rose genome.
Interspecific polymorphisms were also revealed from RFLP patterns among R. roxburghii, R. sterilis, and R. chinensis (Figure 4). On the nucleotide level, a total of 23 nonconservative amino acid changes were observed in R. sterilis, and a polymorphic site in R. chinesis resulted in a change from valine to a stop condon. Further intergenus RFLP survey in Prunus and Malus also revealed presence/absence polymorphism. The absence of interspecific and intergenus cross-hybridization signals and the considerable nucleotide changes confirmed the rapid evolution of NBS-encoding genes.
Positively selected sites were detected in an NBS region:
Types of forces that drive natural selection can been inferred from the ratio (ω) of nonsynonymous substitutions (dN) that cause an amino acid change to synonymous substitutions (dS) that do not. Site-specific models M7 and M8 (described in materials and methods) were used to determine the sites under purifying selection (ω < 1), neutral selection (ω ≈ 1), or diversifying selection (ω > 1). Pairwise values of dS and dN indicated an overall scarcity of amino acid substitutions. The average dS for resistant parents, susceptible parents, and the F1 generation ranged from 0.8 to 1.5 (supplemental Table 1), indicating a high sequence divergence according to the criteria described by Yang et al. (2000). Therefore, the ratios (ω) are reliable. Among the 146 sites, most ω are <1, indicating that most of the sites are under purifying selection. However, one residue, 125 H (histidine), was detected to be under strong diversifying selection (ω > 1) with a posterior probability of 92.4%. Other sites, including 47 P (proline), 57 E (glutamic acid), 107 K (lysine), and 127 L (leucine), are likely to be under diversifying selection with probabilities of 60–80% (Table 1; supplemental Figure 3).
TABLE 1.
Parameter estimates for positively selected sites in NBS-encoding genes in chestnut rose
| Model | Estimates of parametersa | Positively selected sitesb |
|---|---|---|
| M0: one ratio | ω = 0.301 | None |
| M7: β | P = 0.456, q = 1.062 | Not allowed |
| M8: β and ω | p0 = 0.956, p = 0.647, q = 1.363 | 125 H (at P > 0.9) |
| p1 = 0.044, ω = 1.138 | 47 P, 127 L (0.7 < P < 0.8) | |
| 57 E, 107 K (0.6 < P < 0.7) |
ω is the ratio of nonsynonymous substitutions (dN) that cause an amino acid change to the synonymous substitutions (dS) that do not.
P is the probability produced by naive empirical Bayes analysis.
Meiotic instability of NBS genes:
A fragment deletion event was observed during meiosis. The Bk111 gene, cloned from F1 progeny that showed sequence affiliation to the R26 gene from the resistant genotype, lost a 300-bp fragment during meiosis (Figure 5). Estimates of meiotic instability were further investigated by RFLP analysis. The Southern blotting results revealed that the hybridization pattern for F1 individuals can be the combined pattern of both parents, but to some extent, new hybridizing fragments appeared, forming a “recombination pattern” of the two parents (Figure 5). This indicated that during meiosis NBS-encoding genes are not stable and that recombination occurred at these loci and consequently resulted in the appearance of new alleles in the F1 progeny.
Figure 5.—
Meiotic instability of NBS-encoding genes. (A) A new homolog was produced in F1 progeny. (B) Homologs of different sizes were produced in F1 individuals. (C) Deletion was detected in genes from F1 progeny.
DISCUSSION
Previous studies of R genes or RGHs in model plants have accumulated knowledge on the generation of resistance specificities and the evolutionary dynamics of R genes (Bergelson et al. 2001; Bai et al. 2002; Richly et al. 2002; Caicedo and Schaal 2004; Kuang et al. 2004; Xiao et al. 2004; Mondragon-Palomino and Gaut 2005; Bakker et al. 2006; Friedman and Baker 2007). This study, however, focused on a fruit crop, the chestnut rose. Two cloning methods were used: the first, direct PCR amplification with degenerate primers, was believed to have a risk of biased sampling due to preferential amplification. To broaden the sequences, a second method, the overlapping extension approach, was used to capture as many NBS genes from the genome as possible (Xu et al. 2005). Under this strategy, a total of 126 NBS-encoding genes were isolated, fewer than those reported in Arabidopsis and rice. One possible reason for the lower number of NBS genes could be the preferential amplification caused by degenerate primers, although we tried to avoid this as mentioned above. The second reason could be due to the scarce genome information for this fruit tree; no sequence information could be utilized from a public database. Another reason could be the slower diversification rate of NBS genes in wild chestnut rose than in domestic plants such as rice because of the selective pressure. However, on the basis of the 126 cloned genes, this study still provides some interesting knowledge of the characteristics of NBS-encoding genes in chestnut rose, such as genomic organization in a tandem cluster, the proximity to transposon elements, rapid evolution, meiotic instability, etc.
Evolutionary complexities of NBS-encoding genes in chestnut rose:
NBS-encoding genes in chestnut rose exhibited a high level of intraspecific polymorphisms. For example, for the R4 gene, 9.81% of nucleotides were polymorphic, of which 64% were nonconservative amino acid changes, suggesting that this gene is maintained for short time periods.
What are the evolutionary forces shaping the polymorphisms in NBS-encoding genes in the chestnut rose genome? Intraspecific variations in NBS gene copy number and size were observed between resistant and susceptible genotypes (Figure 4). The two chestnut rose cultivars Guinong no. 6 and no. 5 were differentiated and selected as recently as 30 years ago. Figure 4 shows that the signature of unequal crossing-over events can produce gene copy-number variation and size difference (Hammond-Kosack and Jones 1997; McDowell and Simon 2006). Such a process almost certainly leads to rapid gene divergence between different genotypes (gene amplification and reduction, respectively), which is compatible with the opinion that NBS genes evolve rapidly. Moreover, the process of unequal crossing over could be facilitated by the repetitive elements (also found in this study) within the RGH clusters in the chestnut rose genome, as suggested by McDowell and Simon (2006) and Friedman and Baker (2007). Unequal crossing-over events are believed to homogenize genes within a genotype, resulting in paralogs being more similar than orthologs (Michelmore and Meyers 1998). However, in contrast to the above expectations, intergenotype comparison of nucleotide identity between resistant and susceptible genotypes turned out to be higher than that within the resistant genotype, a surprising result with orthologs being more similar than paralogs. The contradiction implies that there have been other processes that have shaped the NBS gene polymorphisms as well as unequal crossing-over events and that the unequal crossing-over events are not predominantly generational gene variations.
Evolutionary analysis and genetic mapping revealed the existence of a “tandem duplicated cluster,” where point mutations were observed among the tandem direct repeats. The accumulated mutations are another evolutionary way to increase the complexity of RGHs and an opportunity to produce a new homolog. Moreover, some RGHs share high homology with Pinus RGHs and produced uniform hybridization band among Rosaceae plants (Figure 4), indicating that these genes are ancient and may be evolutionarily maintained by some form of balancing selection (Tian et al. 2002).
Detecting adaptive evolution by comparing amino acid substitution rates (dN) to synonymous substitution rates (dS) has indicated that positive selection has contributed to the evolution of NBS genes in chestnut rose. For many reported R genes, positive selection has been detected primarily in LRR regions. However, positively selected sites were also detected in the NBS-encoding region; e.g., Mondragon-Palomino et al. (2002) found that in Arabidopsis five positive selected sites were positioned in the NBS region, implying that the NBS domain may function in determining resistance specificity. This could be determined from domain-swapping experiments by replacing the NBS-encoding region of the L10 gene with the equivalent region of L2- or L9-generated new recombinant alleles with novel specificity (Luck et al. 2000). In chestnut rose, the NBS domain may also function in determining the resistance specificity. Further research is required to investigate this hypothesis.
Transposable-like elements appear to have been involved in the evolutionary dynamics of NBS-encoding genes in chestnut rose. A transposable element was isolated proximal to the NBS gene. For most Rosaceae genomes, a large number of gene copies were detected by RFLP analysis. However, on cultivar Guinong no. 1 (intraspecific genotype), peach, and pear, one to three copies were detected. The marked difference in copy number implied that transposable elements could play active roles in the evolutionary history of resistance genes in chestnut rose. In rice, 11 different families of transposable elements were identified at the Xa21 cluster, and the elements appeared to be a major source of variation in this cluster (Song et al. 1998). In addition, in plants, transposable elements are commonly activated by environmental stresses such as pathogen infection (Grandbastien 1998), and it is believed that such activation can increase genomic flexibility with a possible selective advantage.
Together, positive selection, balancing selection, recombination, point mutation, and even transposable elements may constitute the driving forces that shape the complexity, rapid evolution, and even the generation of new resistance specificity of R-gene sequences in chestnut rose.
Meiotic instability of RGH genes:
Sequence pairwise comparison revealed that nucleotide identity averaged 52% within resistant parents, 57% within susceptible parents, and 45% within the F1 generation. Statistical analysis showed that the latter was significantly lower than the former two at the 0.05 level, indicating a higher sequence variation in F1 progeny than in parents. Moreover, some sequences from F1 progeny carried repetitive elements.
Evidence from RFLP markers demonstrated that RGH genes are meiotically unstable. New alleles were observed in F1 plants. To confirm this, we used the RGH sequences from F1 plants to design specific PCR primers and to determine the gene status in two parents. Three types were detected; what merits attention is type III where the RGH gene was detected only in some F1 plants but not in either parent (supplemental Figure 4), suggesting that this allele was newly produced during meiosis. However, these are preliminary data for the estimation of meiotic instability. One classical example was Rp1 complex loci in maize, where the homozygous line for the Rp1 locus was used to generate a large number of testcross progeny. Surprisingly, a high frequency of susceptibles was found in the progeny, indicating the occurrence of meiotically unstable genes (Sudupak et al. 1993). Further research on the Rp1 locus demonstrated that recombination is the primary mechanism of meiotic instability and that such recombination can result in new race specificity (Smith and Hulbert 2005). But for this study, the chestnut rose fruit tree is believed to be highly heterozygous for most gene loci; it is difficult to create a homozygous line to obtain deep insight into the mechanism for meiotic instability. However, further research on sequencing the flanked region around the NBS domain may help us understand the types of recombination and the genetic mechanism for meiotic instability.
Acknowledgments
The authors are grateful to S. Xiao from Center for Biosystems Research of the University of Maryland Biotechnology Institute for his valuable comments. Thanks also go to the anonymous reviewer for critical reviews. This project was supported by the Ministry of Education of China (no. IRT0548), 973 project (2006CB708202), and the National Natural Science Foundation of China (30660115).
References
- Ashfield, T., A. Bocian, D. Held, A. D. Henk, L. F. Marek et al., 2003. Genetic and physical localization of the soybean Rpg1-b disease resistance gene reveals a complex locus containing several tightly linked families of NBS-LRR genes. Mol. Plant Microbe Interact. 16 817–826. [DOI] [PubMed] [Google Scholar]
- Bai, J., L. A. Pennill, J. Ning, S. W. Lee, J. Ramalingam et al., 2002. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res. 12 1871–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker, E. G., C. Toomajian, M. Kreitman and J. Bergelson, 2006. A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell 18 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson, J., M. Kreitman, E. A. Stahl and D. Tian, 2001. Evolutionary dynamics of plant R-genes. Science 292 2281–2284. [DOI] [PubMed] [Google Scholar]
- Caicedo, A. L., and B. A. Schaal, 2004. Heterogeneous evolutionary processes affect R gene diversity in natural populations of Solanum pimpinellifolium. Proc. Natl. Acad. Sci. USA 101 17444–17449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, A. L., B. A. Schaal and B. N. Kunkel, 1999. Diversity and molecular evolution of the RPS2 resistance gene in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calenge, F., C. G. Van De Linden, E. Van De Weg, H. J. Schouten, G. Van Arkel et al., 2005. Resistance gene analogues identified through the NBS-profiling method map close to major genes and QTL for disease resistance in apple. Theor. Appl. Genet. 110 660–668. [DOI] [PubMed] [Google Scholar]
- Cannon, S. B., H. Y. Zhu, A. M. Baumgarten, R. Spangler, G. May et al., 2002. Diversity, distribution, and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene families. J. Mol. Evol. 54 548–562. [DOI] [PubMed] [Google Scholar]
- Chisholm, S. T., G. Coaker, B. Day and B. J. Staskawicz, 2006. Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803–814. [DOI] [PubMed] [Google Scholar]
- Collins, N. C., C. A. Webb, S. Seah, J. G. Ellis, S. H. Hulbert et al., 1998. The isolation and mapping of disease resistance gene analogs in maize. Mol. Plant Microbe Interact. 11 968–978. [DOI] [PubMed] [Google Scholar]
- Dangl, J. L., and J. D. G. Jones, 2001. Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- DeYoung, B. J., and R.W. Innes, 2006. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7 1243–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald, T. M., F. Pellerone, A. F. Adam-Blondon, A. Bouquet, M. R. Thomas et al., 2002. Identification of resistance gene analogs linked to a powdery mildew resistance locus in grapevine. Theor. Appl. Genet. 101 301–308. [DOI] [PubMed] [Google Scholar]
- Friedman, A. R., and B. J. Baker, 2007. The evolution of resistance genes in multi-protein plant resistance systems. Curr. Opin. Genet. Dev. 17 1–7. [DOI] [PubMed] [Google Scholar]
- Grandbastien, M. A., 1998. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 3 181–187. [Google Scholar]
- Grube, R. C., E. R. Radwanski and M. Jahn, 2000. Comparative genetics of disease resistance within the Solanaceae. Genetics 155 873–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack, K. E., and J. D. G. Jones, 1997. Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 575–607. [DOI] [PubMed] [Google Scholar]
- Humphrey, W., A. Dalke and K. Schulten, 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14 33–38. [DOI] [PubMed] [Google Scholar]
- Jones, J. D. G., and J. L. Dangl, 2006. The plant immune system. Nature 444 323–329. [DOI] [PubMed] [Google Scholar]
- Kanazin, V., L. F. Marek and R. C. Shoemaker, 1996. Resistance gene analogs are conserved and clustered in soybean. Proc. Natl. Acad. Sci. USA 93 11746–11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, H. M., T. Vision, J. Liu and S. D. Tanksley, 2000. Comparing sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proc. Natl. Acad. Sci. USA 97 9121–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang, H., S. S. Woo, B. C. Meyers, E. Nevo and R. W. Michelmore, 2004. Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell 16 2870–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang, H., F. Wei, M. R. Marano, U. Wirtz, X. Wang et al., 2005. The R1 resistance gene cluster contains three groups of independently evolving, type I R1 homologues and shows substantial structural variation among haplotypes of Solanum demissum. Plant J. 44 37–51. [DOI] [PubMed] [Google Scholar]
- Lee, S. Y., J. S. Seo, M. Rodriguez-Lanetty and D. H. Lee, 2003. Comparative analysis of superfamilies of NBS-encoding disease resistance gene analogs in cultivated and wild apple species. Mol. Genet. Genomics 269 101–108. [DOI] [PubMed] [Google Scholar]
- Leister, D., 2004. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 20 116–122. [DOI] [PubMed] [Google Scholar]
- Leister, D., J. Kurth, D. A. Laurie, M. Yano, T. Sasaki et al., 1998. Rapid reorganization of resistance gene homologues in cereal genomes. Proc. Natl. Acad. Sci. USA 95 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck, J. E., G. J. Lawrence, P. N. Dodds, K. W. Shepherd and J. G. Ellis, 2000. Regions outside of the leucine-rich repeats of flax rust resistance proteins play a role in specificity determination. Plant Cell 12 1367–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. X., Y. Zhu and C. F. Wang, 1997. The aging retarding effect of ‘Long-Life CiLi’. Mech. Ageing Dev. 96 171–189. [DOI] [PubMed] [Google Scholar]
- Martinez-Zamora, M. G., A. P. Castagnaro and J. C. Diaz-Ricci, 2004. Isolation and diversity analysis of resistance gene analogues (RGAs) from cultivated and wild strawberries. Mol. Genet. Genomics 272 480–487. [DOI] [PubMed] [Google Scholar]
- Marti-Renom, M. A., A. Stuart, A. Fiser, R. Sánchez, F. Melo et al., 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29 291–325. [DOI] [PubMed] [Google Scholar]
- McDowell, J. M., and A. S. A. Simon, 2006. Recent insights into R gene evolution. Mol. Plant Pathol. 7 437–448. [DOI] [PubMed] [Google Scholar]
- McHale, L., X. Tan, P. Koehl and R. W. Michelmore, 2006. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 7 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meer, J., R. Cudmore and K. F. Manly, 2004. Map Manager QTX. http://www.mapmanager.org/mmQTX.html [DOI] [PubMed]
- Meyers, B. C., A. W. Dickerman, R. W. Michelmore, S. Sivaramakrishnan, B. W. Sobral et al., 1999. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20 317–332. [DOI] [PubMed] [Google Scholar]
- Meyers, B. C., A. Kozik, A. Griego, H. Kuang and R. W. Michelmore, 2003. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R. W., and B. C. Meyers, 1998. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8 1113–1130. [DOI] [PubMed] [Google Scholar]
- Mondragon-Palomino, M., and B. S. Gaut, 2005. Gene conversion and the evolution of three leucine-rich repeat gene families in Arabidopsis thaliana. Mol. Biol. Evol. 22 2444–2456. [DOI] [PubMed] [Google Scholar]
- Mondragon-Palomino, M., B. C. Meyers, R. W. Michelmore and B. S. Gaut, 2002. Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 12 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Q., Y. S. Liu, O. Budai-Hadrian, M. Sela, L. Carmel-Goren et al., 2000. a Comparative genetics of nucleotide binding site-leucine rich repeat resistance gene homologues in the genomes of two dicotyledons: tomato and Arabidopsis. Genetics 155 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Q., J. Wendel and R. Fluhr, 2000. b Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J. Mol. Evol. 50 203–213. [DOI] [PubMed] [Google Scholar]
- Plocik, A., J. Layden and R. Kesseli, 2004. Comparative analysis of NBS domain sequences of NBS-LRR disease resistance genes from sunflower, lettuce, and chicory. Mol. Phylogenet. Evol. 31 153–163. [DOI] [PubMed] [Google Scholar]
- Richly, E., J. Kurth and D. Leister, 2002. Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol. Biol. Evol. 19 76–84. [DOI] [PubMed] [Google Scholar]
- Ritter, E., C. Gebhardt and F. Salamini, 1990. Estimation of recombination frequencies and construction of RFLP linkage maps in plants from crosses between heterozygous parents. Genetics 125 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shen, K. A., B. S. Meyers, M. N. Islam-Faridi, D. B. Chin, D. M. Stelly et al., 1998. Resistance gene candidates identified by PCR with degenerate oligonucleotide primers map to clusters of resistance genes in lettuce. Mol. Plant Microbe Interact. 11 815–823. [DOI] [PubMed] [Google Scholar]
- Smith, S. M., and S. H. Hulbert, 2005. Recombination events generating a novel Rp1 race specificity. Mol. Plant Microbe Interact. 18 220–228. [DOI] [PubMed] [Google Scholar]
- Song, W. Y., L.Y. Pi, T. E. Bureau and P. C. Ronald, 1998. Identification and characterization of 14 transponson-like elements in the noncoding regions of members of the Xa21 family of disease resistance genes in rice. Mol. Gen. Genet. 258 449–456. [DOI] [PubMed] [Google Scholar]
- Sudupak, M. A., J. L. Bennetzen and S. H. Hulbert, 1993. Unequal exchange and meiotic instability of disease-resistance genes in the Rp1 region of maize. Genetics 133 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D., H. Araki, E. Stahl, J. Bergelson and M. Kreitman, 2002. Signature of balancing selection in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin, P., 2004. Comparative evolutionary histories of chitinase genes in the genus Zea and family Poaceae. Genetics 167 1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter, L. J., N. Gokturk-Baydar, M. Akkurt, E. Maul, R. Eibach et al., 2007. Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L). Mol. Breed. 20 359–374. [Google Scholar]
- Wen, X. P., and X. X. Deng, 2005. Micropropagation of chestnut rose (Rosa roxburghii Tratt) and genetic stability assessment of the in vitro plants using RAPD and AFLP markers. J. Hortic. Sci. Biotechnol. 80 54–60. [Google Scholar]
- Xiao, S., B. Emerson, K. Ratanasut, E. Patrick, C. O'Neill et al., 2004. Origin and maintenance of a broad-spectrum disease resistance locus in Arabidopsis. Mol. Biol. Evol. 21 1661–1672. [DOI] [PubMed] [Google Scholar]
- Xu, Q., X. P. Wen and X. X. Deng, 2005. Isolation of TIR and nonTIR NBS-LRR resistance gene analogues and identification of molecular markers linked to a powdery mildew resistance locus in chestnut rose (Rosa roxburghii Tratt). Theor. Appl. Genet. 111 819–830. [DOI] [PubMed] [Google Scholar]
- Xu, Q., X. P. Wen and X. X. Deng, 2007. Phylogenetic and evolutionary analysis of NBS-encoding genes in Rosaceae fruit crops. Mol. Phylogenet. Evol. 44 315–324. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: A program package for phylogenetic analysis by maximum likehood. Comput. Appl. Biosci. 13 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., R. Nielsen, N. Goldman and A. M. K. Pedersen, 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, N. D., 2000. The genetic architecture of resistance. Curr. Opin. Plant Biol. 3 285–290. [DOI] [PubMed] [Google Scholar]
- Yu, Y. G., G. R. Buss and M. A. Maroof, 1996. Isolation of a superfamily of candidate disease-resistance genes in soybean based on a conserved nucleotide-binding site. Proc. Natl. Acad. Sci. USA 93 11751–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H., S. B. Cannon, N. D. Young and D. R. Cook, 2002. Phylogeny and genomic organization of the TIR and non-TIR NBS-LRR resistance gene family in Medicago truncatula. Mol. Plant Microbe Interact. 15 529–539. [DOI] [PubMed] [Google Scholar]