Abstract
Plants synthesize an array of natural products that play diverse roles in growth, development, and defense. The plant-specific phenylpropanoid metabolic pathway produces as some of its major products flavonoids, monolignols, and hydroxycinnamic- acid conjugates. The reduced epidermal fluorescence 4 (ref4) mutant is partially dwarfed and accumulates reduced quantities of all phenylpropanoid-pathway end products. Further, plants heterozygous for ref4 exhibit intermediate growth and phenylpropanoid-related phenotypes, suggesting that these mutations are semidominant. The REF4 locus (At2g48110) was cloned by a combined map- and sequencing-based approach and was found to encode a large integral membrane protein that is unique to plants. The mutations in all ref4 alleles cause substitutions in conserved amino acids that are located adjacent to predicted transmembrane regions. Expression of the ref4-3 allele in wild-type and null REF4 plants caused reductions in sinapoylmalate content, lignin content, and growth, demonstrating that the mutant alleles are truly semidominant. Further, a suppressor mutant was isolated that abolishes a WW protein–protein interaction domain that may be important for REF4 function.
MANY mutations have been identified in structural genes that are required for the accumulation of phenylpropanoid-pathway end products, particularly in Arabidopsis and maize. The majority of mutant phenotypes identified in these forward genetic screens segregate as recessive traits. A theoretical basis for this observation has been provided by metabolic control analysis (Kacser and Burns 1981), which suggests that the contribution of a single enzyme's activity within a metabolic pathway is generally small in comparison to the summed activity of all the enzymes in the entire pathway. Thus, this analysis predicts that only in very short metabolic pathways can a null or hypomorphic mutation in one of the biosynthetic enzyme-encoding genes be semidominant.
Although often more difficult to interpret than simple loss-of-function alleles, dominant mutations often lead to interesting insights into the pathways within which the mutated gene operates. In humans, dominant and semidominant diseases are often caused by mutations in transcription factors, transporters, and components of signaling cascades (Jimenez-Sanchez et al. 2001; Kondrashov and Koonin 2004). Mutations in such genes may lead to dominant phenotypes through a number of different mechanisms. The simplest of these is haplo-insufficiency where the wild-type allele of a gene in a +/− heterozygote does not produce enough protein to generate a wild-type phenotype. Although few examples of haplo-insufficiency are known in plants (Weijers et al. 2001), in Antirrhinum flowers, haplo-insufficiency with regard to anthocyanin accumulation is revealed in the presence of null alleles of F3H (Coen et al. 1986; Martin et al. 1991). Further, because these null alleles operate in a biosynthetic pathway, they represent an exception to the predictions of metabolic control analysis and demonstrate that F3H is truly the rate-limiting enzyme in anthocyanin biosynthesis.
Another class of dominant mutations either increases the abundance of or stabilizes mRNA transcripts or their encoded proteins, preventing the normal turnover of these molecules essential for the wild-type phenotype. Although these gain-of-function mutations are now commonly generated synthetically by approaches such as activation tagging (e.g., Sundaresan et al. 1995), examples of such mutations generated via point mutations have been described. One such example is the Arabidopsis (Arabidopsis thaliana L. Heynh) atr1D (altered tryptophan regulation-dominant) mutant that exhibits upregulated transcription of tryptophan biosynthetic genes due to the stabilization of the ATR1 Myb transcription factor mRNA (Bender and Fink 1998; Smolen and Bender 2002). Further, mutations in the miRNA target sequences of REVOLUTA (REV), PHABULOSA (PHB), and PHAVOLUTA (PHV) cause these genes to escape miRNA-mediated transcript degradation, effectively resulting in dominant gain-of-function alleles (Emery et al. 2003; Tang et al. 2003). In the context of protein stability, dominant mutations in ETO2 and ETO3 lead to an increased stability of the encoded 1-aminocyclopropane-1-carboxylic acid synthase proteins (Vogel et al. 1998; Chae et al. 2003), which causes enhanced ethylene production.
Finally, dominance may arise from mutations in signaling cascade components that exhibit binary states. Very specific mutations can lock such components into an “on” or “off” state and thus perturb downstream components of the system. For example, the ethylene receptor ETR1 negatively regulates ethylene signaling in the absence of ethylene (Bleecker et al. 1988; Chang et al. 1993). Only dominant etr1 alleles encoding receptors incapable of binding ethylene were identified in mutant screens because such variants constitutively signal the absence of ethylene even in its presence. Furthermore, since the ethylene receptors are encoded by a small gene family, it is only when several family members are inactivated by loss-of-function mutations that recessive ethylene-hypersensitive phenotypes can be observed (Hua and Meyerowitz 1998; McCourt 1999).
Arabidopsis synthesizes a suite of natural products via the phenylpropanoid pathway, including flavonoids and lignin, as well as hydroxycinnamic acid esters (Chapple et al. 1994). This latter class of compounds includes sinapoylmalate, a metabolite accumulated in Arabidopsis leaves that fluoresces under UV light. Using this phenotype as a genetic marker, we isolated a series of reduced epidermal fluorescence (ref) mutants from an ethyl methane sulphonate (EMS)-mutagenized population (Ruegger and Chapple 2001). Analysis of these mutants, most of which are perturbed in genes encoding phenylpropanoid biosynthetic enzymes, have led to substantial insights into the structure of the phenylpropanoid pathway (Humphreys and Chapple 2002; Stout and Chapple 2004). Here we show that the semidominant ref4 mutant alleles decrease all classes of phenylpropanoids. Further, we show that REF4 encodes a large membrane-localized protein of unknown function that is unique to plants.
MATERIALS AND METHODS
Plant material and growth conditions:
Arabidopsis thaliana L. Heynh was grown at a light intensity of 100 μE m−2 sec−1 at 23° under a photoperiod of 16 hr light/8 hr dark in Redi-Earth potting mix (Scotts-Sierra Horticulture Products, Marysville, OH). The three ref4 alleles (ref4-1–ref4-3) used in this study were identified in an M2 screen of EMS-mutagenized plants of the Columbia ecotype (Ruegger and Chapple 2001). All of the alleles used in this study were backcrossed at least two times to the corresponding wild type prior to analysis.
Analytical methods:
Leaf-derived soluble hydroxycinnamic acid esters were extracted from entire rosettes and subsequently separated and quantified by HPLC as previously described (Hemm et al. 2003). All plants used for these analyses were 20 days old unless otherwise noted. Sinapoylcholine in mature seeds derived from parental plants grown under identical conditions was analyzed similarly, except that a Puresil C18 column (Waters, Milford, MA; 1200 nm pore size, 5 μm particle size) was used. Lignin quantity was analyzed by the Klason method (Kaar and Brink 1991) or the TGA method (Campbell and Ellis 1992) using homogenized stems from plants that had just completed bolting. Lignin quality was analyzed by pyrolysis-GC-MS as previously described (Franke et al. 2002a) and by DFRC (Lu and Ralph 1997).
Map-based cloning of REF4:
The ref4-3 mutant (Columbia background) was crossed to Landsberg erecta to establish a mapping population. F1 individuals were allowed to self-pollinate, and F2 plants were screened for the ref4 phenotype. Due to the dominance of the ref4-3 allele, DNA was extracted only from F2 ref4 mutants that exhibited the most severe ref phenotype for use in PCR-based genotyping experiments. Individuals carrying recombinant chromosomes in the region of the REF4 locus were used to determine a mapping interval for the REF4 gene.
Marker analysis of the REF4 mutant alleles:
A dCAPS marker was created to verify the mutation observed in ref4-1 and ref4-2. The restriction enzyme MseI cuts the mutant PCR products derived from the primers cc1551 (5′-tgtcgggatatcaccctta-3′) and cc1552 (5′-cgggagagtccacgtaatgt-3′). The mutation observed in ref4-3 results in a CAPS polymorphism that eliminates an AvaII restriction site from the wild-type sequence. The PCR primers cc1443 (5′-ctttggttgcccattgatct-3′) and cc1439 (5′-gattggttcccccaattaca-3′) were used to generate PCR products that were then cut with AvaII to verify the EMS-induced mutation. The suppressor mutation was verified by a dCAPS marker using the primers cc1703 (5′-gttgctcaacgcttgctaaatttgctgca-3′) and cc1449 (5′-tgtcccttgatttgtttcagg-3′) where only the wild-type PCR product is cut with PstI.
RNAi of REF4 transcripts:
A vector to trigger RNAi-mediated reduction of REF4 transcript levels was generated by first producing two 362-bp fragments of the REF4 open reading frame by PCR. The first fragment was generated using the primers cc1605 (5′-gaggtaccggaccctcgattggatctct-3′) and cc1608 (5′-ggaattccttggcaagtcaaaacatgga-3′), which introduced terminal KpnI and EcoRI restriction sites, respectively; whereas, the second fragment was generated using the primers cc1602 (5′-gtggatccttggcaagtcaaaacatgga-3′) and cc1603 (5′-gaatcgatggaccctcgattggatctct-3′), which introduced terminal BamHI and a ClaI restriction sites, respectively. These fragments were cloned into pGEM-T Easy (Promega, Madison WI) and were then isolated from this construct by restriction digestion with the appropriate enzymes. The resulting restriction fragments were then subcloned sequentially in sense and antisense orientation into pHANNIBAL (Wesley et al. 2001). The RNAi cassette constructed in pHANNIBAL was then isolated from the vector as a NotI fragment and cloned into the binary vector pART27. This vector was introduced into the Agrobacterium strain C58 pGV3850, which was subsequently introduced into ref4 and wild-type plants by the floral dip method (Clough and Bent 1998).
Quantitative RT–PCR:
RNA was extracted from whole rosette tissue using a hot phenol method, treated with RQ1 DNAse (Promega), and reverse transcribed with ImProm II (Promega). REF4 transcripts were amplified with the primers cc2049 (5′-aagtctgaggcagtggaa cg-3′) and cc2050 (ttgcaagtctccacaatgag); RFR1 transcripts were amplified with the primers cc2053 (5′-acttacttggggcgtggatt-3′) and cc2052 (5′-tttcccatctaaggcactcg-3′). Elongation factor 1-α was chosen as the internal reference gene on the basis of meta-analyses of microarray data (Czechowski et al. 2005), and was amplified with the primers cc2012 (5′-tggtgacgctggtatggtta-3′) and cc2013 (5′-ggtctgcctcatgtccctaa). Each primer set was optimized so that only one amplification product was detected and that the efficiency of the PCR reaction was between 95 and 105%. Quantitative PCR reactions were prepared with a SYBR-green master mix (Applied Biosystems, Foster City, CA) and carried out in a 7000 sequence detection system (Applied Biosystems).
Constructs to express REF4-3:
The full-length open reading frame of REF4 was amplified from an Arabidopsis seedling cDNA library using primers cc1440 (5′-aaggctgaggaagaagacga-3′) and cc1592 (3′-ggaattccgacgtcaagctaatgttgatgg-5′) and cloned into pGEM-T Easy. The Stratagene Quickchange site-directed mutagenesis kit (Stratagene, Cedar Creek TX) was used to generate the ref4-3 allele, using the mutagenic primer cc1679 (5′-agatccgatcgagagtcctgtgccccgca-3′), which also introduced a silent PvuI restriction site to assist in identifying mutagenized plasmids and for subsequent genotyping of transgenic plants. The mutant open reading frame was then introduced into a pBI101 vector (Jefferson et al. 1987) that contained a 2.1-kbp fragment of either the native REF4 promoter or the CaMV 35S promoter element. These constructs were introduced into either wild-type or ref4-4 plants via the floral dip method (Clough and Bent 1998).
ref4-3 supressor screen:
Approximately 75,000 ref4-3 seeds (1.5 g) were mutagenized in a 0.3% solution of EMS for 10 hr. After rinsing eight times with water, the seeds were sown at a density of 1 seed cm−2 and grown in greenhouse conditions. M2 seed was collected and sown at a density of 0.2 seeds cm−2 and screened for plants exhibiting wild-type growth and/or UV fluorescence.
Phylogenic analysis:
Full-length amino acid sequences were aligned and analyzed using the Mega3.1 software packages' neighbor-joining algorithm employing default parameters (Kumar et al. 2004). Distances were computed using 1000 bootstrap replicates.
Statistical analysis:
All statistical analyses were performed using the SAS software package (Cary, NC), with an α-value set at 0.05.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes.
RESULTS
Mutations in REF4 decrease phenylpropanoid accumulation and perturb growth:
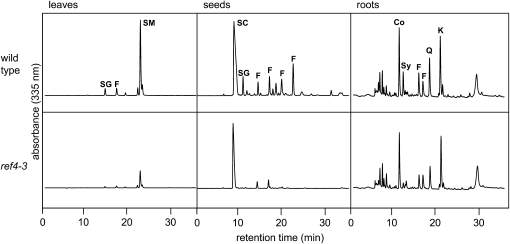
Three independent alleles of ref4 were identified in our original mutant screen (Ruegger and Chapple 2001). To investigate which phenylpropanoids are reduced in the mutant and to what extent, methanolic extracts of leaves, roots, and seeds of ref4-3 were analyzed by HPLC (Figure 1). Fifteen-day-old ref4-3 plants contained significantly less sinapoylmalate than the wild type [Figure 1; pmol mg−1 fresh weight (f.w.) ± SE; Columbia wild type, 1452 ± 146; ref4-3, 289 ± 43; n = 4]. Indeed, of the phenylpropanoid mutants we have identified, only fah1, ref3-2, and ref8 accumulate less sinapoylmalate than ref4-3 (Chapple et al. 1992; Ruegger and Chapple 2001). This reduced sinapoylmalate content was observed at all points in a time-course experiment over several weeks of plant growth, indicating that phenylpropanoid biosynthesis is not simply delayed in the mutant (supplemental Figure 1). Arabidopsis seeds accumulate the hydroxycinnamic acid ester sinapoylcholine and to a lesser extent sinapoylglucose. In mature ref4-3 seeds, significantly less sinapoylcholine was found (pmol mg−1 f.w. ± SE; Columbia wild type, 29.3 ± 5.4; ref4-3, 18.1 ± 0.8; n = 3), and seed sinapoylglucose levels were reduced from 7.1 ± 1.0 μmol g−1 f.w. in Columbia wild type to below the limits of detection in ref4-3. Further, flavonoids putatively identified by their absorption spectra were also observed to be reduced in the mutant (supplemental Figure 2). Finally, light-grown Arabidopsis roots accumulate coniferin and syringin, the 4-O-glucosides of coniferyl alcohol and sinapyl alcohol, respectively (Hemm et al. 2004), and ref4-3 roots had reduced levels of these phenylpropanoids as well (pmol mg−1 f.w. ± SE; coniferin, Columbia wild type, 700 ± 15; ref4-3, 485 ± 77; syringin, Columbia wild type, 278 ± 11; ref4-3, 76 ± 13; n = 3), although, the ref4-3 mutation had a much more modest impact on the accumulation of UV-absorbing compounds in the roots than in aerial tissues. In all samples examined, no novel HPLC peaks were identified that could indicate a biosynthetic reaction in phenylpropanoid metabolism which is blocked in the mutant, as was possible with the sng1, sng2, ref3, and ref8 mutants (Stout and Chapple 2004).
Figure 1.—
HPLC analysis of soluble UV-absorbing compounds in leaves, seeds, and roots of ref4-3 and the wild type. The elution of compounds was monitored by absorbance at 330 nm. Compounds are identified as follows: Co, coniferin; F, flavonoid; K, rha-glc-kaempferol; Q, rha-glc-quercetin; SC, sinapoylcholine; SG, sinapoylglucose; SM, sinapoylmalate; and Sy, syringin.
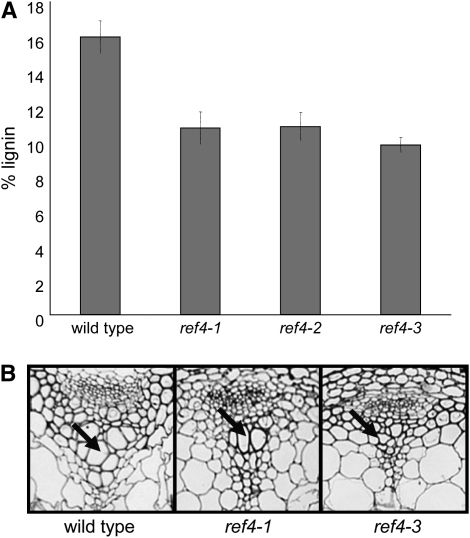
To further characterize the defect in phenylpropanoid metabolism in this mutant, the amount of lignin deposited in ref4 stems was quantified by Klason analysis. Isolated wild-type stem cell walls contained ∼16% lignin by weight (Figure 2A). In comparison, ref4 cell walls contained between 10 and 11% lignin. To examine the impact of the reduced lignin content of the mutant, stem thin sections were stained with toluidine blue O (Figure 2B). Wild-type xylem vessel elements were of normal diameter; whereas, some vessel elements exhibited collapse in ref4-1, and all of the elements were collapsed in ref4-3. Finally, the relative composition of lignin subunits in wild type and ref4-3 was determined by derivatization followed by reductive cleavage (DFRC) analysis. These analyses indicated that ref4-3 does not have an altered ratio of subunits (Table 1, as tested by ANOVA), in agreement with data from alkaline nitrobenzene oxidization (Ruegger and Chapple 2001).
Figure 2.—
Lignin-related phenotypes of ref4. (A) Lignin content as measured by Klason analysis. Bars represent the 95% confidence interval for the mean (n = 3). (B) Toluidine Blue O stained thin sections of stems showing collapsed xylem elements in ref4-3 (arrow).
TABLE 1.
Lignin monomer composition of wild-type and ref4 plants as determined by DFRC analysis
| H mol (%) | G mol (%) | S mol (%) | |
|---|---|---|---|
| Wild type | 1.4 ± 0.6 | 80.4 ± 0.9 | 18.2 ± 0.5 |
| ref4-1 | 1.4 ± 0.2 | 72.3 ± 10.2 | 26.3 ± 10.0 |
| ref4-3 | 2.5 ± 0.8 | 79.1 ± 7.3 | 18.4 ± 6.6 |
H, p-hydroxyphenyl; G, guaiacyl; S, syringyl.
n = 3. Variation represents the standard deviation of the mean.
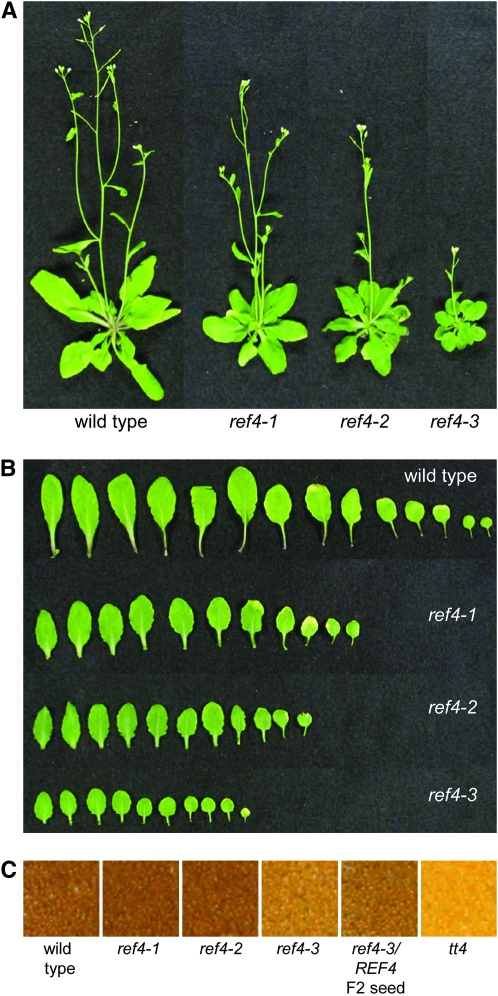
The yellow seeds of the ref4 mutant (Figure 3C) are much like those of the transparent testa mutants (Winkel-Shirley 2001), further indicating that ref4 is impaired in the biosynthesis of flavonoids. Finally, all ref4 alleles lead to slight to moderate reductions in plant stature. The growth of both ref4-1 and ref4-2 are nearest to wild type; whereas, ref4-3 is a dwarfed plant that produces dark-green spatulate leaves (Figure 3, A and B).
Figure 3.—
Visual phenotypes of ref4. (A) The ref4 series of mutants display varying degrees of dwarfism. (B) The leaves of ref4 are more spatulate than wild-type leaves and are darker green in color. (C) The seeds of ref4 exhibit a transparent testa phenotype.
Plants heterozygous for ref4 alleles exhibit a partial ref phenotype:
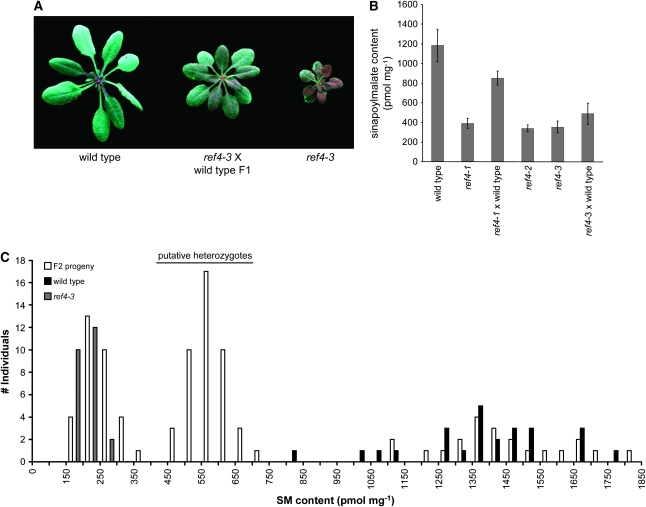
Although we originally reported that the ref4 mutation was recessive (Ruegger and Chapple 2001), we subsequently noticed that REF4/ref4 F1 plants display an intermediate growth phenotype and have a ref phenotype that is less severe than plants homozygous for the mutant allele, indicating that REF4/ref4 heterozygotes may accumulate less sinapoylmalate than the wild type (Figure 4A). This observation was confirmed by subsequent HPLC analysis (Figure 4B) and suggested that either the ref4 alleles we had identified are semidominant or that the wild-type allele is haplo-insufficient in the REF4/ref4 heterozygotes.
Figure 4.—
Analysis of semidominance of the mutant ref4 alleles. (A) Wild-type, ref4-3, and heterozygous plants photographed under UV light. Intermediate growth and ref phenotypes are observable in plants heterozygous for ref4-3. (B) Quantification of sinapoylmalate levels in whole rosettes. Bars represent the 95% confidence interval for the mean (n = 5). (C) Segregation of sinapoylmalate content in F2 individuals derived from a ref4-3 × wild-type cross.
To expand upon this analysis, 98 F2 progeny from a REF4/ref4-3 heterozygote, 25 ref4-3, and 25 wild-type plants were harvested at day 18 and the whole-rosette sinapoylmalate content was assessed by HPLC (Figure 4C). The sinapoylmalate content of the ref4-3 homozygotes ranged from 100 to 250 pmol mg−1 f.w. (mean, 161 ± 5 pmol mg−1 f.w.); whereas, in the wild-type rosettes it ranged from 750 to 1750 pmol mg−1 f.w. (mean, 1356 ± 42 pmol mg−1 fresh weight tissue). The F2 population did not segregate in a typical 3:1 ratio: 32% fell within the ref4 range, 22% within the wild-type range, and 46% between them. This segregation ratio fits a 1:2:1 ratio as determined by a chi-square test (χ2 = 2.3016, P = 0.3157), confirming that the wild-type or mutant alleles are either haplo-insufficient or semidominant, respectively. Further, the accumulation of condensed tannins in the testa of F2 seed also appears to be intermediate between the wild type and ref4-3 (Figure 3C). The overall growth phenotype of the heterozygotes varies between an intermediate phenotype and the wild type dependent upon growth conditions (Figure 4A). The height of the inflorescence stem in the heterozygous plants is 26.2 ± 1.2 cm (n = 25), which is smaller than the wild-type stem height (31.5 ± 1.1 cm, n = 25).
REF4 maps to the bottom of chromosome 2:
To understand how mutations in the REF4 gene affect phenylpropanoid biosynthesis, REF4 was isolated by positional cloning. Using 20 F2 plants from a ref4-3 (Columbia ecotype) × Landsberg erecta cross, a set of Arabidopsis cleaved amplified polymorphic sequence (CAPS) markers spanning the Arabidopsis genome was used to identify an initial map position for the gene near the bottom of chromosome 2. The position of REF4 was delineated further using additional CAPS and simple sequence-length polymorphism markers available on The Arabidopsis Information Resource (TAIR) web site (http://www.arabidopsis.org). Sequence information from the Landsberg erecta database (available at http://www.tigr.org) was used to generate additional cleaved amplified polymorphic sequence markers to screen a mapping population of 1520 plants, eventually narrowing the mapping interval to a 124-kb region spanning the final three BACs on chromosome 2. No bottom marker was found, and thus these results indicated that REF4 is located between marker Cer446007 (11 kb from the centromeric end of BAC T30B22) and the telomere.
All EMS-generated ref4 mutants harbor missense mutations in At2g48110:
In our efforts to isolate other phenylpropanoid genes from Arabidopsis, we have frequently used a transformation-competent cosmid library (Meyer et al. 1996a) to isolate overlapping clones for mutant complementation (Meyer et al. 1996b; Franke et al. 2002b; Nair et al. 2004). Unfortunately, the potentially semidominant nature of the EMS-generated ref4 mutations made this approach problematic, since in this case the phenotype of a ref4/ref4 mutant carrying a REF4 transgene could not be unambiguously predicted. As an alternative approach, we acquired all available T-DNA insertional lines (Alonso et al. 2003) for genes in the REF4 mapping interval (54 insertional lines of 76 total genes), none of which exhibited a ref phenotype, suggesting that either haplo-insufficiency is not the correct explanation for the phenotype of REF4/ref4 heterozygotes or that a REF4 insertional mutant was not represented among this population. In parallel, we tested the hypothesis that the semidominant phenotypes are the result of haplo-insufficiency, by screening for plants that exhibited an intermediate ref phenotype in a fast neutron-mutagenized M1 population. As with the T-DNA population, no mutant with a REF4/ref phenotype was identified in a population of 10,000 M1 plants, suggesting that plants heterozygous for a null REF4 allele were not represented in this population or that the cause of the semidominance in ref4-1, ref4-2, and ref4-3 is not due to haplo-insufficiency.
As a final approach to identify REF4, candidate genes within the REF4 mapping interval were sequenced from each of the three independent ref4 mutants. To prioritize genes for sequencing, several criteria were applied. First, genes of known function within the mapping interval were tentatively eliminated, as were genes that are not expressed (http://www.weigelworld.org) in leaves, stems, seeds, and roots, tissues in which ref4 mutant phenotypes are manifest. Among the remaining genes, genes for which T-DNA lines were not available and those annotated as encoding enzymes that might conceivably have a function in the shikimate or phenylpropanoid pathways were sequenced. Small genes were sequenced simply on the basis of the ease of doing so. Finally, since mutations that lead to disease states in humans and exhibit either haplo-insufficiency or dominance commonly encode regulatory factors, members of signaling cascades, or membrane transporters (Veitia 2002), genes of these classes within the REF4 mapping interval were sequenced.
In total, 28 of 76 genes within the mapping interval were sequenced before a G–A transition was detected in At2g48110 (a 142-kDa expressed protein of unknown function) in ref4-3, which results in a G383S substitution. This mutation was verified to be present in the genomic DNA of the mutant using CAPS marker analysis (supplemental Figure 3A). Similarly, in both ref4-1 and ref4-2 an identical G–A transition was detected that causes a D647N substitution. This mutation was verified using a dCAPS marker (supplemental Figure 3B; Neff et al. 1998). The fact that these two alleles are identical in sequence is consistent with the similar severity of the mutant phenotypes observed in ref4-1 and ref4-2 plants (Figures 2A, 3A, and 4B).
We considered the fact that all three alleles of At2g48110 contain mutations to be strong evidence that we had identified REF4. Further, considering that dominant mutations are rare events that can be engendered by only specific amino acid changes at few positions, the fact that ref4-1 and ref4-2 contain identical mutations, even though they were isolates from independent batches of M2 seed, strongly supported our identification of REF4.
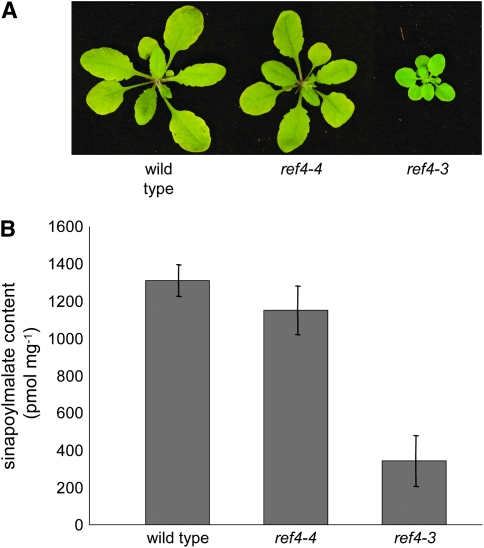
A homozygous insertion line of At2g48110 does not exhibit a ref phenotype:
To further evaluate the proposed semidominant nature of the EMS-generated ref4 alleles, a line that harbored a T-DNA insert in the first exon of the gene (SALK_102505), hereafter referred to as ref4-4, was characterized. Seeds from one kanamycin-resistant hemizygous plant were planted on soil, and the resulting seedlings were assayed for the presence of the T-DNA by PCR. One quarter of the plants were homozygous for the T-DNA insertion in At2g48110, and all of these plants were wild type in growth and accumulated wild-type levels of sinapoylmalate (Figure 5, P = 0.119, α = 0.05). Unfortunately, nonquantitative RT–PCR using intron-spanning primers specific for the 3′ end of the transcript detected a product from ref4-4 plants, leaving open the possibility that this line is not a null allele. For this reason, we obtained and brought to homozygosity two other At2g48110 insertional alleles: SALK_123227 and SALK_037472 (ref4-5 and ref4-6, respectively) in which T-DNAs had integrated into the fifth intron and ninth exon, respectively. Like ref4-4, these other alleles exhibited wild-type growth and leaf fluorescence when observed under UV light. We then analyzed the expression of REF4 in these insertion lines with quantitative RT–PCR to determine whether these plants represent true knockout mutants (supplemental Figure 4). Although the control reactions lacking reverse transcriptase did not generate any products (data not shown), amplification products were measurable in all three REF4 insertion lines analyzed, demonstrating that these lines cannot be considered to be transcriptional null alleles. Indeed, for unknown reasons, REF4 expression in ref4-4 and ref4-5 is 4- and 2-fold higher, respectively, than in the wild type. In contrast, REF4 expression in ref4-6 is >50-fold lower than the wild type, suggesting that this mutant is very likely to be hypomorphic for REF4 function. Further, it should be noted that the insertion in ref4-6 is downstream of the missense mutations identified in our original ref4 alleles. Assuming that these mutations identify amino acid residues that are important for REF4 function, it seems likely that the proteins that would be translated from these truncated mRNAs would be nonfunctional. Taken together, these data strongly suggest that loss-of-function alleles of REF4 do not lead to the phenotypes seen in the EMS-induced ref4 mutants and support the hypothesis that the intermediate phenotype observed in REF4/ref4-3 plants is caused by semidominance of the mutant allele, rather than by haplo-insufficiency.
Figure 5.—
Analysis of a homozygous T-DNA line of REF4 (ref4-4) as compared to the wild type. (A) The ref4-4 rosette is indistinguishable from wild type; whereas, the ref4-3 mutant is substantially smaller. (B) HPLC analysis of whole rosettes indicates that ref4-4 accumulates near wild-type levels of sinapoylmalate. Bars represent the confidence interval for the mean (n = 5).
Downregulation of REF4-3 transcript abundance by RNAi causes a reversion back to wild-type phenotypes:
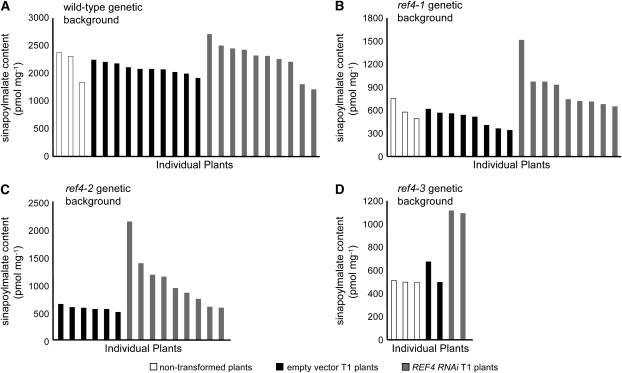
The observation that the ref4 insertional mutants are wild type in appearance suggests that the EMS-generated ref4 alleles we identified are semidominant and that REF4 activity is not essential for normal plant growth and development. This would predict that a reduction of mutant allele expression in a ref4-1, ref4-2, or ref4-3 mutant should cause a reversion to a wild-type phenotype. To test this hypothesis, the levels of At2g48110 transcript in all semidominant ref4 alleles were downregulated using RNAi (Fire et al. 1998).
When ref4 plants were transformed with the RNAi construct by the floral dip method (Clough and Bent 1998), very few kanamycin-resistant T1 seeds were recovered. We later found that ref4 seeds are hypersusceptible to the kanamycin used as a selective agent (data not shown), probably due to their transparent testa phenotype, which allows for a greater influx of exogenous chemicals through the seed coat (Debeaujon et al. 2000). In light of these results, the selection process was repeated at a larger scale, and the mutant T1 seeds were screened on MS plates containing a lower concentration of kanamycin (15 mg liter−1). Even when selecting for kanamycin-resistant plants at this lower concentration, the transformation efficiency of the ref4 plants was ∼10 times lower than for the wild type.
Eighteen-day-old wild-type plants transformed with the empty vector had the same sinapoylmalate content as nontransformed wild-type plants (Figure 6A). Those transformed with the RNAi construct mostly fell within this range, and there was no statistical difference between these plants and the empty vector controls by analysis of variance (P = 0.095). In contrast, all of the ref4 T1 plants transformed with the RNAi vector contained more sinapoylmalate than nontransformed mutant plants and T1 plants transformed with the empty vector (Figure 6, B–D, ref4-1, P = 0.002; ref4-2, P = 0.03; ref4-3, P = 0.028). Although some of the T1 plants carrying the RNAi construct exhibited only modest increases in sinapoylmalate content, others accumulated almost wild-type levels of sinapoylmalate and exhibited a marked increase in blue-green leaf fluorescence when observed under UV light (data not shown). Taken together, these data strongly suggest that At2g48110 encodes REF4, and that the mutations in ref4-1, ref4-2, and ref4-3 are semidominant over the wild-type allele.
Figure 6.—
RNAi-mediated downregulation of REF4 transcript accumulation leads to increased sinapoylmalate content in plants carrying semidominant ref4 alleles. (A) The RNAi transgene in a wild-type background causes no change in the sinapoylmalate content as compared to soil-grown plants and plants transformed with the empty vector. (B–D) The expression of a REF4 RNAi transgene in a ref4-1 (B), ref4-2 (C), and ref4-3 (D) background causes an increase in sinapoylmalate content.
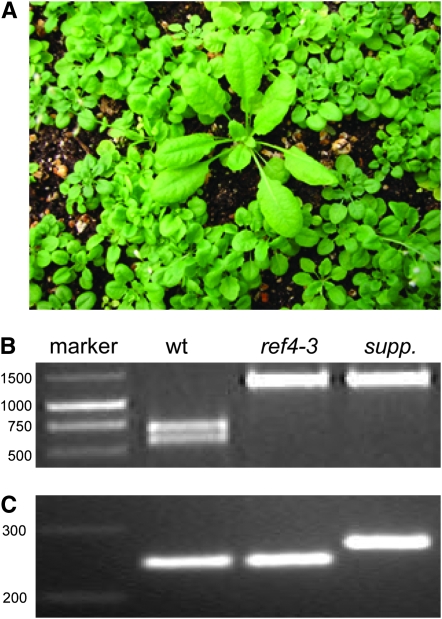
An intragenic suppressor mutation in the ref4-3 mutant causes a reversion back to wild-type phenotypes:
As a parallel approach to obtain proof of the identity of the REF4 gene, we began the isolation of suppressors by treating 1.5 grams of ref4-3 seed (∼75,000 seeds) with EMS. The resulting M1 seeds were grown to maturity for bulk harvest of M2 seeds. Among many plants identified as putative suppressors, one particularly promising plant was identified from the M2 population, which exhibited wild-type growth (Figure 7A), wild-type blue-green fluorescence when observed under UV light, and wild-type accumulation of anthocyanins during senescence. REF4 was sequenced from this plant, and both the original mutation causing the ref4 phenotype (Figure 7B) and a new intragenic C–T mutation was identified. This suppressor mutation, which results in the substitution of a proline to leucine (P919L), was verified using dCAPS marker analysis (Figure 7C). These data further support the hypothesis that At2g48110 is REF4.
Figure 7.—
Analysis of an EMS-derived intragenic suppressor mutant of ref4-3. (A) CAPS marker analysis demonstrating that the suppressor is homozygous for the original REF4 mutation that causes the mutant phenotypes in ref4-3. (B) dCAPS marker analysis verifying the intragenic mutation detected in the suppressor mutant. (C) Visual phenotype of the suppressor mutant within the M2 mutant population.
Wild-type plants transformed with ref4-3 exhibit defects in growth and phenylpropanoid metabolism:
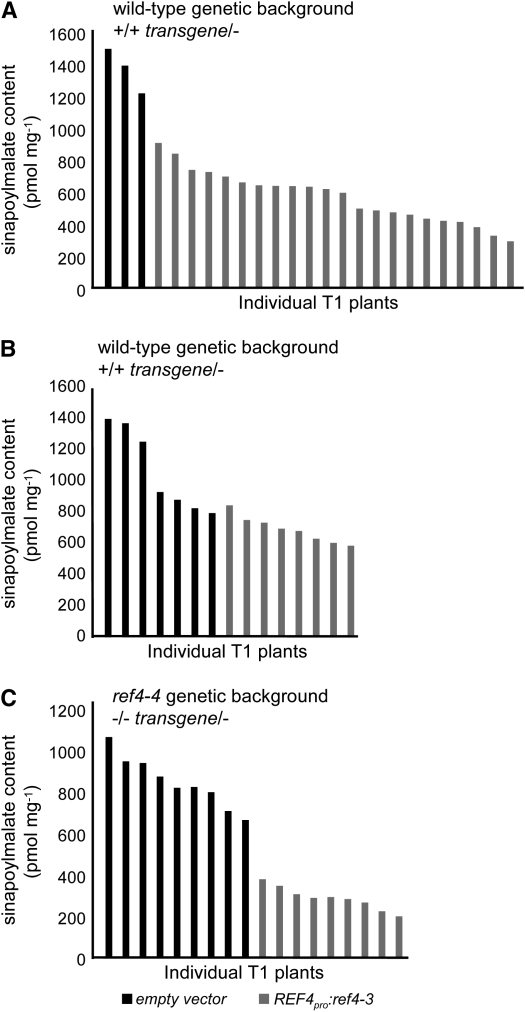
The observation that ref4-4 did not exhibit a ref phenotype, coupled with the identification of an intragenic suppressor mutation of ref4-3 that may inactivate the protein, suggest that the intermediate phenotype seen in REF4/ref4 heterozygotes is not due to haplo-insufficiency and that our original ref4 alleles are semidominant. To test this hypothesis, and to determine whether the single mutation found in the ref4-3 allele conferred both the reduced epidermal fluorescence and the reduced growth phenotypes, the mutant ref4-3 allele was expressed in wild-type plants.
When driven by its native promoter in wild-type plants, the ref4-3 allele caused a significant reduction in sinapoylmalate content compared to plants transformed with the empty vector (Figure 8A; P = 3.7 × 10−8). Interestingly, T1 wild-type plants transformed with the ref4-3 allele driven by the 35S promoter exhibited a more modest decrease in sinapoylmalate content (Figure 8B; P = 1.8 × 10−3), suggesting that the REF4 promoter may drive expression in cell types that are not effectively targeted by the 35S promoter. Finally, the T-DNA insertional mutant ref4-4 transformed with the ref4-3 allele driven by the REF4 promoter exhibited the highest reduction in sinapoylmalate content in T1 plants (Figure 8C; P-value = 4.1 × 10−9). The greater efficacy of the transgene in the mutant background is consistent with the hypothesis that ref4-4 is hypomorphic for REF4 function. These data provide unequivocal proof that At2g48110 is REF4 and that the ref4 alleles we identified initially are semidominant.
Figure 8.—
Effect of the introduction of a ref4-3 transgene on sinapoylmalate content in T1 transgenic plants. (A) Effect of the ref4-3 transgene driven by the native promoter in a wild-type genetic background. (B) Effect of the ref4-3 transgene driven by the 35S promoter in a wild-type genetic background. (C) Effect of the ref4-3 transgene driven by the native promoter in a putatively null ref4-4 background.
Further effects of the ref4-3 transgene on plant growth and phenylpropanoid metabolism in a wild-type genetic background were assessed in the T2 generation. Among a population of 70 T2 plants, only 15 exhibited wild-type fluorescence when observed under UV light, again demonstrating the dominance of the transgene (χ2 = 0.4762, P = 0.4902). To ensure that the ref phenotype of the remaining plants was due to the presence of the transgene, all individuals were genotyped by PCR. Only the DNA from plants exhibiting the ref phenotype generated a PCR product corresponding to the size of the cDNA-based transgene. Further, these products also tested positively for the presence of a PvuI restriction site that was cointroduced in the transgene along with the ref4-3 mutation (data not shown).
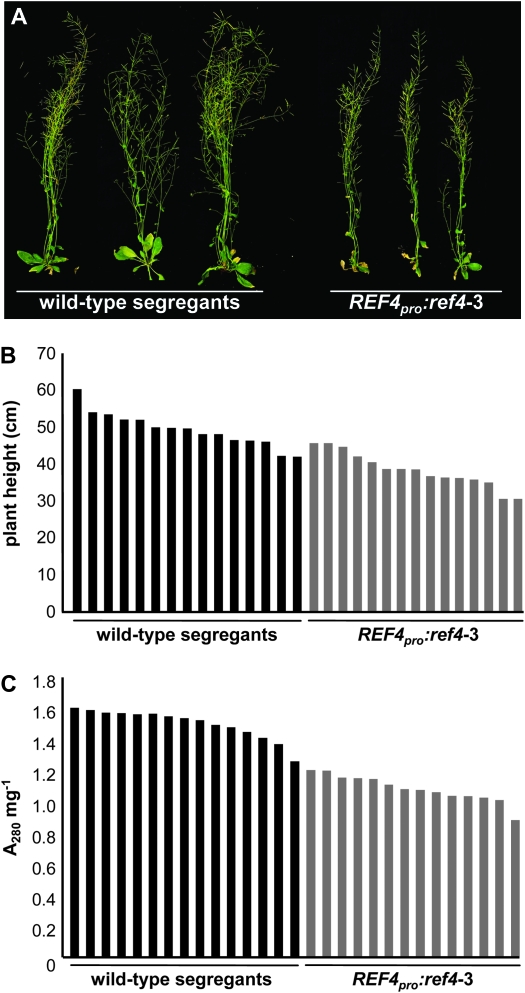
Plants carrying the REF4pro:ref4-3 transgene were slightly smaller than the wild-type segregants, demonstrating that the transgene had an effect on plant growth (Figure 9, A and B). The lignin content of these plants was assessed using the thioglycolic acid assay (Figure 9C). The 15 wild-type plants generated A280 mg−1 values between 1.3 and 1.6, which is typical for plants that deposit wild-type quantities of lignin (Ruegger and Chapple 2001). In contrast, all of the plants that carried the ref4-3 transgene generated values ranging from 0.9 to 1.2, demonstrating that the presence of the semidominant ref4-3 allele lowered the lignin content in the transgenic plants (analysis of variance, P < 0.001).
Figure 9.—
Effect of the introduction of a REF4pro:ref4-3 transgene on plant growth and lignin deposition in T2 transgenic plants carrying the transgene and corresponding wild-type segregants. (A) Visual phenotypes of T2 plants. (B) Effect of the transgene on plant growth of individual plants. (C) Thioglycolic acid analysis of lignin content in individual plants.
REF4 is expressed widely:
To determine in which organs REF4 is expressed, RT–PCR was conducted using RNA extracted from various tissue types. These data are consistent with publicly available microarray data (Schmid et al. 2005) and indicate that REF4 appears to be basally expressed in all plant organs and at developmental stages (data not shown).
in Silico analysis of REF4:
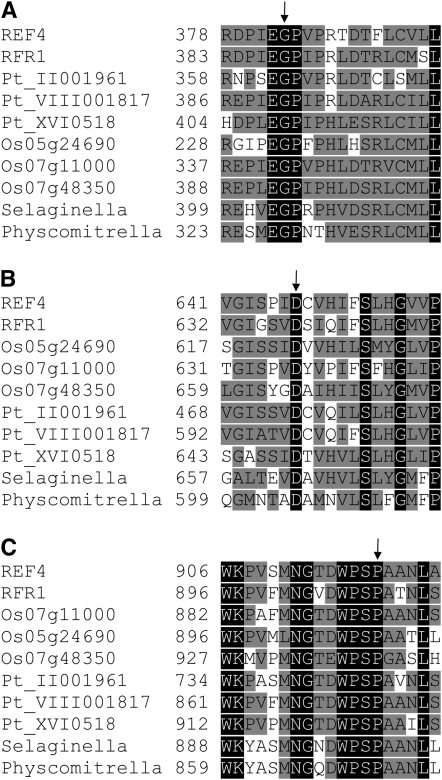
Using the REF4 sequence and the tBLASTn algorithm to query the nonredundant database returned a single homolog in Arabidopsis, At3g23590 [hereafter referred to as REF4 resembling 1 (RFR1)], which is 52% identical to REF4. Oryza sativa contains three homologs: Os07g11000, Os07g48350, and Os05g24690 that are between 32 and 52% identical to REF4. Similarly, the poplar (Populus trichocarpa) genome contains three homologs: Pt_II001961, Pt_VIII001817, and Pt_XVI0518. Full-length REF4 homologs were also identified in the genomes of the lycophyte Selaginella moellendorffii and the bryophyte Physcomitrella patens. Many homologous ESTs, predominantly derived from angiosperms, were also detected in the databases. No homologs could be identified in Chlamydomonas, fungi, animals, or prokaryotes. Alignments of REF4 and its homologs (Figure 10) reveal that the G383 residue substituted in ref4-3, the D647 residue substituted in ref4-1 and ref4-2, and the P919 residue substituted in the suppressor mutant are conserved among REF4 homologs in all plant lineages, suggesting that these residues may be important for the function of REF4 and its counterparts in other species.
Figure 10.—
Alignments of regions of REF4 and its homologs. (A) Region containing the G383 residue substituted in ref4-3. (B) Region containing the D647 residue substituted in ref4-1 and ref4-2. (C) Region containing the P919 residue substituted in the intragenic suppressor mutant. Numbers indicate amino acid positions within the proteins. Arrows indicate substituted residues. Black shading indicates complete conservation; gray shading indicates residues conserved in half or more of the sequences.
A phylogenetic tree was constructed using these sequences to assess potential relationships among these proteins (supplemental Figure 5), which was rooted to the Physcomitrella sequence on the basis of our current understanding of plant phylogeny (Qiu et al. 2005). The sequences for REF4, RFR1, two of the poplar homologs, and one of the rice homologs, grouped together in one clade; the remaining rice homologs and poplar homologs grouped as a sister clade, with the Selaginella and Physcomitrella homologs in a position that reflects the more distant relation of these species to seed-bearing plants.
On the TAIR web site (http://www.arabidopsis.org), At2g48110 is annotated as functioning as a ribosomal structural constituent, likely due to the presence of a ribosomal S10 protein superfamily domain as identified by InterProScan. However, this domain was not identified when the REF4 amino acid sequence was used as a query against this same database using the default parameters. Further, alignments of S10 proteins with the identified domain in REF4 did not show any substantial degree of similarity (data not shown). It thus likely appears that REF4 does not serve in this function, especially in light of the fact that S10 proteins are small proteins of ∼100 amino acid residues in length.
Online database searches were performed to identify other protein motifs that might reveal the function of REF4 (Falquet et al. 2002; Puntervoll et al. 2003; Quevillon et al. 2005). Although no informative large-scale domains could be recognized within REF4, many short peptide motifs were identified, two of which were completely conserved across REF4 and its homologs: a class IV WW protein-interaction domain (DWPSPA) (Sudol and Hunter 2000), which is a submotif within a proline-directed serine kinase phosphorylation site (DWPSPAA) (Lu et al. 2002). Interestingly the substitution of the conserved P919 in the suppressor mutant abolishes both of these motifs, consistent with the hypothesis that they may be important for REF4 function. REF4 and its homologs also contain a conserved 22-amino-acid sequence that contains an absolutely conserved tyrosine phosphorylation consensus motif RX3D/EX3Y.
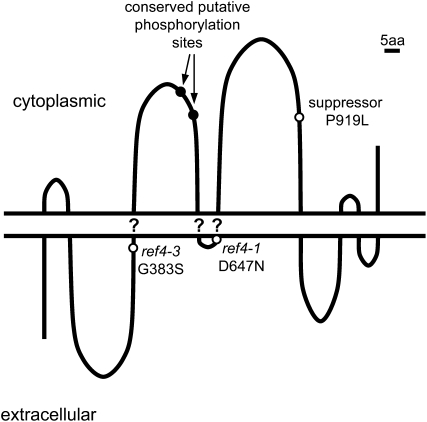
The PSORT subcellular localization algorithm identifies a number of putative membrane-spanning domains in REF4 and predicts that the protein and its homologs are localized to the plasma membrane. Unfortunately, neither REF4 nor RFR1 has been identified in any of the proteomic studies performed to date (e.g., Alexandersson et al. 2004; Marmagne et al. 2004; Dunkley et al. 2006; Morel et al. 2006; Hartman et al. 2007; Heazlewood et al. 2007; Lanquar et al. 2007; Mitra et al. 2007). The Aramemnon database for plant membrane spanning proteins (Schwacke et al. 2003) annotates REF4 and its homologs as having between 10 to 12 transmembrane regions (TMRs), although for any individual protein, less than half of these score above a cutoff score of 0.5. In contrast, alignments of the protein sequences show that the predicted TMRs are found in the same relative position between the proteins, and in many cases, TMRs with scores just below 0.5 in one protein often align with higher scoring TMRs in another protein. Although these data are not conclusive, it is tempting to speculate that REF4 contains 9 TMRs (Figure 11). Interestingly, the D602 residue mutated in ref4-1 and ref4-2 is immediately N-terminal to the fifth predicted TMR; the G338 residue mutated in ref4-3 is immediately N-terminal to the third predicted TMR. The P919 mutation in the intragenic suppressor is in a loop between putative TMRs 5 and 6.
Figure 11.—
Model of REF4 topology. Question marks indicate putative transmembrane regions that are less strongly supported by prediction algorithms and sequence alignments.
DISCUSSION
Forward genetic screens, most often conducted in Arabidopsis and maize, have led to the isolation of many genes involved in plant secondary metabolism (Stout and Chapple 2004; Halkier and Gershenzon 2006; Lepiniec et al. 2006). Although conventional biochemical approaches have also contributed greatly to our understanding of plant metabolism, an important attribute of genetic screens is that they do not depend upon prior knowledge of a system or pathway and thus can identify genes of unknown function that are relevant to the biological process of interest. This report is an example of the value of this type of unbiased approach, which resulted in the association of phenylpropanoid phenotypes with a gene that had previously been annotated as a gene of unknown function. The ref4 mutant was isolated from a genetic screen on the basis of reduced sinapoylmalate content, and a combination of map-based cloning and sequencing of genes from all three ref4 alleles identified G–A transitions in the gene At2g48110. Definitive proof of the identity of the REF4 gene was provided by (1) reduction in sinapoylmalate content following introduction of the ref4-3 allele into the wild type and a ref4 insertional mutant, (2) an increase in sinapoylmalate content when the At2g48110 transcript was reduced in ref4 using RNAi, and (3) the isolation of an At2g48110 intragenic suppressor mutant. Taken together, these data show that At2g48110 encodes REF4 and that the EMS-generated mutant alleles of REF4 are semidominant over the wild-type allele.
ref4 exhibits reduced phenylpropanoid accumulation:
Metabolic analyses of ref4 showed that the content of all major phenylpropanoids, including flavonoids, lignin, and sinapate esters, are reduced in the mutant. These phenotypes must be the result of either decreased synthesis or enhanced turnover of one or more intermediates within the shikimic acid or phenylpropanoid pathways.
Formally, a reduction in flux through any step of the shikimate pathway (Herrmann and Weaver 1999), or a diversion of pathway intermediates to other metabolic fates, could lead to the phenotypes observed in ref4. Although enhanced turnover of phenylpropanoid intermediates could also explain these phenotypes (see below), a model involving downregulation of phenylpropanoid metabolism would depend on the altered activity of one or more enzymes common to the synthesis of all phenylpropanoid end products, specifically phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-(hydroxy)cinnamoyl-coenzymeA ligase (4CL). These three enzymes collectively lead to the biosynthesis of p-coumaroyl CoA, which is the branch-point precursor for both flavonoid biosynthesis and monolignol/sinapate ester biosynthesis in Arabidopsis. The impact of reducing the activity of these enzymes has already been assessed in many studies. The Arabidopsis pal1/pal2 double-null mutant deposits less lignin than the wild type, is male sterile, yet exhibits wild-type growth characteristics (Rohde et al. 2004). The ref3 mutants harbor lesions in C4H (J. Stout, A. Schilmiller and C. Chapple, unpublished data), which cause a number of phenotypic consequences (Ruegger and Chapple 2001). Like ref4, these plants are dwarfed and have a seed tt phenotype. In contrast, these mutants do not display the spatulate leaf phenotype of ref4 (Ruegger and Chapple 2001) and only ref3 mutants accumulate cinnamoylmalate, suggesting that ref4 is not deficient in C4H activity. Antisense-mediated suppression of 4CL1 in Arabidopsis leads to decreases in lignin content but causes no alterations in plant growth and development (Lee et al. 1997), but it is now known that there are four 4CL genes present in the Arabidopsis genome (Raes et al. 2003), and the consequences of concurrent downregulation of all 4CL genes is unknown. Taken together, the observation that ref4 does not completely phenocopy plants that are deficient in PAL, C4H, or 4CL suggests that perturbation of these enzymes is not the cause of the ref4 phenotypes. On the other hand, it is important to note that two or more of these enzymes may be misregulated either transcriptionally or posttranscriptionally in ref4, leading to phenylpropanoid phenotypes that cannot be predicted from analyzing plants that are deficient in only one of these enzymes.
It is possible that multiple genes of the shikimic acid and/or phenylpropanoid pathways could be misregulated in ref4 at the transcriptional level, perhaps through changes in the expression of one or more transcription factors. Possible targets for such a misregulation include the MYB-class transcription factor AtMYB15, which has been shown to activate the shikimic acid pathway (Chen et al. 2006). Although this gene has been studied primarily in the context of wound-inducibility, it may also be required for the basal expression of shikimate pathway genes, and perturbation of this activity by the mutant ref4 alleles may thus restrict the supply of phenylalanine for phenylpropanoid biosynthesis. Alternatively, multiple elements of the phenylpropanoid pathway may be downregulated at the transcriptional level, for example, by reduced expression of the MYB-class transcriptional activator PAP1 (Borevitz et al. 2000; Tohge et al. 2005), or by increased expression of the transcriptional repressor AtMYB4 (Jin et al. 2000).
Another mechanism that may result in the reduction in phenylpropanoids in ref4 is alterations in posttranslational regulation of the enzymes of either pathway. In plants, both the provision of carbon via the Calvin cycle and the activity of two shikimate pathway enzymes (DAHP-synthase and shikimate kinase) are activated by reduced thioredoxin (Schmidt and Schultz 1987; Entus et al. 2002; Balmer et al. 2003). Thus the channeling of carbon into aromatic amino acid biosynthesis is tightly coupled to the redox potential of the cell. If mutations in ref4 alter the redox status of the cell, the synthesis of phenylalanine that can be utilized by the phenylpropanoid pathway could be decreased to rate-limiting levels.
The ref4 phenotypes could also be explained by misregulation of phenylpropanoid turnover. Little is known about the breakdown of these molecules; although it has been shown that in some species, anthocyanin catabolism is a regulated process that is enzymatically catalyzed (Vaknin et al. 2005). Analysis of some Arabidopsis mutants has suggested that active turnover and catabolism of phenylpropanoid intermediates may be common. For example, whereas the ref3 mutant accumulates cinnamoylmalate, and sng1 and sng2 mutants accumulate sinapoylglucose in leaves and seeds, respectively (Lorenzen et al. 1996; Shirley et al. 2001), the ferulate 5-hydroxylase (F5H)-deficient fah1 mutant does not accumulate substantial quantities of F5H substrates or their conjugates (Hemm et al. 2003). These observations suggest that either specific phenylpropanoids, such as guaiacyl-substituted compounds, can trigger feedback inhibition of earlier enzymes of the pathway or that these phenylpropanoid-pathway intermediates are substrates for catabolic pathways. Given that REF4 is a putative transmembrane protein, it may function to transport phenylpropanoid-pathway intermediates, including those accumulated in fah1, into the peroxisome for degradation. The proteins encoded by the semidominant mutant alleles may be constitutively active, which would account for the reduction of phenylpropanoids in ref4.
ref4 is dwarfed:
Of the ref mutants isolated in our initial genetic screen, ref3-2, ref4-3, and ref8 all exhibit severe dwarfism, and RNAi-mediated reduced hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (HCT) activity in tobacco also leads to dwarfing (Hoffmann et al. 2004). Further analysis in Arabidopsis plants with reduced HCT activity showed that this dwarfing may be attributed to hyperaccumulation of flavonoids, which are negative regulators of auxin transport (Buer and Muday 2004; Peer et al. 2004; Besseau et al. 2007). The ref8 mutant is purple in color, suggesting that it too hyperaccumulates anthocyanins, and considering that the C3′H functions immediately downstream of HCT, a similar mechanism may account for the ref8 dwarf phenotype. In contrast, the C4H-deficient ref3-2 mutant is also severely dwarfed, even though its lesion in phenylpropanoid metabolism occurs before the biosynthesis of p-coumaryl CoA and thus the mutant presumably does not hyperaccumulate flavonoids. Similarly, ref4-3 is dwarfed, even though it too contains fewer flavonoids (Figure 1; supplemental Figure 2). Together, these data suggest that an auxin-independent mechanism of dwarfing may lead to the developmental phenotypes seen in ref4 mutants. Alternatively, the growth phenotypes seen in ref4 may be independent of perturbations in both auxin and phenylpropanoid metabolism and may arise through an as yet unidentified mechanism. Indeed, many suppressor mutants were isolated in our ref4-3 suppressor screen that exhibited wild-type growth, yet still appeared ref when examined under UV light, demonstrating that the phenylpropanoid and dwarf phenotypes of the ref4-3 mutant can be genetically disentangled.
REF4 and its homologs may have overlapping function:
With the evidence currently in hand, we cannot exclude the possibility that the phenylpropanoid phenotypes observed in the ref4 mutants are the result of neomorphic mutations (Muller 1934; Wilkie 1994). On the other hand, it may be important to note that REF4 homologs appear to be restricted to land plants, consistent with a role for REF4-like proteins in a plant-specific pathway such as phenylpropanoid metabolism. Although a function cannot be inferred on the basis of their sequences, REF4 and its homologs may have a conserved function in plants since ∼10% of amino acid residues (144 of 1322 in REF4) are identical among plant lineages representing >400 million years of divergent evolution. Among these are the amino acids substituted in ref4 mutants and the ref4 suppressor, providing genetic evidence of the importance of these conserved residues in REF4 function.
Phylogenetic analysis of REF4 and its homologs revealed that REF4, RFR1, two of the poplar homologs, and one of the rice homologs, group into a single clade. This suggests that the Arabidopsis and poplar proteins may be orthologous and functionally redundant, which may explain why no mutant phenotypes were observed in the ref4-4 T-DNA line. One of the poplar homologs and two of the rice homologs group into a sister clade, suggesting a gene duplication event that occurred prior to the monocot/dicot divergence. Interestingly, Arabidopsis does not contain a homolog that falls within this clade, suggesting that the orthologous gene in the Arabidopsis lineage has been lost.
Dominant mutations in REF4 may suggest putative gene function:
The finding that two missense alleles of REF4 exhibit semidominance provides clues to the possible function of the wild-type protein. As stated by the metabolic control analysis, mutant alleles of metabolic enzymes rarely exhibit dominance. This, together with the fact that REF4 does not contain any known enzyme-like domains, and that the phenylpropanoid pathway leading to the production of sinapoylmalate is now well characterized (Humphreys and Chapple 2002), indicates that it is unlikely that REF4 encodes an enzyme. Instead, genes whose mutant alleles exhibit dominance are more likely to encode transcription factors, transporters, or components of signaling cascades (Kondrashov and Koonin 2004). Assuming that REF4 is a membrane-localized protein, it is unlikely that it functions as a transcription factor. In contrast, as a putative transmembrane protein, REF4 may encode a transporter of shikimic acid or phenylpropanoid-pathway intermediates or end products, potentially targeting them to the peroxisome for catabolism, as discussed previously. Alternatively, REF4 may function as a component in a signaling cascade. It is well established that light, interactions with pathogens, and oxylipin signaling induce phenylpropanoid metabolism (Hemm et al. 2004; Chen et al. 2006; Fujiwara et al. 2006). The wild-type function of REF4 could be to attenuate this signaling in response to external cues, with the dominant mutations preventing modulation of this effect, leading to constitutive downregulation of phenylpropanoid metabolism. Interestingly, dominant mutations in a G-protein-coupled receptor are primarily the result of amino acid substitutions in, or immediately adjacent to, TMRs (Dosil et al. 1998), as is the case in mutant REF4 alleles. If G-protein-coupled receptors can serve as a model for REF4, missense mutations in REF4 could lock the encoded protein into a constitutively active state. Furthermore, this signaling could be mediated through protein–protein interactions via the REF4 WW-interaction motif. Consistent with this model, the suppressor mutation that abolishes this motif would eliminate this interaction, effectively blocking the signal and the repressive effect of semidominant ref4 alleles.
In conclusion, although the function of REF4 is still unknown, this research clearly shows that dominant mutations in REF4 lead to a decreased accumulation of phenylpropanoid end products. The identification of REF4 as an effector of phenylpropanoid metabolism may lead to new insights into the regulation of secondary metabolism in plants. Furthermore, if these alleles, or analogous mutations in REF4 homologs, operate similarly in other species, semidominant REF4 alleles will add another important tool to the “lignin modification toolbox.” Lignin significantly impedes the utilization of cellulosic plant material for the production of biofuels (Coughlan 1992; Chen and Dixon 2007). Even modest decreases in the lignin content of biofuel feedstock brought about by semidominant REF4 alleles could increase the efficiency of cellulosic biofuel production. Further, the reduced-lignin phenotype brought about by such REF4 transgenes may ultimately be more stable than utilizing RNAi-based strategies over successive generations or years in annual and perennial crops, respectively.
Acknowledgments
We thank Mike Zanis for helpful conversations about REF4 phylogenetic analysis and Joe Ogas both for providing the cDNA library from which we amplified REF4 and for suggesting the ref4 RNAi experiment. This work was supported by a grant from the Division of Energy Biosciences, U. S. Department of Energy. This is journal paper no. 2008-18292 of the Purdue University Agricultural Experiment Station.
References
- Alexandersson, E., G. Saalbach, C. Larsson and P. Kjellbom, 2004. Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol. 45 1543–1556. [DOI] [PubMed] [Google Scholar]
- Alonso, J. M., A. N. Stepanova, T. J. Leisse, C. J. Kim, H. Chen et al., 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Balmer, Y., A. Koller, G. Del Val, W. Manieri, P. Schurmann et al., 2003. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc. Natl. Acad. Sci. USA 100 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, J., and G. R. Fink, 1998. A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 95 5655–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau, S., L. Hoffmann, P. Geoffroy, C. Lapierre, B. Pollet et al., 2007. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker, A. B., M. A. Estelle, C. Somerville and H. Kende, 1988. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241 1086–1089. [DOI] [PubMed] [Google Scholar]
- Borevitz, J. O., Y. J. Xia, J. Blount, R. A. Dixon and C. Lamb, 2000. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer, C. S., and G. K. Muday, 2004. The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16 1191–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, M. M., and B. E. Ellis, 1992. Fungal elicitor-mediated responses in pine cell-cultures. 1. induction of phenylpropanoid metabolism. Planta 186 409–417. [DOI] [PubMed] [Google Scholar]
- Chae, H. S., F. Faure and J. J. Kieber, 2003. The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C., S. F. Kwok, A. B. Bleecker and E. M. Meyerowitz, 1993. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262 539–544. [DOI] [PubMed] [Google Scholar]
- Chapple, C. C. S., T. Vogt, B. E. Ellis and C. R. Somerville, 1992. An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple, C., B. W. Shirley, M. Zook, R. Hammerschmidt and C. S. Somerville, 1994. Secondary metabolism in Arabidopsis, pp. 989–1030 in Arabidopsis, edited by E. M. Meyerowitz and C. R. Somerville. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Chen, F., and R. A. Dixon, 2007. Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotech. 25 759–761. [DOI] [PubMed] [Google Scholar]
- Chen, H., A. D. Jones and G. A. Howe, 2006. Constitutive activation of the jasmonate signaling pathway enhances the production of secondary metabolites in tomato. FEBS Lett. 580 2540–2546. [DOI] [PubMed] [Google Scholar]
- Clough, S. J., and A. F. Bent, 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Coen, E. S., R. Carpenter and C. Martin, 1986. Transposable elements generate novel spatial patterns of gene expression in Antirrhinum majus. Cell 47 285–296. [DOI] [PubMed] [Google Scholar]
- Coughlan, M. P., 1992. Enzymatic-hydrolysis of cellulose: an overview. Bioresour. Technol. 39 107–115. [Google Scholar]
- Czechowski, T., M. Stitt, T. Altmann, M. K. Udvardi and W. R. Scheible, 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon, I., K. M. Leon-Kloosterziel and M. Koornneef, 2000. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 122 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosil, M., L. Giot, C. Davis and J. B. Konopka, 1998. Dominant-negative mutations in the G-protein-coupled α-factor receptor map to the extracellular ends of the transmembrane segments. Mol. Cell. Biol. 18 5981–5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley, T. P. J., S. Hester, I. P. Shadforth, J. Runions, T. Weimar et al., 2006. Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. USA 103 6518–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, J. F., S. K. Floyd, J. Alvarez, Y. Eshed, N. P. Hawker et al., 2003. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13 1768–1774. [DOI] [PubMed] [Google Scholar]
- Entus, R., M. Poling and K. M. Herrmann, 2002. Redox regulation of Arabidopsis 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase. Plant Physiol. 129 1866–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. A. Sigrist et al., 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver et al., 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 806–811. [DOI] [PubMed] [Google Scholar]
- Franke, R., M. R. Hemm, J. W. Denault, M. O. Ruegger, J. M. Humphreys et al., 2002. a Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J. 30 47–59. [DOI] [PubMed] [Google Scholar]
- Franke, R., J. M. Humphreys, M. R. Hemm, J. W. Denault, M. O. Ruegger et al., 2002. b The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 30 33–45. [DOI] [PubMed] [Google Scholar]
- Fujiwara, M., K. Umemura, T. Kawasaki and K. Shimamoto, 2006. Proteomics of Rac GTPase signaling reveals its predominant role in elicitor-induced defense response of cultured rice cells. Plant Physiol. 140 734–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier, B. A., and J. Gershenzon, 2006. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57 303–333. [DOI] [PubMed] [Google Scholar]
- Hartman, N. T., F. Sicilia, K. S. Lilley and P. Dupree, 2007. Proteomic complex detection using sedimentation. Anal. Chem. 79 2078–2083. [DOI] [PubMed] [Google Scholar]
- Heazlewood, J. L., R. E. Verboom, J. Tonti-Filippini, I. Small and A. H. Millar, 2007. SUBA: the Arabidopsis subcellular database. Nucleic Acids Res. 35 D213–D218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemm, M. R., S. D Rider, J. Ogas, D. J. Murry and C. Chapple, 2004. Light induces phenylpropanoid metabolism in Arabidopsis roots. Plant J. 38 765–778. [DOI] [PubMed] [Google Scholar]
- Hemm, M. R., M. O. Ruegger and C. Chapple, 2003. The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, K. M., and L. M. Weaver, 1999. The Shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 473–503. [DOI] [PubMed] [Google Scholar]
- Hoffmann, L., S. Besseau, P. Geoffroy, C. Ritzenthaler, D. Meyer et al., 2004. Silencing of hydroxycinnamoy-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16 1446–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, J., and E. M. Meyerowitz, 1998. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94 261–271. [DOI] [PubMed] [Google Scholar]
- Humphreys, J. M., and C. Chapple, 2002. Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 5 224–229. [DOI] [PubMed] [Google Scholar]
- Jefferson, R., T. A. Kavanagh and M. Bevan, 1987. GUS-fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Sanchez, G., B. Childs and D. Valle, 2001. Human disease genes. Nature 409 853–855. [DOI] [PubMed] [Google Scholar]
- Jin, H. L., E. Cominelli, P. Bailey, A. Parr, F. Mehrtens et al., 2000. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaar, W. E., and D. L. Brink, 1991. Simplified analysis of acid-soluble lignin. J. Wood Chem. Technol. 11 465–477. [Google Scholar]
- Kacser, H., and J. A. Burns, 1981. The molecular basis of dominance. Genetics 97 639–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, F., and E. V. Koonin, 2004. A common framework for the understanding of the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 20 287–291. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5 150–163. [DOI] [PubMed] [Google Scholar]
- Lanquar, V., L. Kuhn, F. Lelièvre, M. Khafif, C. Espagne et al., 2007. 15N-metabolic labeling for comparative plasma membrane proteomics in Arabidopsis cells. Proteomics 7 750–754. [DOI] [PubMed] [Google Scholar]
- Lee, D., K. Meyer, C. Chapple and C. J. Douglas, 1997. Antisense suppression of 4-coumarate:coenzyme A ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell 9 1985–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiniec, L., I. Debeaujon, J. M. Routaboul, A. Baudry, L. Pourcel et al., 2006. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57 405–430. [DOI] [PubMed] [Google Scholar]
- Lorenzen, M., V. Racicot, D. Strack and C. Chapple, 1996. Sinapic acid ester metabolism in wild type and a sinapoylglucose-accumulating mutant of Arabidopsis. Plant Physiol. 112 1625–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, F. C., and J. Ralph, 1997. Derivatization followed by reductive cleavage (DFRC method), a new method for lignin analysis: protocol for analysis of DFRC monomers. J. Agric. Food Chem. 45 2590–2592. [Google Scholar]
- Lu, P. J., X. Z. Zhou, Y. C. Liou, J. P. Noel and K. P. Lu, 2002. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J. Biol. Chem. 277 2381–2384. [DOI] [PubMed] [Google Scholar]
- Marmagne, A., M.-A. Rouet, M. Ferro, N. Rolland, C. Alcon et al., 2004. Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol. Cell Proteomics 3 675–691. [DOI] [PubMed] [Google Scholar]
- Martin, C., A. Prescott, S. Mackay, J. Bartlett and E. Vrijlandt, 1991. Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J. 1 37–49. [DOI] [PubMed] [Google Scholar]
- McCourt, P., 1999. Genetic analysis of hormone signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 219–243. [DOI] [PubMed] [Google Scholar]
- Meyer, K., G. Benning and E. Grill, 1996. a Cloning of plant genes based on genetic map location, pp. 137–154 in Genome Mapping in Plants, edited by A. H. paterson. Academic Press, New York/Landes Bioscience, Austin, TX.
- Meyer, K., J. C. Cusumano, C. Somerville and C. C. S. Chapple, 1996. b Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monooxygenases. Proc. Natl. Acad. Sci. USA 93 6869–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, S. K., J. A. Gantt, J. F. Ruby, S. D. Clouse and M. B. Goshe, 2007. Membrane proteomic analysis of Arabidopsis thaliana using alternative solubilization techniques. J. Proteome Res. 6 1933–1950. [DOI] [PubMed] [Google Scholar]
- Morel, J., S. Claverol, S. Mongrand, F. Furt, J. Fromentin et al., 2006. Proteomics of plant detergent-resistant membranes. Mol. Cell Proteomics 5 1396–1411. [DOI] [PubMed] [Google Scholar]
- Muller, H. J., 1934. Further studies on the nature and causes of gene mutations, pp. 213–255 in Proceedings of the Sixth International Congress of Genetics, edited by D. Jones. Brooklyn Botanical Gardens, Brooklyn, New York. Available at: http://www.genetics.org/cgi/content/full/148/4/1419/T1.
- Nair, R. B., K. L. Bastress, M. O. Ruegger, J. W. Denault and C. Chapple, 2004. The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16 544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M. M., J. D. Neff, J. Chory and A. E. Pepper, 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14 387–392. [DOI] [PubMed] [Google Scholar]
- Peer, W. A., A. Bandyopadhyay, J. J. Blakeslee, S. I. Makam, R. J. Chen et al., 2004. Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntervoll, P., R. Linding, C. Gemund, S. Chabanis-Davidson, M. Mattingsdal et al., 2003. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 31 3625–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Y. L., O. Dombrovska, J. Lee, L. B. Li, B. A. Whitlock et al., 2005. Phylogenetic analyses of basal angiosperms based on nine plastid, mitochondrial, and nuclear genes. Int. J. Plant Sci. 166 815–842. [Google Scholar]
- Quevillon, E., V. Silventoinen, S. Pillai, N. Harte, N. Mulder et al., 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33 W116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes, J., A. Rohde, J. H. Christensen, Y. Van De Peer and W. Boerjan, 2003. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133 1051–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, A., K. Morreel, J. Ralph, G. Goeminne, V. Hostyn et al., 2004. Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16 2749–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger, M., and C. Chapple, 2001. Mutations that reduce sinapoylmalate accumulation in Arabidopsis thaliana define loci with diverse roles in phenylpropanoid metabolism. Genetics 159 1741–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, C. L., and G. Schultz, 1987. Stimulation by thioredoxin of shikimate kinase from spinach chloroplasts. Physiol. Plant 70 65–67. [Google Scholar]
- Schmid, M., T. S. Davison, S. R. Henz, U. J. Pape, M. Demar et al., 2005. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Schwacke, R., A. Schneider, E. Van Der Graaff, K. Fischer, E. Catoni et al., 2003. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley, A. M., C. M. McMichael and C. Chapple, 2001. The sng2 mutant of Arabidopsis is defective in the gene encoding the serine carboxypeptidase-like protein sinapoylglucose: choline sinapoyltransferase. Plant J. 28 83–94. [DOI] [PubMed] [Google Scholar]
- Smolen, G., and J. Bender, 2002. Arabidopsis cytochrome P450 cyp83B1 mutations activate the tryptophan biosynthetic pathway. Genetics 160 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout, J., and C. Chapple, 2004. The phenylpropanoid pathway in Arabidopsis: lessons learned from mutants in sinapate ester biosynthesis, pp. 39–68 in Recent Advances in Phytochemistry, edited by J. T. Romeo. Academic Press, New York.
- Sudol, M., and T. Hunter, 2000. NeW Wrinkles for an old domain. Cell 103 1001–1004. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V., P. Springer, T. Volpe, S. Haward, J. D. Jones et al., 1995. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9 1797–1810. [DOI] [PubMed] [Google Scholar]
- Tang, G., B. J. Reinhart, D. P. Bartel and P. D. Zamore, 2003. A biochemical framework for RNA silencing in plants. Genes Dev. 17 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge, T., Y. Nishiyama, M. Y. Hirai, M. Yano, J. I. Nakajima et al., 2005. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42 218–235. [DOI] [PubMed] [Google Scholar]
- Vaknin, H., A. Bar-akiva, R. Ovadia, A. Nissim-Levi, I. Forer et al., 2005. Active anthocyanin degradation in Brunfelsia calycina (yesterday-today-tomorrow) flowers. Planta 222 19–26. [DOI] [PubMed] [Google Scholar]
- Veitia, R., 2002. Exploring the etiology of haploinsufficiency. BioEssays 24 175–184. [DOI] [PubMed] [Google Scholar]
- Vogel, J. P., K. E. Woeste, A. Theologis and J. J. Kieber, 1998. Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. USA 95 4766–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers, D., M. Franke-Van Dijk, R.-J. Vencken, A. Quint, P. Hooykaas et al., 2001. An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128 4289–4299. [DOI] [PubMed] [Google Scholar]
- Wesley, S. V., C. A. Helliwell, N. A. Smith, M. Wang, D. T. Rouse et al., 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Wilkie, A. O., 1994. The molecular basis of genetic dominance. J. Med. Genet. 31 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley, B., 2001. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]