Abstract
Although intensively studied, the biological purpose of sleep is not known. To identify candidate genes affecting sleep, we assayed 136 isogenic P-element insertion lines of Drosophila melanogaster. Since sleep has been negatively correlated with energy reserves across taxa, we measured energy stores (whole-body protein, glycogen, and triglycerides) in these lines as well. Twenty-one insertions with known effects on physiology, development, and behavior affect 24-hr sleep time. Thirty-two candidate insertions significantly impact energy stores. Mutational genetic correlations among sleep parameters revealed that the genetic basis of the transition between sleep and waking states in males and females may be different. Furthermore, sleep bout number can be decoupled from waking activity in males, but not in females. Significant genetic correlations are present between sleep phenotypes and glycogen stores in males, while sleep phenotypes are correlated with triglycerides in females. Differences observed in male and female sleep behavior in flies may therefore be related to sex-specific differences in metabolic needs. Sleep thus emerges as a complex trait that exhibits extensive pleiotropy and sex specificity. The large mutational target that we observed implicates genes functioning in a variety of biological processes, suggesting that sleep may serve a number of different functions rather than a single purpose.
UNLIKE many behaviors, the biological purpose of sleep remains elusive. Yet sleep is thought to serve an important physiological function for two reasons: it has been observed in a wide variety of taxa (Amlaner and Ball 1994; Hartse 1994; Zepelin 1994), and animals deprived of sleep suffer severe physical consequences, including death (Rechtschaffen et al. 1989; Rechtschaffen and Bergmann 2002). Diverse hypotheses have been postulated about the function of sleep. A number of proposals stem from the negative correlation observed between sleep and body weight across mammalian species (Zepelin 1994). Since animals with lower energy reserves tend to sleep more, sleep might serve the purpose of reducing total caloric expenditure, thereby conserving energy (Zepelin 1994; Berger and Phillips 1995). Studies of sleep deprivation in humans tend to support the idea that sleep plays a role in metabolism: sleep loss results in impaired glucose tolerance (Spiegel et al. 1999) and changes in appetite-stimulating/suppressing hormones such as leptin and ghrelin (Spiegel et al. 2004a,b). Furthermore, a widespread association between short sleep times and obesity has been observed in humans (reviewed in Cizza et al. 2005). However, few calories are saved by sleeping vs. quiet waking (Zepelin 1994), implying that the loss of consciousness is central to the function of sleep. A number of hypotheses therefore argue that sleep is primarily for the benefit of the brain. Sleep may restore brain glycogen, an important source of metabolic energy (Benington and Heller 1995). In addition, sleep may be required for synaptic plasticity (Tononi and Cirelli 2003). Waking activities that result in cellular changes in the brain increase the strength of wake-active synapses; slow-wave activity during sleep may serve to downscale synaptic strength, thus improving performance (Tononi and Cirelli 2003). Or sleep may stimulate neurons that were not sufficiently activated in the course of waking, as a means of preserving information (Krueger et al. 1995). Sleep has also been associated with the process of memory consolidation in mammals (Smith 1995); while sleep does not appear to affect all types of memory, nondeclarative memories such as motor learning tasks are enhanced by a night of sleep (Stickgold 2005). Each theory cited above has been supported to some degree by experimental evidence. Sleep thus emerges as a complex behavior that may serve more than one purpose.
Quiescence in the fruit fly Drosophila melanogaster possesses all of the behavioral characteristics of mammalian sleep (Hendricks et al. 2000; Shaw et al. 2000). Flies exhibit a diurnal sleep–wake cycle regulated by the circadian clock. Videotape analysis shows that flies sometimes change posture during sleep (standing with the head drooping down) and prefer to sleep near a food source (Hendricks et al. 2000). Like mammals, flies that are asleep are less responsive to outside stimuli than normal (Hendricks et al. 2000; Shaw et al. 2000). These observations enable the use of Drosophila, a classic genetic model organism, to rapidly identify candidate genes involved in this elusive behavior.
Thus far, few candidate genes that alter sleep phenotypes in flies have been identified. The neurotransmitters serotonin and dopamine have been implicated in sleep. Serotonin appears to promote sleep through the d5-HT1A receptor, while increased extracellular dopamine lowers the arousal threshold and may promote waking (Kume et al. 2005; Yuan et al. 2006). Flies bearing mutations of the molecular circadian clock genes cycle and Clock sleep less in both standard day/night cycles and in constant darkness (Hendricks et al. 2003). Alterations in cAMP signaling in a specific region of the fly brain affect sleep duration (Hendricks et al. 2001; Joiner et al. 2006). Heterozygous null mutations in the immune response gene Relish reduce day and night sleep in females and night sleep in males (Williams et al. 2007). In addition, mutations in Shaker, the α-subunit of a voltage-dependent potassium channel, and Hyperkinetic, the β-subunit of the same channel, reduce sleep (Cirelli et al. 2005a; Bushey et al. 2007). Intriguingly, background modifiers mitigated the effect of Shaker mutant alleles on sleep in older stocks; outcrosses to two wild-type backgrounds restored the short-sleeping phenotype (Cirelli et al. 2005a).
To minimize genetic background effects when comparing across lines, we examined sleep in a collection of 136 P-element insertion lines created in an isogenic background (Bellen et al. 2004). Theoretically, each line differs only by the P-element insertion and can be compared to the isogenic parent line as a control. A line carrying a single P-element insertion in the first exon of Calreticulin showed a reduction in sleep by as much as 289.2 min (4.82 hr)/24 hr. In contrast, a line carrying an insertion in the third exon of malic enzyme displayed an increase in sleep of as much as 424.8 min (7.08 hr). Precise excisions of the P-element tagging Calreticulin increased the short-sleeping phenotype back to wild type, while precise excisions of the insertion in malic enzyme reverted the long-sleeping phenotype. Furthermore, measurements of whole-body energy stores (protein, triglycerides, and glycogen) in all 136 lines enabled us to quantify the relationship between sleep and energy stores. Significant mutational genetic correlations between sleep and energy storage parameters are present and sex specific. The wide variety of biological processes attributed to sleep candidate genes and the extensive pleiotropy observed in the lines tested suggest that sleep has more than one function.
MATERIALS AND METHODS
Drosophila stocks:
A random subset of 136 P-element lines previously tested for PNS developmental phenotypes, starvation resistance, and life span, created as part of the Berkeley Drosophila Genome Disruption Project (Bellen et al. 2004), were used to assay sleep and energy storage phenotypes. Each line has a single transposable P[GT1] insertion in an otherwise isogenic w1118; Canton-S background. For the phenotypic assays, lines were divided into blocks of 20. The isogenic w1118; Canton-S parent served as a control line and was assayed in each block.
Flies were reared and maintained on standard medium in a 25°, 12-hr light/dark cycle incubator. For all assays, adult virgins were collected and maintained at 30 flies to a single-sex vial until the time of assay. This protocol ensured that each insertion line was exposed to identical levels of social enrichment, which can alter sleep (Ganguly-Fitzgerald et al. 2006). Specifically, flies held in vials containing <30 flies have lower daytime sleep than when they are held in vials containing 30 flies or more (Ganguly-Fitzgerald et al. 2006). Since we were particularly interested in mutations that reduce sleep, we purposely maintained our flies in groups of 30 to a vial to bias our study toward finding increased daytime sleep. Furthermore, maintaining virgins at constant density provided equal access to food. Sixteen flies of each sex were assayed per line, which gives a statistical power of 80% to detect a 1.75-hr difference between the insertion line and the control based on pilot studies. Flies were 4–7 days old at the time of the sleep assay. We measured energy stores in a separate group of virgin flies that were age matched to those in the sleep assay.
Baseline sleep and activity assays:
Activity and sleep behavior were monitored using the Drosophila Activity Monitoring System (Trikinetics, Waltham, MA) (Ho and Sehgal 2005). With this system, each fly is loaded into an activity monitor tube. An activity count is recorded by a computer each time a fly crosses an infrared beam that bisects the monitor tube. Activity counts were recorded at 1-min intervals. Seven continuous days of sleep and activity were recorded for each P-element insertion line. After 7 days, flies were visually examined; any flies that died during the course of the experiment were removed from the analysis. Sleep was defined as any period ≥5 min without an activity count, as previously determined (Hendricks et al. 2000; Shaw et al. 2000; Huber et al. 2004; Ho and Sehgal 2005). An in-house C++ program was used to calculate duration of sleep in minutes per day, numbers of sleep bouts per day, average sleep bout in minutes, and the number of activity counts per waking minute (or waking activity). As males sleep more during the day than females (Huber et al. 2004), sleep times and bout numbers were divided into daytime/nighttime sleep and daytime/nighttime bout number.
Measurement of energy stores:
Flies were weighed and homogenized on ice in 0.01 m KH2PO4, 1 mm EDTA, pH 7.4, buffer as previously described (Clark and Keith 1988), using 25 μl of homogenizing buffer per fly. Homogenates were used immediately to measure protein, glycogen, and triglycerides. Each assay is colorimetric; spectrophotometer measurements were made using a Perkin-Elmer V3 plate reader (Waltham, MA). Protein in micrograms per fly was determined via Bradford's method (Bradford 1976) with BSA used for the protein standard curve. Total glycogen in micrograms per fly was measured as previously described (Clark and Keith 1988). Briefly, glycogen from the homogenates was broken down into glucose by adding 0.1 unit/ml amyloglucosidase enzyme slurry (Sigma, St. Louis) to 1.5-μl samples of homogenate in a 96-well plate. Total glucose was then determined using the PGO enzymes kit (Sigma) (Clark and Keith 1988). This measure is effectively the amount of whole-body glycogen, as free glucose is estimated at <5% of the amount of glycogen stored (Clark and Keith 1988). Glucose concentrations were determined using a glucose standard curve run on the same plate. Known concentrations of glycogen were used to assess the expected recovery of glycogen (Zimmerman et al. 2004); if <95% of glycogen was recovered, the samples were rerun. Triglyceride measurements were determined using an enzymatic assay kit (serum triglyceride determination kit, Sigma) (McGowan et al. 1983). The true serum triglyceride in micrograms per fly was determined from blank and glycerol standards run with each plate. Homogenates were then stored at −80°, and measurements were repeated the next day.
Statistical analysis of P-element insertion lines:
For each fly, all measures of sleep, activity, and energy stores were first expressed as a deviation from the contemporaneous w1118; Canton-S line mean assayed in each experimental block. This calculation has two benefits. First, the variation between experimental blocks due to random environmental fluctuations is mitigated. Second, some measures of sleep and activity (average sleep bout length and waking activity) are not normally distributed; however, when computed as deviations from the control mean, their distribution is normal.
Mutational effects were evaluated using analysis of variance (ANOVA). Two-way ANOVA models were performed for each trait using the model y = μ + L + S + (L × S) + E, where μ is the overall mean, L is the random effect of line, S is the fixed effect of sex, and E is the among-fly variance. A reduced version of this model was also performed for each sex separately. For glycogen and triglyceride measures, body weight and protein were added into the ANOVA model as covariates; for protein measures, body weight was included as a covariate in the ANOVA model. To account for the removal of dead flies from the data set, variance components were estimated using the restricted maximum-likelihood method, which accounts for unbalanced data. The total variance for each trait was estimated as the sum of the L, L × S, and E components in the combined-sex model and of L and E in the reduced model.
Broad-sense mutational heritability, 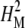 , was estimated for each trait as σ2G/σ2P, where σ2G is the genetic variance component and σ2P is the phenotypic variance (Falconer and Mackay 1996). σ2G was estimated as σ2L + σ2LS and σ2P as σ2L + σ2LS + σ2E from the line, line × sex, and environmental variance estimates of the combined-sex ANOVA, while σ2G = σ2L and σ2P = σ2L + σ2E from the reduced ANOVAS for each sex (Sambandan et al. 2006).
, was estimated for each trait as σ2G/σ2P, where σ2G is the genetic variance component and σ2P is the phenotypic variance (Falconer and Mackay 1996). σ2G was estimated as σ2L + σ2LS and σ2P as σ2L + σ2LS + σ2E from the line, line × sex, and environmental variance estimates of the combined-sex ANOVA, while σ2G = σ2L and σ2P = σ2L + σ2E from the reduced ANOVAS for each sex (Sambandan et al. 2006).
To identify candidate insertions for retesting, confidence limits were computed as ±zασP/(n)1/2, where zα is the value of the normal distribution at significance level α (0.05), σP is computed from the total phenotypic variance estimate determined above, and n is the number of flies assayed per line (Norga et al. 2003; Harbison et al. 2004). Confidence intervals were calculated at the 95, 99, and 99.9% level. Candidate lines of interest were chosen from lines that exceeded these thresholds.
Twenty-two of the most extreme short- and long-sleeping lines were retested using the same protocol as for the original test. Results were pooled for both tests and analyzed using the ANOVA model y = μ + G + S + T + (G × S) + (G × T) + (T × S) + (G × S × T) + E, where G, S, and T are the fixed effects of genotype (parental control or P-element insertion), sex, and experimental test (original screen or retest), and E is the residual among-fly variance. Insertion lines having significant (P < 0.05) G or G × S terms were interpreted as candidate genes for 24-hr sleep.
Partial Pearson product-moment correlations were determined for each phenotypic measurement as cov12/(σ2L1 × σ2L2)1/2, where cov12 is the covariance between traits 1 and 2, σ2L1 is the estimate of line variance for trait 1, and σ2L2 is the estimate of line variance for trait 2. The SAS CORR procedure was used to estimate the covariance matrix between traits. The restricted maximum-likelihood estimate of σ2Line from the ANOVAs described above was used to estimate the line variance for each trait. The P-values in Tables 4, 5, and 7 represent how significantly different each correlation is from zero. For correlations involving glycogen and triglycerides, protein and body weight were included as covariates; correlations involving protein included body weight as a covariate (Clark and Keith 1988). All statistical analyses were carried out using the SAS software package (SAS Institute, Cary, NC).
TABLE 4.
Genetic correlations between sleep phenotypes for males and females
| 24-hr sleep (min) | Nighttime sleep (min) | Daytime sleep (min) | |
|---|---|---|---|
| Male correlations | |||
| 24-hr bout number | −0.2920 | −0.1771 | −0.3553 |
| (0.0006) | (0.0392) | (<0.0001) | |
| Nighttime bout number | −0.2989 | −0.1629 | −0.3835 |
| (0.0004) | (0.0581) | (<0.0001) | |
| Daytime bout number | −0.1997 | −0.1400 | −0.2225 |
| (0.0198) | (0.1040) | (0.0092) | |
| Average bout length (min) | 0.8505 | 0.6792 | 0.8583 |
| (<0.0001) | (<0.0001) | (<0.0001) | |
| Female correlations | |||
| 24-hr bout number | 0.4544 | 0.3211 | 0.5152 |
| (<0.0001) | (0.0001) | (<0.0001) | |
| Nighttime bout number | −0.1646 | −0.1179 | −0.1849 |
| (0.0555) | (0.1716) | (0.0312) | |
| Daytime bout number | 0.7663 | 0.5428 | 0.8674 |
| (<0.0001) | (<0.0001) | (<0.0001) | |
| Average bout length (min) | 0.8072 | 0.8319 | 0.6260 |
| (<0.0001) | (<0.0001) | (<0.0001) | |
P-values are listed in parentheses. Correlations significantly different from zero (P < 0.05) are indicated by italics.
TABLE 5.
Genetic correlations between sleep and waking activity for males and females
| Waking activity (counts/min)
|
||
|---|---|---|
| Males | Females | |
| 24-hr sleep (min) | −0.3825 | −0.4770 |
| (<0.0001) | (<0.0001) | |
| Nighttime sleep (min) | −0.2854 | −0.4141 |
| (0.0008) | (<0.0001) | |
| Daytime sleep (min) | −0.4076 | −0.4556 |
| (<0.0001) | (<0.0001) | |
| Average bout length (min) | −0.1966 | −0.1184 |
| (0.0218) | (0.1698) | |
| 24-hr bout number | −0.0596 | −0.5477 |
| (0.4907) | (<0.0001) | |
| Nighttime bout number | 0.0342 | −0.1796 |
| (0.6926) | (0.0364) | |
| Daytime bout number | −0.1395 | −0.6125 |
| (0.1053) | (<0.0001) | |
P-values are listed in parentheses. Correlations significantly different from zero (P < 0.05) are indicated by italics.
TABLE 7.
Correlations between sleep phenotypes and energy stores in males and females
| Glycogen (μg/fly)
|
Triglycerides (μg/fly)
|
|||
|---|---|---|---|---|
| Sleep parameter | Males | Females | Males | Females |
| 24-hr sleep (min) | 0.2376 (0.0053) | 0.0272 (NS) | 0.0431 (NS) | 0.0554 (NS) |
| Nighttime sleep (min) | 0.2322 (0.0065) | 0.0835 (NS) | 0.1132 (NS) | 0.1081 (NS) |
| Daytime sleep (min) | 0.2128 (0.0129) | −0.0674 (NS) | −0.0028 (NS) | 0.0532 (NS) |
| 24-hr bout number | −0.3861 (<0.0001) | −0.0368 (NS) | −0.1352 (NS) | −0.2096 (0.0143) |
| Nighttime bout number | −0.4321 (<0.0001) | 0.0275 (NS) | −0.1116 (NS) | −0.3088 (0.0003) |
| Daytime bout number | −0.2195 (0.0102) | −0.0767 (NS) | −0.0587 (NS) | −0.0829 (NS) |
| Average bout length (min) | 0.3279 (<0.0001) | 0.0999 (NS) | 0.1152 (NS) | 0.3328 (<0.0001) |
| Waking activity (counts/min) | −0.0977 (NS) | 0.0046 (NS) | −0.0412 (NS) | −0.0002 (NS) |
Values are listed as the correlation, with the P-value given in parentheses. Correlations significantly different from zero (P < 0.05) are in italics.
Assessment of transcript level in whole flies:
Flies from lines BG02566, BG01628, BG01037, and BG01565 were reared as described above along with w1118; Canton-S controls. Flies were harvested at the “lights on” time of their day–night cycle. Three replicate RNA isolations were prepared for each line and sex. RNA was extracted using Triazol reagent (Sigma-Aldrich, St. Louis) according to the manufacturer's instructions. RNA was converted to cDNA using the Hi Capacity cDNA archive kit (Applied Biosystems, Foster City, CA). Primer Express 2.0 software (Applied Biosystems) was used to design transcript-specific primer pairs to ensure that RNA transcripts rather than genomic DNA would be amplified. RNA levels were measured using the following primer pairs for each gene: Calreticulin, 5′-GGATCGTTCACATGATGTGGTG-3′ and 5′-CGTATCCTCCCAGTTTTCGTTG-3′; malic enzyme isoform A, 5′-AAACTTTTGGACCCACGCC-3′ and 5′-TTCTCTTCGTGTAACAGCCGAG-3′; malic enzyme isoform B, 5′-AAATGGCACGCCGGTTTATC-3′ and 5′-CGGCACTTTGCGTTGTGATT-3′; βνintegrin, 5′-TGGTGCACGGACAAGGAATAC-3′ and 5′-ACATCTAGGACCGGCTGGTTCT-3′; and Defense repressor I, 5′-GGCCAAAAGATGTGGTGCAT-3′ and 5′-TGATGTTCATTGCGCGACA-3′. SYBR Green chemistry (Applied Biosystems) was used for the quantitative PCR reaction in an ABI 7000 thermal cycler (Applied Biosystems) under default PCR protocol conditions. Resultant RNA quantities were normalized to actin measured in each respective sample. Samples were compared to the contemporaneous w1118; Canton-S control using the Kruskal–Wallis nonparametric test (SAS Institute). RNA samples were also obtained from selected Calreticulin revertant lines and assayed as described above.
Construction and verification of revertant lines:
The P[GT1] construct was mobilized by crossing w, isoCanton-S; isoCanton-S; P[GT1] females to w/Y; wg[Sp-1]/CyO; ry[506] Sb[1] P[ry[+t7.2]=Δ2-3]99B/TM6 males. To maintain background integrity, third chromosome balancer stock constructed from the w1118; Canton-S parent (gift of Akihiko Yamamoto and Trudy Mackay) was used to create these revertants (w, isoCanton-S; isoCanton-S; H/TM3). Male offspring with the genotype w, isoCanton-S; CyO/isoCanton-S; P[GT1]/ry[506] Sb[1] P[ry[+t7.2]=Δ2-3]99B were mated to w, isoCanton-S; isoCanton-S; H/TM3 females. Single males without the P[GT1] insertion were mated to w, isoCanton-S; isoCanton-S; H/TM3 females, and the resulting progeny were used to make homozygous P[−] excision stock.
PCR was used to identify those P[−] lines that were precise excisions. Putative Calreticulin precise excisions were verified with DNA sequencing. Primers were chosen to surround the P-element insertion region and produce a PCR product of ∼500 bp. PCR products were run on a 1.5% agarose gel, and PCR product sizes were verified with a DNA ladder. Primers used for the Calreticulin revertants were 5′-CCTGGCCGGTGAAAAAGA-3′ and 5′-TCCTTTCGTTATTCATTGAAGG-3′ to amplify a 391-bp region containing the P-element insertion site inside the first exon. For the malic enzyme revertants, primers 5′-ATCAGCGCATTTCAAAGGTT-3′ and 5′-GTTGCTGTTTCTCTTCGTGTAA-3′ were used to amplify a 497-bp region in the third exon surrounding the P-element insertion site.
Statistical analysis of revertant lines:
The revertant lines were assayed for sleep phenotypes as described above. A mixed ANOVA model was used to determine whether the revertant line sleep was the same as that of the w1118; Canton-S parent using y = μ + G + B(G) + E, where μ is the overall mean, G is the genotype (P insertion or wild type), B is the random effect of experimental block, and E represents the within-fly environmental variance.
RESULTS
Identification of insertions that impact sleep phenotypes:
High mutational variance was present for sleep and activity parameters; for the combined-sex ANOVAs, the main effect of line was highly significant (P < 0.0001) in every case, with the exception of daytime bout number (P = 0.0580; Table 1). With the exception of 24-hr sleep time, the main effect of sex was also highly significant for each trait. Mutational heritability estimates for both sexes combined are presented in Table 1. Mutational heritability was high for sleep time—0.45, 0.38, and 0.43 for 24-hr, nighttime, and daytime sleep, respectively (Table 1). Heritability for sleep bout number was lower: 0.29, 0.24, and 0.33 for 24-hr, nighttime, and daytime bout number, respectively.
TABLE 1.
Analysis of variance and broad-sense mutational heritabilities pooled over sexes
| Source | d.f. | MS | F | P | σ2 | H2M |
|---|---|---|---|---|---|---|
| 24-hr sleep (min) | ||||||
| Sex | 1 | 17,367.650 | 0.26 | 0.6078 | — | 0.45 |
| Line | 135 | 392,439.950 | 5.46 | <0.0001 | 12,091.70 | |
| Line × sex | 134 | 72,022.460 | 3.66 | <0.0001 | 3,891.80 | |
| Error | 3597 | 19,680.100 | — | — | 19,678.90 | |
| Nighttime sleep (min) | ||||||
| Sex | 1 | 2,789,073.880 | 95.99 | <0.0001 | — | 0.38 |
| Line | 135 | 127,961.060 | 4.02 | <0.0001 | 3,657.90 | |
| Line × sex | 134 | 31,887.210 | 3.70 | <0.0001 | 1,673.50 | |
| Error | 3597 | 8,615.820 | — | — | 8,615.80 | |
| Daytime sleep (min) | ||||||
| Sex | 1 | 2,366,343.690 | 121.85 | <0.0001 | — | 0.43 |
| Line | 135 | 111,730.850 | 5.26 | <0.0001 | 3,376.30 | |
| Line × sex | 134 | 21,296.810 | 3.63 | <0.0001 | 1,078.00 | |
| Error | 3597 | 5,874.840 | — | — | 5,874.10 | |
| 24-hr sleep bout number | ||||||
| Sex | 1 | 2,968.426 | 22.83 | <0.0001 | — | 0.29 |
| Line | 135 | 301.183 | 2.11 | <0.0001 | 6.19 | |
| Line × sex | 134 | 143.289 | 4.16 | <0.0001 | 8.19 | |
| Error | 3597 | 34.445 | — | — | 34.48 | |
| Nighttime sleep bout number | ||||||
| Sex | 1 | 834.361 | 21.2 | <0.0001 | — | 0.24 |
| Line | 135 | 127.700 | 3.00 | <0.0001 | 3.11 | |
| Line × sex | 134 | 42.624 | 2.71 | <0.0001 | 1.93 | |
| Error | 3597 | 15.748 | — | — | 15.77 | |
| Daytime sleep bout number | ||||||
| Sex | 1 | 6,944.478 | 88.49 | <0.0001 | — | 0.33 |
| Line | 135 | 114.519 | 1.31 | 0.0580 | 0.96 | |
| Line × sex | 134 | 87.521 | 6.63 | <0.0001 | 5.46 | |
| Error | 3597 | 13.203 | — | — | 13.20 | |
| Average bout length (min) | ||||||
| Sex | 1 | 10,043.693 | 27.13 | <0.0001 | — | 0.35 |
| Line | 135 | 1,546.851 | 3.83 | <0.0001 | 43.38 | |
| Line × sex | 134 | 404.618 | 3.31 | <0.0001 | 21.93 | |
| Error | 3597 | 122.141 | — | — | 122.26 | |
| Waking activity (counts/min) | ||||||
| Sex | 1 | 2.326 | 4.01 | 0.0472 | — | 0.32 |
| Line | 135 | 1.711 | 2.69 | <0.0001 | 0.0413 | |
| Line × sex | 134 | 0.064 | 3.85 | <0.0001 | 0.0358 | |
| Error | 3597 | 0.166 | — | — | 0.1655 | |
| Glycogen (μg/fly) | ||||||
| Sex | 1 | 1,471.409 | 14.75 | 0.0002 | — | 0.56 |
| Line | 135 | 207.674 | 1.71 | 0.0009 | 38.86 | |
| Line × sex | 135 | 121.343 | 1.91 | <0.0001 | 51.69 | |
| Error | 256 | 63.642 | — | — | 69.99 | |
| Triglycerides (μg/fly) | ||||||
| Sex | 1 | 9.381 | 3.61 | 0.0589 | — | 0.60 |
| Line | 135 | 5.849 | 1.92 | <0.0001 | 0.6104 | |
| Line × sex | 135 | 3.059 | 1.73 | <0.0001 | 2.4210 | |
| Error | 256 | 1.758 | — | — | 1.9920 | |
| Protein (μg/fly) | ||||||
| Sex | 1 | 96.850 | 10.72 | 0.0012 | — | 0.56 |
| Line | 135 | 15.802 | 1.53 | 0.0070 | 2.2238 | |
| Line × sex | 135 | 10.332 | 1.65 | 0.0003 | 7.8902 | |
| Error | 257 | 6.244 | — | — | 7.9836 | |
MS, mean square. σ2, variance component as calculated by restricted maximum-likelihood method. H2M, broad-sense mutational heritability.
The line × sex interaction term was highly significant for all sleep traits (PLine×Sex < 0.0001; Table 1), which indicates that the insertions have sex-specific effects on each trait. As it has been previously reported that males and females have different sleep patterns (Huber et al. 2004), the remainder of our analysis focused on each sex separately. Reduced ANOVA models for each sex indicated that the insertions had highly significant effects on each sleep trait measured (PLine < 0.0001; Table 2). Furthermore, mutational broad-sense heritabilities for each sex separately were quite similar to estimates made with sexes combined (Table 2).
TABLE 2.
Analysis of variance and broad-sense mutational heritability for each sex separately
| Sex | Source | d.f. | MS | F | P | σ2 | H2M |
|---|---|---|---|---|---|---|---|
| 24-hr sleep (min) | |||||||
| Female | Line | 135 | 283,453.930 | 12.73 | <0.0001 | 19,131.40 | 0.46 |
| Error | 1780 | 22,265.960 | — | — | 22,259.10 | ||
| Male | Line | 134 | 187,512.140 | 10.94 | <0.0001 | 12,822.00 | 0.43 |
| Error | 1817 | 17,146.930 | — | — | 17,154.60 | ||
| Nighttime sleep (min) | |||||||
| Female | Line | 135 | 98,108.160 | 9.93 | <0.0001 | 6,375.90 | 0.39 |
| Error | 1780 | 9,875.890 | — | — | 9,872.30 | ||
| Male | Line | 134 | 64,305.544 | 8.71 | <0.0001 | 4,265.00 | 0.37 |
| Error | 1817 | 7,381.420 | — | — | 7,385.20 | ||
| Daytime sleep (min) | |||||||
| Female | Line | 135 | 80,597.200 | 11.27 | <0.0001 | 5,217.70 | 0.42 |
| Error | 1780 | 7,153.810 | — | — | 7,152.30 | ||
| Male | Line | 134 | 54,301.430 | 11.75 | <0.0001 | 3,663.90 | 0.44 |
| Error | 1817 | 4,621.920 | — | — | 4,622.80 | ||
| 24-hr sleep bout number | |||||||
| Female | Line | 135 | 260.946 | 6.77 | <0.0001 | 17.55 | 0.31 |
| Error | 1780 | 38.539 | — | — | 38.59 | ||
| Male | Line | 134 | 186.125 | 6.12 | <0.0001 | 11.33 | 0.37 |
| Error | 1817 | 30.435 | — | — | 30.43 | ||
| Nighttime sleep bout number | |||||||
| Female | Line | 135 | 102.779 | 5.41 | <0.0001 | 6.17 | 0.24 |
| Error | 1780 | 19.008 | — | — | 19.04 | ||
| Male | Line | 134 | 67.816 | 5.40 | <0.0001 | 3.96 | 0.24 |
| Error | 1817 | 12.554 | — | — | 12.56 | ||
| Daytime sleep bout number | |||||||
| Female | Line | 135 | 141.624 | 8.69 | <0.0001 | 9.11 | 0.36 |
| Error | 1780 | 16.289 | — | — | 16.29 | ||
| Male | Line | 134 | 61.438 | 6.04 | <0.0001 | 3.73 | 0.27 |
| Error | 1817 | 10.180 | — | — | 10.18 | ||
| Average bout length (min) | |||||||
| Female | Line | 135 | 618.703 | 6.97 | <0.0001 | 38.74 | 0.30 |
| Error | 1780 | 88.828 | — | — | 88.78 | ||
| Male | Line | 134 | 1,393.964 | 9.01 | <0.0001 | 92.94 | 0.38 |
| Error | 1817 | 154.775 | — | — | 154.84 | ||
| Activity per waking minute (counts/min) | |||||||
| Female | Line | 135 | 1.005 | 8.17 | <0.0001 | 0.0692 | 0.36 |
| Error | 1780 | 0.123 | — | — | 0.1231 | ||
| Male | Line | 134 | 1.388 | 6.7 | <0.0001 | 0.0840 | 0.29 |
| Error | 1817 | 0.207 | — | — | 0.2070 | ||
| Glycogen (μg/fly) | |||||||
| Female | Line | 135 | 215.977 | 3.29 | <0.0001 | 123.224 | 0.65 |
| Error | 128 | 65.596 | — | — | 67.629 | ||
| Male | Line | 135 | 108.790 | 1.85 | 0.0003 | 57.769 | 0.44 |
| Error | 126 | 58.884 | — | — | 72.397 | ||
| Triglycerides (μg/fly) | |||||||
| Female | Line | 135 | 7.169 | 2.93 | <0.0001 | 4.8649 | 0.66 |
| Error | 129 | 2.446 | — | — | 2.5337 | ||
| Male | Line | 135 | 1.469 | 1.34 | 0.0489 | 1.2004 | 0.45 |
| Error | 125 | 1.096 | — | — | 1.4578 | ||
| Protein (μg/fly) | |||||||
| Female | Line | 135 | 19.263 | 2.25 | <0.0001 | 15.870 | 0.63 |
| Error | 130 | 8.561 | — | — | 9.496 | ||
| Male | Line | 135 | 5.766 | 1.54 | 0.0074 | 4.373 | 0.40 |
| Error | 126 | 3.746 | — | — | 6.468 | ||
MS, mean square. σ2, variance component calculated using restricted maximum-likelihood method. H2M, broad-sense mutational heritability.
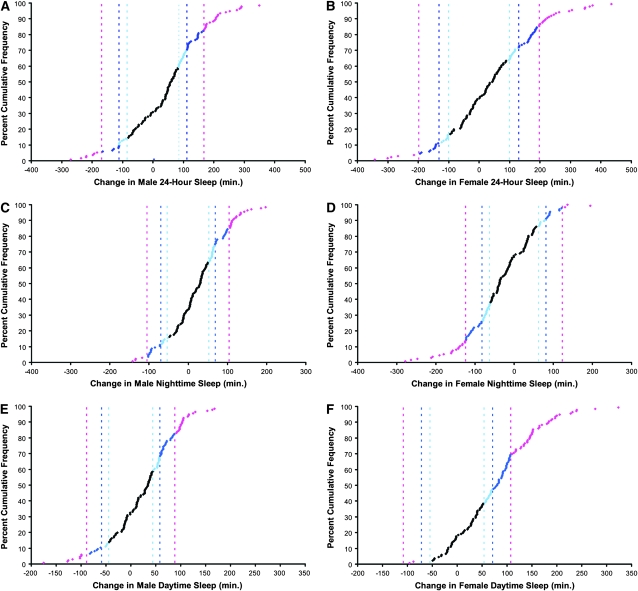
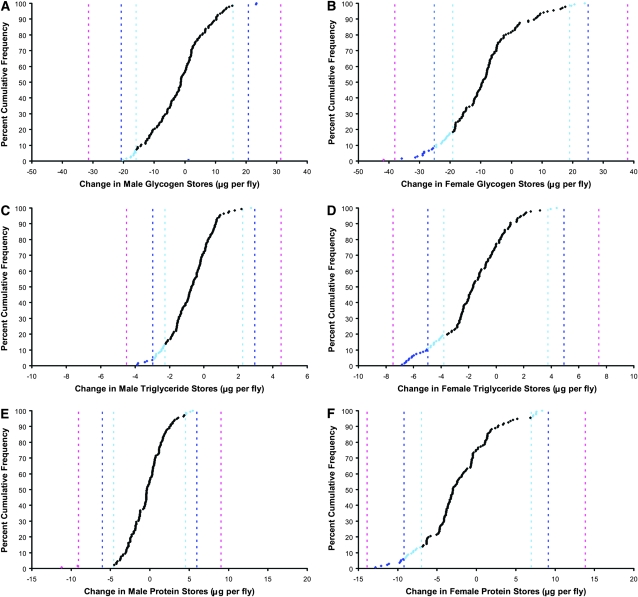
The distribution of mutational effects for 24-hr sleep time, nighttime sleep, and daytime sleep for each sex is given in Figure 1, which illustrates the 95, 99, and 99.9% confidence interval limits for the deviation from the parental line mean for each trait by sex. Mutations were more likely to increase 24-hr sleep than decrease it in both males and females (Figure 1, A and B). However, when 24-hr sleep is broken into nighttime or daytime sleep, a different pattern emerges. The effect of mutations on male nighttime and daytime sleep was similar to the effect seen on 24-hr sleep: mutations tended to increase sleep (Figure 1, C and E). However, while the net effect of mutations on 24-hr sleep in females was an increase overall, nighttime sleep generally decreased, while daytime sleep increased. This effect may be partially explained by the fact that females sleep little during the day (2.9 hr on average in this experiment) as compared to males (7.0 hr on average in this experiment). A mutation might therefore be more likely to increase daytime sleep in females. Why the mutations tended to reduce nighttime sleep in females is unclear, however. On average, females slept 7.59 hr at night, which is well below the total 12-hr nighttime period.
Figure 1.—
Distribution of mutational effects on sleep. Pink, 99.9% C.I. threshold; dark blue, 99% C.I. threshold; light blue, 95% C.I. threshold. (A) Male 24-hr sleep time: 99.9% C.I. threshold, 168.4 min; 99% C.I. threshold, 111.5 min; 95% C.I. threshold, 84.8 min. (B) Female 24-hr sleep time: 99.9% C.I. threshold, 197.9 min; 99% C.I. threshold, 131.0 min; 95% C.I. threshold, 99.7 min. (C) Male nighttime sleep: 99.9% C.I. threshold, 105.0 min; 99% C.I. threshold, 69.5 min; 95% C.I. threshold, 52.9 min. (D) Female nighttime sleep: 99.9% C.I. threshold, 124.0 min; 99% C.I. threshold, 82.1 min; 95% C.I. threshold, 62.5 min. (E) Male daytime sleep: 99.9% C.I. threshold, 88.6 min; 99% C.I. threshold, 58.6 min; 95% C.I. threshold, 44.6 min. (F) Female daytime sleep: 99.9% C.I. threshold, 108.2 min; 99% C.I. threshold, 71.6 min; 95% C.I. threshold, 54.5 min.
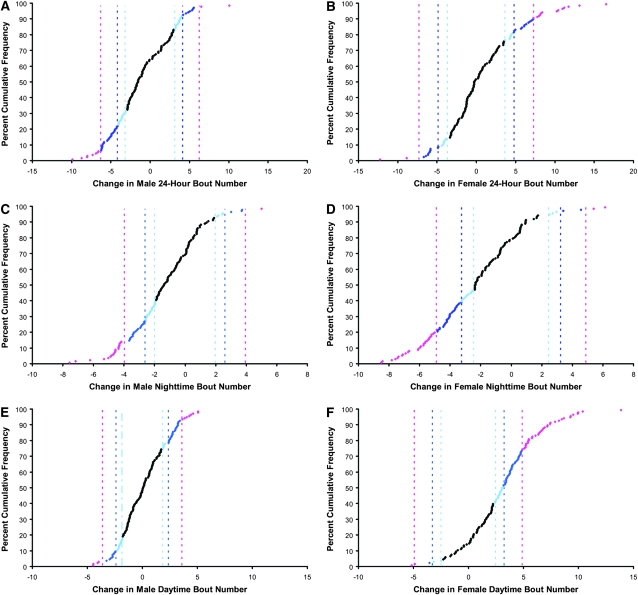
The distribution of sleep bout number mutational effects is given in Figure 2. Mutations tended to decrease the number of sleep bouts in a 24-hr period for males, while increasing it for females (Figure 2, A and B). Mutations tended to decrease nighttime bout number in both sexes (Figure 2, C and D). The effect of a mutation on male daytime bout number was equally likely to be positive or negative, while female daytime bout number tended to increase (Figure 2, E and F).
Figure 2.—
Distribution of mutational effects on sleep bout number. Pink, 99.9% C.I. threshold; dark blue, 99% C.I. threshold; light blue, 95% C.I. threshold. (A) Male 24-hr bout number: 99.9% C.I. threshold, 6.3; 99% C.I. threshold, 4.2; 95% C.I. threshold, 3.2. (B) Female 24-hr bout number: 99.9% C.I. threshold, 7.3; 99% C.I. threshold, 4.8; 95% C.I. threshold, 3.7. (C) Male nighttime bout number: 99.9% C.I. threshold, 4.0; 99% C.I. threshold, 2.6; 95% C.I. threshold, 2.0. (D) Female nighttime bout number: 99.9% C.I. threshold, 4.9; 99% C.I. threshold, 3.2; 95% C.I. threshold, 2.5. (E) Male daytime bout number: 99.9% C.I. threshold, 3.6; 99% C.I. threshold, 2.4; 95% C.I. threshold, 1.8. (F) Female daytime bout number: 99.9% C.I. threshold, 4.9; 99% C.I. threshold, 3.3; 95% C.I. threshold, 2.5.
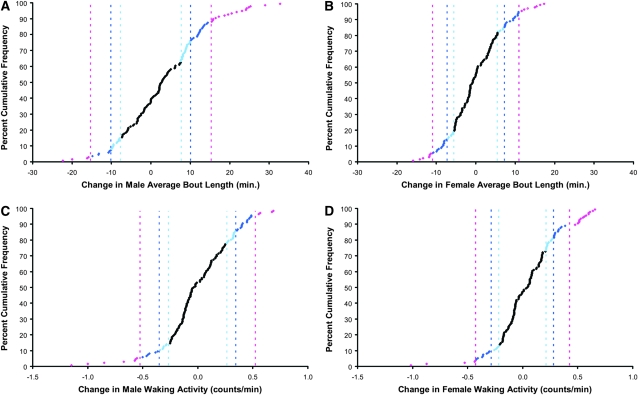
The effect of mutation on average bout length and waking activity is given in Figure 3. Effects were greater and tended to increase average bout length in males (Figure 3, A and B). These mutations had a slight tendency to increase waking activity in both males and females (Figure 3, C and D).
Figure 3.—
Distribution of mutational effects on average sleep bout length and waking activity. Pink, 99.9% C.I. threshold; dark blue, 99% C.I. threshold; light blue, 95% C.I. threshold. (A) Male average sleep bout length: 99.9% C.I. threshold, 15.3 min; 99% C.I. threshold, 10.1 min; 95% C.I. threshold, 7.7 min. (B) Female average sleep bout length: 99.9% C.I. threshold, 11.0 min; 99% C.I. threshold, 7.3 min; 95% C.I. threshold, 5.5 min. (C) Male activity per waking minute: 99.9% C.I. threshold, 0.5247 counts/min; 99% C.I. threshold, 0.3474 counts/min; 95% C.I. threshold, 0.2643 counts/min. (D) Female activity per waking minute: 99.9% C.I. threshold, 0.4266 counts/min; 99% C.I. threshold, 0.2824 counts/min; 95% C.I. threshold, 0.2149 counts/min.
Supplemental Table 1 lists the deviations from sleep trait means for all lines tested. Our primary interest was 24-hr sleep duration; thus, 22 short- and long-sleeping were selected lines for retesting. Of the 22 lines retested, only one, BG01635, failed the retest criteria. The remaining 21 mutations remained significant after retest. Table 3 lists the retested lines, the nearest gene and cytological location of the P element where known, and the effect of the insertion on 24-hr sleep. Supplemental Table 2 lists 24-hr sleep deviations from the control mean for the retest. Of the insertions with significant genotype-by-sex (G × S) effects, one, BG01986, had a female-specific sleep phenotype, while the effects of the mutation in lines BG01007, BG01491, and BG02466 were specific to males. The shortest-sleeping line for both males and females was BG02566, which has a P element in the first exon of the gene Calreticulin. Flies from this line slept 4.8 fewer hours (289.2 min) than the w1118; Canton-S control. Male flies with an insertion in the third exon of malic enzyme had the longest 24-hr sleep: 4.0 hr (240 min) more than w1118; Canton-S. Sleep also increased in BG01628 females as compared to the control: 2.45 hr (147 min). The greatest increase in female 24-hr sleep was in line BG01907: 7.08 hr (424.8 min). The location of the P element in line BG01907 is unknown. Eleven of the 22 retested insertions had block effects (initial vs. retest) that were significant (P < 0.05): BG01007, BG01277, BG01376, BG01491, BG01536, BG01635, BG01744, BG01986, BG02439, BG02565, and BG02602 (supplemental Table 2). These results suggest that the effects of some mutations are modified by changes in the environment (i.e., genotype-by-environment interaction).
TABLE 3.
Effects of significant retested P-element insertion lines on 24-hr sleep time
| Line | Nearest gene | Cytological location | P[GT1] position relative to nearest gene | Sleep effect (min) (♀ + ♂) | Significant G × S? | ♀ effect (min) | ♂ effect (min) |
|---|---|---|---|---|---|---|---|
| BG00080 | Unknown | — | Unknown | 222.6**** | Yes** | 293.4**** | 156.6**** |
| BG01007 | CG8776 | 49B9 | 28 kb in gene (first intron) | 30.0 (NS) | Yes** | −69.0 (NS) | 118.2*** |
| BG01037 | βνintegrin | 39A1 | 170 bp in gene (first exon) | −254.4**** | No | −282.0**** | −228.0**** |
| BG01245 | Sema-5C | 68F2 | 200 bp from 5′-end of gene | 108.6*** | No | 87.0* | 130.2** |
| BG01277 | CG11940 | 18F3-4 | 500 bp in gene (first exon) | 158.4**** | No | 192.0**** | 127.2*** |
| BG01376 | Chd64 | 64A7 | 70 bp in gene (first exon) | 215.4**** | No | 198.6**** | 231.0**** |
| BG01491 | tramtrack | 100D1 | 12 kb in gene (third exon) | 101.4**** | Yes* | 30.0 (NS) | 158.4**** |
| BG01536 | Beadex | 17C3 | 58 bp from 5′-end of gene | 217.2**** | No | 243.6**** | 194.4**** |
| BG01565 | Defense repressor I | 58E9 | 22 kb in gene (first intron) | −120.6**** | No | −139.2*** | −100.8* |
| BG01628 | Malic enzyme | 87C7 | 2.5 kb in gene (third exon) | 193.2**** | No | 147.0*** | 240.0**** |
| BG01744 | Unknown | — | Sequence not available | −112.8**** | No | −121.8** | −103.8** |
| BG01907 | Unknown | — | Unknown | 293.4**** | Yes** | 424.8**** | 178.8*** |
| BG01986 | CG12477 | 75D7 | 185 bp in gene (first exon) | 108.0**** | Yes*** | 198.0**** | 12.6 (NS) |
| BG02003 | 6-phosphofructo-2-kinase | 18C8 | 2.5 kb in gene (second intron) | −130.8*** | No | −105.6* | −155.4* |
| BG02009 | taiman | 30A2 | 10 kb in gene (first intron) | 150.0**** | No | 187.2**** | 115.8*** |
| BG02419 | escargot | 35D1 | 1 kb from 5′-end of gene | −141.6**** | No | −193.8*** | −85.8* |
| BG02439 | CG32556 | 16C1 | 300 bp from 5′-end of gene | −160.2**** | No | −183.6**** | −135.6*** |
| BG02466 | Unknown | — | Sequence not available | −115.8**** | Yes** | −60.6 (NS) | −165.6**** |
| BG02565 | β4GalNAcTA | 50E6 | 13 bp in gene (first exon) | −77.4*** | No | −81.6** | −73.8* |
| BG02566 | Calreticulin | 85E1 | 100 bp in gene (first exon) | −289.2**** | No | −288.6**** | −289.2**** |
| BG02602 | CG14782 | 2B1 | 75 bp in gene (first intron) | 162.6**** | No | 184.8**** | 141.0*** |
Boldface type with underlining indicates that the insertion also has significant effects on energy stores. Underlining indicates that alleles of this insertion have significant effects on energy stores. See text for details. NS, nonsignificant; *0.01 < P ≤ 0.05; **0.001 < P ≤ 0.01; ***0.0001 < P ≤ 0.001; ****P < 0.0001.
Genetic correlations among sleep phenotypes for each sex:
The genetic correlation between sleep time and the number of sleep bouts was determined for males and females separately (Table 4). Sleep time was strongly correlated with 24-hr bout number in both males and females. In males, significant negative correlations were observed between 24-hr/daytime sleep and any measure of sleep bout number (24 hr, nighttime, or daytime). However, the correlation between nighttime sleep and nighttime bout number and nighttime sleep and daytime bout number was not significant. The negative correlations suggest that the longer a male fly sleeps, the fewer sleep bouts he will have, meaning that his increased sleep is more consolidated.
In contrast, the correlation between sleep and bout number for females tended to be positive. The strongest correlations were between 24-hr/daytime bout number and all three measures of sleep time. We observed nonsignificant negative correlations between sleep and nighttime bout number in females. Generally, increased sleep in females implies increased numbers of sleep bouts, indicating that longer sleep might be less consolidated. These observations suggest that mutations affecting sleep bout number can have opposing effects on male and female sleep. Furthermore, the relationship between nighttime sleep and nighttime bout number may be fundamentally different from the relationship between bout number and daytime sleep; nighttime sleep and nighttime bout number are not significantly correlated in either males or females.
Correlation between sleep parameters and waking activity:
To better examine the genetic relationship between sleep and activity, we decoupled sleep time from activity by calculating waking activity as the number of beam crossings per minute spent awake (Andretic and Shaw 2005). Table 5 shows the correlations between sleep phenotypes and waking activity. Male and female sleep times were significantly negatively correlated with waking activity. Thus, in the select group of lines examined in this study, more active flies tended to sleep less and less active flies tended to sleep more. Average sleep bout length was negatively correlated with waking activity, although the correlation was not significant in females. While waking activity was not correlated with bout number in males, it was strongly negatively correlated in females. The lack of correlation between waking activity and sleep bout number indicates that sleep consolidation can be genetically perturbed independently of activity in males. Mutations affecting female sleep bout number, however, may potentially impact waking activity.
Correlation between male and female sleep phenotypes:
The genetic correlation between males and females, rGS (the cross-sex genetic correlation), was calculated for each sleep trait (supplemental Table 3). With the exception of daytime bout number, all male and female sleep phenotypes were significantly and positively correlated, but not unity. Thus, some, but not all, genes that influence sleep in males overlap with genes that influence sleep in females. The variance due to the line × sex interaction term from the initial combined-sex ANOVA was partitioned into two groups (supplemental Table 3): the contribution due to differences in male and female among-line variance components [(σLM − σLF)2] and the contribution due to sex-specific effects of the insertions [σLM × σLF (1 − rGS)] (Robertson 1959). For each sleep phenotype, the relative contribution due to sex-specific effects of the insertions accounted for the greatest percentage of the line × sex variance, underscoring the tendency of P-element insertions to have sex-specific phenotypic effects (Harbison et al. 2004). Effects tended to be sex specific rather than sex antagonistic (significant effects in both sexes, but of opposite sign; see supplemental Table 1 for examples of sex-antagonistic effects).
Transcript expression for selected insertion lines:
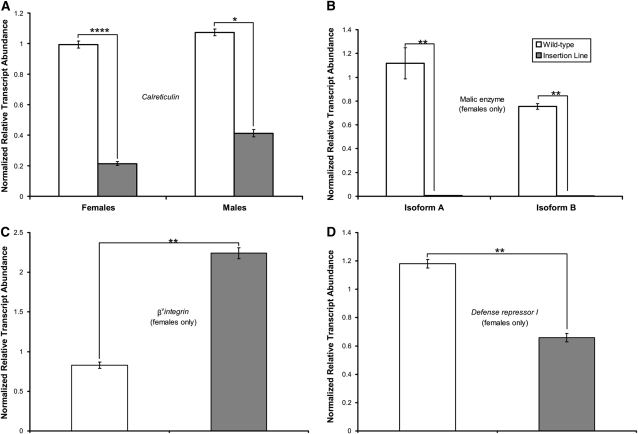
Transcript abundance relative to the w1118; Canton-S parental line was examined in four insertion lines. Comparisons were made using age- and sex-matched lines harvested at the same circadian time of day. Gene transcript levels were normalized to actin transcript levels in each sample and compared to w1118;Canton-S using the nonparametric Kruskal–Wallis test. Transcript abundance was examined in BG02566 (Calreticulin), BG01037 (βνintegrin), and BG01565 (Defense repressor I), which had reduced sleep; transcript level was also assayed in an insertion line that increased sleep, BG01628 (malic enzyme). Whole-body Calreticulin expression was reduced to 21.5% of wild-type levels in females and to 38.5% of wild-type levels in males (Figure 4A). We assayed transcript abundance in females only for lines BG01037 (βνintegrin), BG01565 (Defense repressor I), and BG01628 (malic enzyme). Malic enzyme has two transcript isoforms, A and B; both were reduced to near zero in female mutants as compared to the wild-type parent (Figure 4B). Female βνintegrin RNA actually increased relative to w1118; Canton-S by 170% (Figure 4C). Finally, Defense repressor I transcripts were moderately reduced (by 44%) in females as compared to the wild-type parent (Figure 4D). Thus, transcript levels in all four insertion lines examined were significantly different from the w1118, Canton-S parent.
Figure 4.—
Comparison of transcript level in selected insertion lines and w1118; Canton-S parent. *P < 0.05, **P < 0.01, and ****P < 0.0001 by Kruskal–Wallis test. (A) BG02566 (Calreticulin). (B) BG01628 (malic enzyme). (C) BG01037 (βνintegrin). (D) BG01565 (Defense repressor I).
Reversion lines:
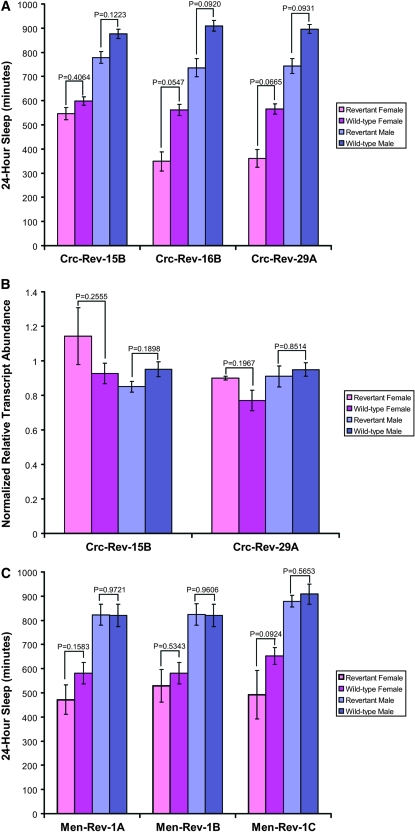
A source of transposase was introduced to excise the P-element insertion from BG02566 (Calreticulin) and BG01628 (malic enzyme). Importantly, the isogenic w1118; Canton-S background was maintained through the use of a w1118; Canton-S third chromosome balancer stock (see materials and methods). Three precise excision lines from the Calreticulin crosses showed increased sleep that was not significantly different from that of the w1118; Canton-S parent (Figure 5A). With the exception of line 15B females, we observed block effects in these revertant lines, unlike the original BG02566 insertion line. We assayed transcript levels in two of these lines and found that they were the same as wild type (Figure 5B). Furthermore, sequence data from the region reveal that all three revertant lines are precise excisions (data not shown). The P-element insertion in the malic enzyme gene was also mobilized. Three revertant lines restored the sleep phenotype to wild-type levels in both males and females (Figure 5C). Each of these revertant lines originated from males from the same culture vial and likely had the same excision. We did not observe significant block effects in these lines. The PCR product of the region surrounding the P element in these three lines was of the expected size (i.e., identical to wild type), but the region was not sequenced nor followed up with RNA assays.
Figure 5.—
Sleep and RNA phenotypes of Calreticulin and malic enzyme precise excision lines. (A) Twenty-four-hour sleep in Calreticulin revertants. (B) Relative transcript abundance in Calreticulin revertants. (C) Twenty-four-hour sleep in malic enzyme revertants.
Identification of insertions that impact energy stores:
Like sleep, a high degree of variation among the P-element insertion lines for protein, glycogen, and triglycerides was observed; deviations from the parental mean for each line are provided in supplemental Table 4. Line effects were highly significant for all traits whether sexes were combined or analyzed separately; see the ANOVA results in Tables 1 and 2. Broad-sense mutational heritability (H2M) was very high for all three traits, but it was particularly high in females. For females, H2M was 0.65 for glycogen, 0.66 for triglycerides, and 0.63 for protein. In contrast, H2M for males was 0.44 for glycogen, 0.45 for triglycerides, and 0.40 for protein. Thus, the genetic variance components (additive, dominance, and/or interaction) were greater in females than in males. Indeed, females were more sensitive to the effects of a single-gene perturbation than males. The distribution of mutational effects for each trait by sex in Figure 6 shows a more pronounced effect on females. However, for both sexes, the insertions tended to decrease rather than increase energy stores. Table 6 shows the putative locations of P-element insertions having at least one effect greater than the 99% confidence interval in the initial screen and lists the effect of significant insertions on both males and females. As Table 6 shows, most of these genes had sex-specific effects on energy stores, and the majority of those effects were observed in females.
Figure 6.—
Distribution of mutational effects on energy stores. Pink, 99.9% C.I. threshold; dark blue, 99% C.I. threshold; light blue, 95% C.I. threshold. (A) Male glycogen: 99.9% C.I. threshold, 31.39; 99% C.I. threshold, 20.78; 95% C.I. threshold, 15.81. (B) Female glycogen: 99.9% C.I. threshold, 38.01; 99% CI threshold, 25.16; 95% CI threshold, 19.15. (C) Male triglycerides: 99.9% C.I. threshold, 4.49; 99% C.I. threshold, 2.97; 95% C.I. threshold, 2.26. (D) Female triglycerides: 99.9% C.I. threshold, 7.48; 99% C.I. threshold, 4.95; 95% C.I. threshold, 3.77. (E) Male protein: 99.9% C.I. threshold, 9.06; 99% C.I. threshold, 6.00; 95% C.I. threshold, 4.56. (F) Female protein: 99.9% C.I. threshold, 13.86; 99% C.I. threshold, 9.17; 95% C.I. threshold, 6.98.
TABLE 6.
P-element insertion lines having at least one effect >99% C.I. on glycogen, triglycerides, or protein in the initial test
| Line | Nearest gene | Cytological location | P[GT1] position relative to nearest gene | Phenotype | ♀ effect (min) | ♂ effect (min) |
|---|---|---|---|---|---|---|
| BG00357 | Guanine nucleotide exchange factor | 64B13 | 1.1 kb from 5′-end | Protein | 1.38 (NS) | −9.17*** |
| BG00737 | Heat shock protein 27 | 67B3 | 10 bp from 5′-end | Protein | −11.11** | −1.83 (NS) |
| BG01020 | Unknown | Glycogen | −41.73*** | −19.15** | ||
| Triglycerides | −5.52** | −0.02 (NS) | ||||
| Protein | −12.85** | 0.67 (NS) | ||||
| BG01127 | muscleblind | 54B1 | 150 bp in gene (first intron) | Triglycerides | −6.55** | −0.42 (NS) |
| Protein | −7.74* | 0.53 (NS) | ||||
| BG01130 | alan shepard | 64C11 | 15 bp from 5′-end | Glycogen | −10.22 (NS) | 23.45** |
| Triglycerides | 0.46 (NS) | −3.83** | ||||
| BG01214 | sugarless | 65D5 | 100 bp from 5′-end | Glycogen | −25.64** | −0.16 (NS) |
| Triglycerides | −2.57 (NS) | −3.87** | ||||
| BG01296 | CG33691 | 6E2 | 300 bp from 5′ end | Glycogen | −28.49** | −11.00 (NS) |
| Triglycerides | −6.82** | −0.86 (NS) | ||||
| BG01340 | Unknown | Triglycerides | −5.09** | 0.35 (NS) | ||
| Protein | −9.76** | 1.88 (NS) | ||||
| BG01420 | CG14709 | 86E11 | 85 bp from 5′-end | Protein | −9.59** | −0.72* |
| BG01442 | Tenascin major | 79E3 | 60 bp from 5′-end | Glycogen | −28.46** | 8.95 (NS) |
| BG01478 | Heat shock RNAω | 93D4 | 600 bp from 5′-end | Protein | −9.31** | −3.13 (NS) |
| BG01575 | Unknown | Triglycerides | −6.64** | −1.34 (NS) | ||
| BG01602 | brother of odd with | 24C3 | 10 bp from 5′-end | Glycogen | −30.71** | −14.43 (NS) |
| entrails limited | Triglycerides | −5.79** | −1.33 (NS) | |||
| BG01623 | bunched | 33E7 | 39 kb in gene (fifth intron) | Triglycerides | −4.50* | 0.11 (NS) |
| Protein | −9.63** | 1.61 (NS) | ||||
| BG01635 | Unknown | Glycogen | −22.47* | −7.40 (NS) | ||
| Triglycerides | −6.36** | −0.97 (NS) | ||||
| Protein | −8.92* | 1.18 (NS) | ||||
| BG01662 | Laminin A | 65A9 | 60 bp from 5′-end | Glycogen | −25.92** | 4.81 (NS) |
| BG01672 | CG14591 | 42A8 | 25 bp from 3′-end | Glycogen | −22.93* | −1.89 (NS) |
| Triglycerides | −6.30** | −0.83 (NS) | ||||
| BG01822 | IGF-II mRNA-binding protein | 9F2 | 17 kb in gene (seventh intron) | Triglycerides | −6.52** | −0.43 (NS) |
| BG01836 | CG6854 | 71B4 | 600 bp in gene (first intron) | Glycogen | −21.87* | −1.04 (NS) |
| Triglycerides | −5.98** | 2.73* | ||||
| BG01887 | Unknown | Triglycerides | −1.90 (NS) | −3.01** | ||
| BG01888 | mir-279 | 99A1 | 1.4 kb from 5′-end | Glycogen | −27.05** | −9.10 (NS) |
| Triglycerides | −5.32** | −0.19 (NS) | ||||
| BG01914 | sugarless | 65D5 | 100 bp in gene (first exon) | Triglycerides | −4.74* | −2.19 (NS) |
| Protein | −12.11** | −3.05 (NS) | ||||
| BG01916 | no ocelli | 35B2 | 300 bp from 5′-end | Glycogen | −28.96** | −16.79* |
| Triglycerides | −4.77* | 0.05 (NS) | ||||
| BG01971 | escargot | 35D1 | 250 bp from 5′-end | Glycogen | −0.15 (NS) | −16.25* |
| Triglycerides | −1.08 (NS) | −2.63* | ||||
| Protein | −4.67 (NS) | −11.20*** | ||||
| BG01989 | visgun | 67C5 | 30 bp in gene (first exon) | Glycogen | −35.78** | −17.57* |
| Triglycerides | −5.02** | −0.36 (NS) | ||||
| BG02009 | taiman | 30A2 | 10 kb in gene (first intron) | Glycogen | −15.00 (NS) | −16.68* |
| Triglycerides | −6.12** | −0.90 (NS) | ||||
| BG02077 | Rtnl1 | 25B10 | 9.4 kb in gene (fourth intron) | Glycogen | −18.44 (NS) | −16.48* |
| Triglycerides | −2.67 (NS) | −3.32** | ||||
| BG02201 | Unknown | Glycogen | 23.96* | 23.24** | ||
| Triglycerides | 4.40* | 2.18 (NS) | ||||
| BG02207 | liliputian | 23C2 | 22 kb in gene (third intron) | Triglycerides | −3.24 (NS) | −3.44** |
| BG02210 | lamina ancestor | 64C13 | 550 bp in gene (first intron) | Glycogen | −31.43** | −15.35 (NS) |
| Triglycerides | −4.04* | −2.79* | ||||
| Protein | −9.03* | −4.27 (NS) | ||||
| BG02297 | escargot | 35D1 | 200 bp from 5′-end | Glycogen | −24.60** | 1.56 (NS) |
| Triglycerides | −4.53* | −2.79* | ||||
| Protein | −11.33** | −4.52 (NS) | ||||
| BG02491 | Unknown | Glycogen | −28.99** | 1.56 (NS) | ||
| Triglycerides | −6.11** | −0.35 (NS) |
Boldface type with underlining indicates that the insertion also has significant effects on sleep. Underlining indicates that alleles of this insertion have significant effects on sleep. See text for details. NS, nonsignificant; *significant at 95% C.I.; **significant at 99% C.I., ***significant at 99.9% C.I.
Although this screen was small, two genes were implicated twice: BG01214 and BG01914, which putatively map to sugarless, had significant effects on energy stores. The BG01214 P element, which is inserted 100 bp from the 5′-end of sugarless, decreased glycogen by 25.64 μg/fly in females and decreased triglycerides by 3.87 μg/fly in males. The BG01914 insertion, which is 100 bp within the first exon of sugarless, affected females only, decreasing triglycerides and protein by 4.74 and 12.11 μg/fly, respectively. Two alleles of escargot, BG01971 and BG02297, also had energy storage phenotypes. The BG01971 insertion, which is located 250 bp from the 5′-end of escargot, affected glycogen, triglycerides, and protein in males only, decreasing them by 16.25, 2.63, and 11.20 μg/fly, respectively. Interestingly, the BG02297 insertion is located 200 bp from the 5′-end of escargot and decreased glycogen and protein—but only in females. Triglycerides were significantly decreased in both sexes. A third insertion putatively affecting escargot, BG02419, was also tested in this study. This insertion maps 1 kb from the 5′-end of the gene and exhibited reduced 24-hr sleep in males and females, but no significant changes in energy stores were observed for this insertion.
One insertion listed in Table 6 also had a significant effect on 24-hr sleep: BG02009, which putatively maps to the first intron of taiman, had significantly increased sleep in both males (115.8 min) and females (187.2 min). This insertion also resulted in a decrease of 16.68 μg/fly of glycogen in males and a decrease of 6.12 μg/fly of triglycerides in females, relative to w1118; Canton-S.
Genetic correlations between energy stores:
The genetic correlations among glycogen, triglycerides, and protein were computed for both males and females. Genetic correlations between the energy storage traits were positive and significant, but they were not unity, indicating that some, but not all, genes are common to all three traits. The correlation between triglycerides and glycogen was 0.4198 (P < 0.0001) for males and 0.2989 (P = 0.0004) for females. The correlation between triglycerides and protein was 0.5395 (P < 0.0001) and 0.5561 (P < 0.0001) for males and females, respectively. In addition, the correlation between glycogen and protein was 0.7348 (P < 0.0001) for males and 0.1873 (P = 0.0290) for females. Interestingly, the correlations tended to be lower in females than in males. Thus, although mutational heritability was high for energy storage traits in females, implying high genetic variation among the insertion lines, the covariance between traits was relatively low, suggesting that the genetic variation observed for each trait stems from a different set of insertions.
The cross-sex genetic correlations for energy stores were also calculated (supplemental Table 3). Male and female energy stores were positively correlated, although the correlations were not unity. As with the cross-sex genetic correlations on sleep phenotypes, much of the line × sex variance component reflects sex-specific effects of the insertions on energy stores.
Correlations between sleep and energy stores:
To assess whether genes impacting endogenous sleep also affect triglyceride and glycogen stores, we calculated the mutational genetic correlations among sleep phenotypes, glycogen, and triglycerides. Table 7 shows the correlations for both males and females. These correlations show a striking sex-specific pattern. Glycogen stores were positively correlated with all measures of sleep duration in males. The significant positive correlation implies that males that sleep longer have higher levels of glycogen. Glycogen stores were also negatively correlated with sleep bout number and positively correlated with average sleep bout length in males. Thus, males with fragmented sleep tend to have lower levels of glycogen.
In female flies, however, the pattern was different. Neither glycogen nor triglycerides were correlated with sleep time in females. Instead, triglycerides were negatively correlated with 24-hr and nighttime bout number, but not with daytime bout number. In addition, triglycerides were positively correlated with average bout length in females. Females with fragmented sleep, therefore, would tend to have lower levels of triglycerides. These results indicate that a genetic connection is present between endogenous sleep and energy stores in flies and that it is sex specific. Importantly, neither males nor females exhibited significant correlations between energy storage measures and waking activity, indicating that these correlations are specific to sleep phenotypes.
DISCUSSION
P-element insertional mutagenesis was used to identify 21 novel candidate genes for 24-hr sleep time. Twenty-one of the 22 candidate insertions retested had significant effects on sleep upon retest, owing partly to the use of a high statistical power assay and partly to the relatively high broad-sense mutational heritability (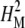 ) of 24-hr sleep time (see Tables 1 and 2). In wild-type lines, broad-sense heritability for sleep phenotypes appears to be higher than these estimates (S. T. Harbison and T. F. C. Mackay, unpublished results). Furthermore, human twin studies have estimated the narrow-sense heritability of 24-hr sleep time as 0.303 (De Castro 2002) and 0.44 (Partinen et al. 1983), and heritability of the number of wake-ups (i.e., 24-hr sleep bout number) as 0.256 (De Castro 2002). By definition, however, the broad-sense mutational heritability estimates in this study contain dominance and interaction variance (Falconer and Mackay 1996) and are likely higher than a narrow-sense estimate would be. Thirty-two candidate insertions that putatively disrupt genes impacting energy stores were also identified. Unlike sleep, broad-sense mutational heritability for energy stores was sexually dimorphic: females had much higher H2M than males for protein, glycogen, and triglycerides (Table 2). Thus, high levels of genetic variation for both sleep and energy storage phenotypes are present; female energy stores were more sensitive to genetic effects than were males.
) of 24-hr sleep time (see Tables 1 and 2). In wild-type lines, broad-sense heritability for sleep phenotypes appears to be higher than these estimates (S. T. Harbison and T. F. C. Mackay, unpublished results). Furthermore, human twin studies have estimated the narrow-sense heritability of 24-hr sleep time as 0.303 (De Castro 2002) and 0.44 (Partinen et al. 1983), and heritability of the number of wake-ups (i.e., 24-hr sleep bout number) as 0.256 (De Castro 2002). By definition, however, the broad-sense mutational heritability estimates in this study contain dominance and interaction variance (Falconer and Mackay 1996) and are likely higher than a narrow-sense estimate would be. Thirty-two candidate insertions that putatively disrupt genes impacting energy stores were also identified. Unlike sleep, broad-sense mutational heritability for energy stores was sexually dimorphic: females had much higher H2M than males for protein, glycogen, and triglycerides (Table 2). Thus, high levels of genetic variation for both sleep and energy storage phenotypes are present; female energy stores were more sensitive to genetic effects than were males.
Candidate insertions identified for 24-hr sleep time and energy stores have effects on genes involved in physiology, development, and behavior. Each P-element insertion is assumed to disrupt the function of the nearest identified candidate gene. To prove that the P-element insertions disrupt sleep phenotypes, they will need to be excised/reverted, tested by complementation to deficiencies or mutant alleles, and exhibit phenotypic rescue when a wild-type construct is introduced. The first step toward this proof has been taken through the creation of precise excision lines that restored the normal sleep phenotype from two P-element lines: BG02566 (Calreticulin) and BG01628 (malic enzyme).
We observed reduced sleep in males and females of the BG02566 (Calreticulin) line; precise excision of the P-element insertion restored wild-type sleep. Calreticulin has many proposed functions, including a role in neural development (Prokopenko et al. 2000; Norga et al. 2003), protein folding, and calcium signaling and homeostasis (Krause and Michalak 1997; Johnson et al. 2001). Calreticulin mutant flies have normal exploratory behavior and geotaxis (Stoltzfus et al. 2003), but are sensitive to ether anesthesia (Gamo et al. 1998). Furthermore, Calreticulin mutants exhibit impaired olfactory avoidance (Stoltzfus et al. 2003; Sambandan et al. 2006), yet respond normally to attractants (Stoltzfus et al. 2003). Calreticulin transcripts also increase in sleep-deprived flies relative to flies in recovery sleep after sleep deprivation (Williams et al. 2007). Another molecular chaperone, BiP, which acts in the same molecular pathway as Calreticulin (Johnson et al. 2001), was recently shown to be involved in sleep homeostasis. BiP protein increases after sleep deprivation and decreases with recovery sleep; however, transgenic animals overexpressing BiP or producing a dominant negative construct of BiP have wild-type sleep (Naidoo et al. 2007). The P[GT1] insertion in the first exon of Calreticulin reduced, but did not totally eliminate, whole-body transcript levels in both males and females. Other mutant alleles of Calreticulin tested in flies are not null mutations (Gamo et al. 1998; Stoltzfus et al. 2003); indeed, it may not be possible to create a Calreticulin null mutation in flies as homozygous Calreticulin null mice die in the early embryonic stages (Mesaeli et al. 1999). Future studies will determine what role Calreticulin has in sleep and waking.
Some of the candidate insertions identified have previously been implicated in metabolic pathways. We observed an increase in sleep in males and females of line BG01628, which has a P[GT1] insertion in the third exon of malic enzyme. Malic enzyme encodes a malate dehydrogenase, which functions in the citric acid cycle and malate metabolism. In a study of transcript abundance in the heads of wild-type flies, malic enzyme transcript increased significantly during both spontaneous wakefulness and sleep deprivation relative to sleep (Cirelli et al. 2005b). Malic enzyme whole-body transcript expression was essentially zero in BG01628 females. Sleep was reduced to wild-type levels when this P element was precisely excised, suggesting that malic enzyme has a role in sleep and waking. In addition to malic enzyme, other metabolic pathways were implicated in sleep: glycolysis (Horton et al. 1996) (BG02003, 6-phosphofructo-2-kinase), electron transport (BG01007, CG8776, and BG02419, escargot), and glucosamine metabolism (Haines and Irvine 2005) (BG02565, β4GalNAcTA). One insertion line had effects on both 24-hr sleep and energy stores in this experiment: the insertion in this line, BG02009, is located in the first intron of taiman. taiman is thought to be involved in signal transduction, activates transcription by binding to the ecdysone receptor, and is required for migration of border cells in the developing oocyte (Starz-Gaiano and Montell 2004). Both males and females of the BG02009 insertion line had increased 24-hr sleep; however, males had decreased glycogen, while females had decreased triglycerides. The observation that genes involved in metabolism may also have a function in sleep is intriguing as recent human studies have linked short sleep times to obesity and diabetes (reviewed in Cizza et al. 2005), indicating a potential molecular relationship between the two.
escargot encodes a zinc-finger protein with a role in the fusion of tracheal tubes (Zelzer and Shilo 2000; Ghabrial et al. 2003) and PNS development (Norga et al. 2003). Line BG02419, which has the P-element insertion 1 kb from the 5′-end of escargot, had significantly reduced sleep in both males and females (Table 3). We also observed a significant decrease in 24-hr sleep in BG01971 females at the 95% confidence interval; this insertion maps to 250 bp from the 5′-end of escargot. However, a significant effect on 24-hr sleep was not present in BG02297 flies, which have an insertion 200 bp from the 5′-end of escargot. Interestingly, male-specific decreases were observed in all three energy storage parameters in line BG01971, and line BG02297 had largely female-specific decreases (Table 6). No significant energy storage phenotypes were observed in BG02419. If each of these phenotypes maps to escargot, this observation would underscore the importance of the location and orientation of transposable element insertions in their effects on complex trait phenotypes (Rollmann et al. 2006). Merely changing the orientation of a single P-element insertion within an intergenic region can alter behavioral and physiological traits such as taste preference and life span (Rollmann et al. 2006).
To our knowledge, this is the first study to systematically analyze the genetic correlation among multiple sleep measures in flies. Numbers of sleep bouts can be thought of as the number of transitions from sleeping to waking states; abnormal numbers of transitions from waking to sleep are the hallmark of some sleep disorders in humans, such as sleep apnea and narcolepsy. Significant correlations between sleep times and sleep bout number exist in both males and females; however, the significant correlations were negative in males and tended to be positive in females. Thus, mutations that increase sleep bout number in both sexes may decrease sleep time in males and increase sleep time in females. Two interpretations of this result are possible. First, genetic perturbations that affect the transition between sleep and waking states will have opposing effects in males and females. Alternatively, the genetic basis of the forces that promote sleep and stimulate waking might be different in male and female flies. Interestingly, studies of human twins have revealed a positive correlation (0.183) between sleep time and number of wake-ups (bout number) (De Castro 2002), similar to the pattern observed in female flies.
We also examined the correlation between waking activity and sleep phenotypes (Table 5). The negative correlations that we observed between waking activity and sleep suggest a genetic relationship between both measures. The correlations were not unity, however; thus, mutations affecting sleep will not necessarily affect waking activity. A recent study of the gene Hyperkinetic illustrates this point: two separate Hyperkinetic alleles reduced sleep without affecting waking activity (Bushey et al. 2007). Furthermore, a recent study of EMS-induced mutants suggests that the correlation between waking activity and sleep is weak (Wu et al. 2008). Some of the lines screened in our study have been associated with effects on starvation resistance (Harbison et al. 2004); we speculate that genes affecting a physiological measure of this sort are more likely to affect both sleep and waking activity.
The mutational correlation between triglycerides and glycogen has been previously assessed in male flies and was high and positive (Clark et al. 1995). In our study, high positive correlations between triglycerides and glycogen were also found for both males and females. Yet the genetic correlations between males and females for energy stores were relatively low, implying that genes affecting energy stores are different in males and females (supplemental Table 3). Recent QTL studies support this notion, as sex-specific QTL for triglycerides (De Luca et al. 2005) and X-linked QTL for glycogen (Montooth et al. 2003) have been identified.
Mutations that affect endogenous sleep affect body metabolic stores as well. Male flies had positive genetic correlations between glycogen and sleep time. This would suggest that genes involved in male sleep behavior impact glycogen synthesis, storage, or expenditure. Glycogen stores in males were also negatively correlated with sleep bout number and positively correlated with sleep bout length. Taken together, these data imply that genes affecting glycogen levels in males affect total sleep time and sleep consolidation. This finding contrasts with that for females, whose glycogen stores were not correlated with any measure of sleep time; instead, triglycerides were negatively correlated with 24-hr and nighttime sleep bout number and positively correlated with average bout length, indicating that triglycerides are associated with sleep consolidation. For females, therefore, triglyceride levels may have a more critical relationship to sleep patterns, while male sleep is linked to glycogen.
Our correlation data suggest that the differences in male and female sleep patterns may be related to sex-specific differences in metabolic needs (Montooth et al. 2003; Morgan et al. 2003; De Luca et al. 2005). The relatively high genetic correlations that we observed between sleep and energy stores may reflect the fact that large metabolic changes occur with high probability with the introduction of single P-element insertions in flies, implying that large numbers of genes influence metabolism (Clark et al. 1995). We found that our correlations were driven by insertions having effects that were greater than or less than the 95% confidence interval. Few insertions had large effects on both sleep and energy stores simultaneously, however. Thus, the high correlations that we observed may also indicate that large numbers of genes impact sleep behavior.
Our correlations suggest that increases in whole-body glycogen and triglycerides are associated with an increase in sleep or sleep consolidation. This is in contrast to the human association data mentioned previously, which generally indicate that decreased sleep is associated with obesity (reviewed in Cizza et al. 2005), although several studies have noted a link between considerably longer sleep times and obesity (Kripke et al. 2002; Taheri et al. 2004) and diabetes (Patel et al. 2004). The decreased sleep in the human data could have resulted from chronic sleep deprivation rather than endogenous sleep need. Further studies will determine which genes play a combined role in endogenous sleep and energy stores, and whether or not these pathways have been conserved across taxa.
Pleiotropy has become a recurring theme in behavioral analyses. Four other studies examined these same P-element insertion lines for bristle number (Norga et al. 2003), starvation resistance (Harbison et al. 2004), olfaction (Sambandan et al. 2006), and aggression (Edwards et al. 2006). Table 8 shows the overlap between candidate alleles for these traits, energy stores (this study), and the alleles identified for 24-hr sleep in this study. Table 8 provides clear evidence for pleiotropy as lines bearing the same mutation (i.e., alleles) have effects on as many as four different phenotypes. Additional evidence for pleiotropy stems from the genetic correlation analysis. The relatively high correlations observed between sleep phenotypes suggest that many of the same genes will affect different aspects of sleep. However, none of the correlations between sleep phenotypes are unity, implying that multiple molecular pathways impact sleep. Thus, when considering sleep, specificity of the candidate genes appears to be the exception, not the rule (Greenspan 2001). However, sleep can be dissected from other phenotypes in a pleiotropic gene. The escargot insertion lines suggest that both sleep and energy stores can be affected by different locations of the P element and that the effects of the insertion location can be sex specific. Thus, although many genes are pleiotropic, subtle mutations allow the identification and separation of polymorphisms that are specific to each phenotype (Sokolowski 2001). This separation was previously achieved for life-history traits in natural populations of D. melanogaster: independent polymorphisms in the gene Catsup have effects on life span, bristle number, and locomotion (Carbone et al. 2006) and may be maintained by balancing selection. A similar situation between sleep and other important traits in natural populations, if present, would potentially explain why genetic variation for sleep exists.
TABLE 8.
Sleep alleles from this study that also have bristle number, olfaction, starvation resistance, aggression, and energy storage phenotypes
| Line | Nearest gene | Effect on sleep (this study) | Bristle no.a | Olfactory avoidanceb | Starvation resistancec | Aggressiond | Energy stores (this study) |
|---|---|---|---|---|---|---|---|
| BG00080 | Unknown | ↑ | ↑ | ||||
| BG01037 | βνintegrin | ↓ | ↓ | ||||
| BG01245 | Sema-5C | ↑ | ↓ | ↑♂ | |||
| BG01491 | tramtrack | ↑ | ↑ | ↓ | ↓♂ | ||
| BG01536 | Beadex | ↑ | ↑ | ||||
| BG02009 | taiman | ↑ | ↓G♂, ↓T♀ | ||||
| BG02439 | CG32556 | ↓ | ↓ | ||||
| BG02566 | Calreticulin | ↓ | ↓ | ↓ |
↑, effect was increased relative to control. ↓, effect was decreased relative to control. G, glycogen; T, triglycerides.
As stated previously, an isogenic background was employed to study the effects of single-gene mutations. Although the number of lines surveyed was small (136), the number of insertions having quantitative effects (from 0.5 to 1σ) (Falconer and Mackay 1996) on 24-hr sleep was quite large—21. This finding suggests the presence of a large mutational target for sleep; only a few such mutations of opposite effect would be needed to mask a more severe mutation, as was observed in the background of long-standing mutations in Shaker (Cirelli et al. 2005a). This phenomenon may explain why the function of sleep, which up until recently was studied in largely wild-type populations of humans and mammals, remains elusive.
Acknowledgments
The authors thank Trudy Mackay for the P-element insertion lines, Akihiko Yamamoto for the third chromosome balancer stock used in the construction of revertant lines, and Mike Magwire for the annotation of the P-element lines. S.T.H. is the recipient of a National Sleep Foundation Pickwick Fellowship and National Institutes of Health training grant T32 HL07713. A.S. is an investigator of the Howard Hughes Medical Institute.
References
- Amlaner, C. J., and N. J. Ball, 1994. Avian sleep, pp. 81–94 in Principles and Practice of Sleep Medicine, edited by M. H. Kryger, T. Roth and W. C. Dement. W. B. Saunders, Philadelphia.
- Andretic, R., and P. J. Shaw, 2005. Essentials of sleep recordings in Drosophila: moving beyond sleep time. Methods Enzymol. 393 759–772. [DOI] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP Gene Disruption Project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benington, J. H., and H. C. Heller, 1995. Restoration of brain energy metabolism as the function of sleep. Prog. Neurobiol. 45 347–360. [DOI] [PubMed] [Google Scholar]
- Berger, R. J., and N. H. Phillips, 1995. Energy conservation and sleep. Behav. Brain Res. 69 65–73. [DOI] [PubMed] [Google Scholar]
- Bradford, M. M., 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Bushey, D., R. Huber, G. Tononi and C. Cirelli, 2007. Drosophila hyperkinetic mutants have reduced sleep and impaired memory. J. Neurosci. 27 5384–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone, M. A., K. W. Jordan, R. F. Lyman, S. T. Harbison, J. Leips et al., 2006. Phenotypic variation and natural selection at Catsup, a pleiotropic quantitative trait gene in Drosophila. Curr. Biol. 16 912–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli, C., D. Bushey, S. Hill, R. Huber, R. Kreber et al., 2005. a Reduced sleep in Drosophila Shaker mutants. Nature 434 1087–1092. [DOI] [PubMed] [Google Scholar]
- Cirelli, C., T. M. LaVaute and G. Tononi, 2005. b Sleep and wakefulness modulate gene expression in Drosophila. J. Neurochem. 94 1411–1419. [DOI] [PubMed] [Google Scholar]
- Cizza, G., M. Skarulis and E. Mignot, 2005. A link between short sleep and obesity: building the evidence for causation. Sleep 28 1289–1296. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., and L. E. Keith, 1988. Variation among extracted lines of Drosophila melanogaster in triacylglycerol and carbohydrate storage. Genetics 119 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., L. Wang and T. Hulleberg, 1995. P-element-induced variation in metabolic regulation in Drosophila. Genetics 139 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro, J. M., 2002. The influence of heredity on self-reported sleep patterns in free-living humans. Physiol. Behav. 76 479–486. [DOI] [PubMed] [Google Scholar]
- De Luca, M., N. Yi, D. B. Allison, J. Leips and D. M. Ruden, 2005. Mapping quantitative trait loci affecting variation in Drosophila triacylglycerol storage. Obes. Res. 13 1596–1605. [DOI] [PubMed] [Google Scholar]
- Edwards, A. C., S. M. Rollmann, T. J. Morgan and T. F. C. Mackay, 2006. Quantitative genomics of aggressive behavior in Drosophila melanogaster. PLoS Genet. 2 1386–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics, Ed. 4. Addison-Wesley/Longman, Harlow, UK.
- Gamo, S., K. Dodo, H. Matakatsu and Y. Tanaka, 1998. Molecular genetical analysis of Drosophila ether sensitive mutants. Toxicol. Lett. 100 329–337. [DOI] [PubMed] [Google Scholar]
- Ganguly-Fitzgerald, I., J. Donlea and P. J. Shaw, 2006. Waking experience affects sleep need in Drosophila. Science 313 1775–1781. [DOI] [PubMed] [Google Scholar]
- Ghabrial, A., S. Luschnig, M. M. Metzstein and M. A. Krasnow, 2003. Branching morphogenesis of the Drosophila tracheal system. Annu. Rev. Cell Dev. Biol. 19 623–647. [DOI] [PubMed] [Google Scholar]
- Greenspan, R. J., 2001. The flexible genome. Nat. Rev. Genet. 2 383–387. [DOI] [PubMed] [Google Scholar]
- Haines, N., and K. D. Irvine, 2005. Functional analysis of Drosophila β1,4-N-acetylgalactosaminyltransferases. Glycobiology 15 335–346. [DOI] [PubMed] [Google Scholar]
- Harbison, S. T., A. H. Yamamoto, J. J. Fanara, K. K. Norga and T. F. C. Mackay, 2004. Quantitative trait loci affecting starvation resistance in Drosophila melanogaster. Genetics 166 1807–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartse, K., 1994. Sleep in insects and nonmammalian vertebrates, pp. 95–104 in Principles and Practice of Sleep Medicine, edited by M. H. Kryger, T. Roth and W. C. Dement. W. B. Saunders, Philadelphia.
- Hendricks, J. C., S. M. Finn, K. A. Panckeri, J. Chavkin, J. A. Williams et al., 2000. Rest in Drosophila is a sleep-like state. Neuron 25 129–138. [DOI] [PubMed] [Google Scholar]
- Hendricks, J. C., J. A. Williams, K. A. Panckeri, D. Kirk, M. Tello et al., 2001. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat. Neurosci. 4 1108–1115. [DOI] [PubMed] [Google Scholar]
- Hendricks, J. C., S. Lu, K. Kume, J. C.-P. Yin, Z. Yang et al., 2003. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J. Biol. Rhythms 18 12–25. [DOI] [PubMed] [Google Scholar]
- Ho, K. S., and A. Sehgal, 2005. Drosophila melanogaster: an insect model for fundamental studies of sleep. Methods Enzymol. 393 772–793. [DOI] [PubMed] [Google Scholar]
- Horton, H. R., L. A. Moran, R. S. Ochs, J. D. Rawn and K. G. Scrimgeour, 1996. Principles of Biochemistry. Prentice-Hall, Upper Saddle River, NJ.
- Huber, R., S. L. Hill, C. Holladay, M. Biesiadecki, G. Tononi et al., 2004. Sleep homeostasis in Drosophila melanogaster. Sleep 27 628–639. [DOI] [PubMed] [Google Scholar]
- Johnson, S., M. Michalak, M. Opas and P. Eggleton, 2001. The ins and outs of calreticulin: from the ER lumen to the extracellular space. Trends Cell Biol. 11 122–129. [DOI] [PubMed] [Google Scholar]
- Joiner, W. J., A. Crocker, B. H. White and A. Sehgal, 2006. Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441 757–760. [DOI] [PubMed] [Google Scholar]
- Krause, K.-H., and M. Michalak, 1997. Calreticulin. Cell 88 439–443. [DOI] [PubMed] [Google Scholar]
- Kripke, D. F., L. Garfinkel, D. L. Wingard, M. R. Klauber and M. R. Marler, 2002. Mortality associated with sleep duration and insomnia. Arch. Gen. Psychiatry 59 131–136. [DOI] [PubMed] [Google Scholar]
- Krueger, J. M., F. Obal, K. Kapas and J. Fang, 1995. Brain organization and sleep function. Behav. Brain Res. 69 177–185. [DOI] [PubMed] [Google Scholar]
- Kume, K., S. Kume, S. K. Park, J. Hirsh and F. R. Jackson, 2005. Dopamine is a regulator of arousal in the fruit fly. J. Neurosci. 25 7377–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan, M. W., J. D. Artiss, D. R. Strandbergh and B. Zak, 1983. A perioxidase-coupled method for the colorimetric determination of serum triglycerides. Clin. Chem. 29 538–542. [PubMed] [Google Scholar]
- Mesaeli, N., K. Nakamura, E. Zvaritch, P. Dickie, E. Dziak et al., 1999. Calreticulin is essential for cardiac development. J. Cell Biol. 144 857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth, K. L., J. H. Marden and A. G. Clark, 2003. Mapping determinants of variation in energy metabolism, respiration and flight in Drosophila. Genetics 165 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, L. M., S. M. Hampton, M. Gibbs and J. Arendt, 2003. Circadian aspects of postprandial metabolism. Chronobiol. Int. 20 795–808. [DOI] [PubMed] [Google Scholar]
- Naidoo, N., V. Casiano, J. Cater, J. E. Zimmerman and A. I. Pack, 2007. A role for the molecular chaperone protein BiP/GRP78 in Drosophila sleep homeostasis. Sleep 30 557–565. [DOI] [PubMed] [Google Scholar]
- Norga, K. K., M. C. Gurganus, C. L. Dilda, A. H. Yamamoto, R. F. Lyman et al., 2003. Quantitative analysis of bristle number in Drosophila mutants identifies genes involved in neural development. Curr. Biol. 13 1388–1397. [DOI] [PubMed] [Google Scholar]
- Partinen, M., J. Kaprio, M. Koskenvuo, P. Putkonen and H. Langinvainio, 1983. Genetic and environmental determination of human sleep. Sleep 6 179–185. [DOI] [PubMed] [Google Scholar]
- Patel, S. R., N. T. Ayas, M. R. Malhotra, D. P. White, E. S. Schernhammer et al., 2004. A prospective study of sleep duration and mortality risk in women. Sleep 27 440–444. [DOI] [PubMed] [Google Scholar]
- Prokopenko, S. N., Y. He, Y. Lu and H. J. Bellen, 2000. Mutations affecting the development of the peripheral nervous system in Drosophila: a molecular screen for novel proteins. Genetics 156 1691–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtschaffen, A., and B. M. Bergmann, 2002. Sleep deprivation in the rat: an update of the 1989 paper. Sleep 25 18–24. [DOI] [PubMed] [Google Scholar]
- Rechtschaffen, A., B. M. Bergmann, C. A. Everson, C. A. Kushida and M. A. Gilliland, 1989. Sleep deprivation in the rat: X. Integration and discussion of the findings. Sleep 12 68–87. [PubMed] [Google Scholar]
- Robertson, A., 1959. The sampling variance of the genetic correlation coefficient. Biometrics 15 469–485. [Google Scholar]
- Rollmann, S. M., M. M. Magwire, T. J. Morgan, E. D. Ozsoy, A. H. Yamamoto et al., 2006. Pleiotropic fitness effects of the Tre1-Gr5a region in Drosophila melanogaster. Nat. Genet. 38 824–829. [DOI] [PubMed] [Google Scholar]
- Sambandan, D., A. H. Yamamoto, J. J. Fanara, T. F. C. Mackay and R. R. H. Anholt, 2006. Dynamic genetic interactions determine odor-guided behavior in Drosophila melanogaster. Genetics 174 1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, P. J., C. Cirelli, R. J. Greenspan and G. Tononi, 2000. Correlates of sleep and waking in Drosophila melanogaster. Science 287 1834–1837. [DOI] [PubMed] [Google Scholar]
- Smith, C., 1995. Sleep states and memory processes. Behav. Brain Res. 69 137–145. [DOI] [PubMed] [Google Scholar]
- Sokolowski, M. B., 2001. Drosophila: genetics meets behaviour. Nat. Rev. Genet. 2 879–890. [DOI] [PubMed] [Google Scholar]
- Spiegel, K., R. Leproult and E. Van Cauter, 1999. Impact of sleep debt on metabolic and endocrine function. Lancet 354 1435–1439. [DOI] [PubMed] [Google Scholar]
- Spiegel, K., R. Leproult, M. L'Hermite-Baleriaux, G. Copinschi, P. Penev et al., 2004. a Leptin levels are dependent on sleep duration: relationships with sympathovagal balance, carbohydrate regulation, cortisol, and thyrotropin. J. Clin. Endocrinol. Metab. 89 5762–5771. [DOI] [PubMed] [Google Scholar]
- Spiegel, K., E. Tasali, P. Penev and E. Van Cauter, 2004. b Brief communication: sleep curtailment in healthy young men is associated with decreased leptin levels, elevated ghrelin levels, and increased hunger and appetite. Ann. Intern. Med. 141 846–850. [DOI] [PubMed] [Google Scholar]
- Starz-Gaiano, M., and D. J. Montell, 2004. Genes that drive invasion and migration in Drosophila. Curr. Opin. Genet. Dev. 14 86–91. [DOI] [PubMed] [Google Scholar]
- Stickgold, R., 2005. Sleep-dependent memory consolidation. Nature 437 1272–1278. [DOI] [PubMed] [Google Scholar]
- Stoltzfus, J. R., W. J. Horton and M. S. Grotewiel, 2003. Odor-guided behavior in Drosophila requires calreticulin. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 189 471–483. [DOI] [PubMed] [Google Scholar]
- Taheri, S., L. Lin, D. Austin, T. Young and E. Mignot, 2004. Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLoS Med. 1 210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi, G., and C. Cirelli, 2003. Sleep and synaptic homeostasis: a hypothesis. Brain Res. Bull. 62 143–150. [DOI] [PubMed] [Google Scholar]
- Williams, J. A., S. Sathyanarayanan, J. C. Hendricks and A. Sehgal, 2007. Interaction between sleep and the immune response in Drosophila: a role for the NFkB Relish. Sleep 30 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M. N., K. Koh, Z. Yue, W. J. Joiner and A. Sehgal, 2008. A genetic screen for sleep and circadian mutants reveals mechanisms underlying regulation of sleep in Drosophila. Sleep 31 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Q., W. J. Joiner and A. Sehgal, 2006. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr. Biol. 16 1051–1062. [DOI] [PubMed] [Google Scholar]
- Zelzer, E., and B.-Z. Shilo, 2000. Cell fate choices in Drosophila tracheal morphogenesis. BioEssays 22 219–226. [DOI] [PubMed] [Google Scholar]
- Zepelin, H., 1994. Mammalian sleep, pp. 69–80 in Principles and Practice of Sleep Medicine, edited by M. H. Kryger, T. Roth and W. C. Dement. W. B. Saunders, Philadelphia.
- Zimmerman, J. E., M. Mackiewicz, R. J. Galante, L. Zhang, J. Cater et al., 2004. Glycogen in the brain of Drosophila melanogaster: diurnal rhythm and the effect of rest deprivation. J. Neurochem. 88 32–40. [DOI] [PubMed] [Google Scholar]