Abstract
Proteins of the RAD52 epistasis group play an essential role in repair of some types of DNA damage and genetic recombination. In Schizosaccharomyces pombe, Rad22 (a Rad52 ortholog) has been shown to be as necessary for repair and recombination events during vegetative growth as its Saccharomyces cerevisiae counterpart. This finding contrasts with previous reports where, due to suppressor mutations in the fbh1 gene, rad22 mutants did not display a severe defect. We have analyzed the roles of Rad22 and Rti1, another Rad52 homolog, during meiotic recombination and meiosis in general. Both proteins play an important role in spore viability. During meiotic prophase I, they partially colocalize and partially localize to Rad51 foci and linear elements. Genetic analysis showed that meiotic interchromosomal crossover and conversion events were unexpectedly not much affected by deletion of either or both genes. A strong decrease of intrachromosomal recombination assayed by a gene duplication construct was observed. Therefore, we propose that the most important function of Rad22 and Rti1 in S. pombe meiosis is repair of double-strand breaks with involvement of the sister chromatids. In addition, a novel mating-type-related repair function of Rad22 specific to meiosis and spore germination is described.
HOMOLOGOUS recombination (HR) is an important process in DNA metabolism. It is the preferential pathway for the repair of various DNA lesions, including single-stranded gaps, double-stranded breaks (DSBs), and collapsed replication forks (reviewed in Rothstein et al. 2000). It is also essential for crossing over (CO) and hence the proper segregation of chromosomes during meiosis I (reviewed in Roeder 1997). In Saccharomyces cerevisiae, HR has been shown to involve replication protein A (RPA) and the proteins of the RAD52 epistasis group Mre11, Rad50, Rad51, Rad52, Rad54, Rad55, Rad57, Rad59, Rdh54 (also known as Tid1), and Xrs2 (reviewed in Symington 2002). The corresponding genes are well conserved. Mutants exhibit an increased sensitivity to DNA-damaging agents and defects in mitotic and meiotic recombination. Rad51 is the eukaryotic homolog of Escherichia coli RecA and is responsible for homologous DNA pairing and strand exchange through nucleofilament formation, the central activity of HR (Shinohara et al. 1992; Sung and Robberson 1995; Sung et al. 2003; Sauvageau et al. 2005). In Schizosaccharomyces pombe meiosis, it is required for high spore viability, as well as CO and gene conversion (Muris et al. 1997; Grishchuk and Kohli 2003). Dmc1, another RecA homolog, is meiosis specific and also important for recombination and progression through meiosis (Bishop et al. 1992; Grishchuk and Kohli 2003).
In S. cerevisiae, the rad52 mutants show the most severe phenotype of the epistasis group (Malkova et al. 1996). Rad52 interacts with Rad51 to promote the strand exchange activity of Rad51 in the presence of RPA (Sung 1997a; Benson et al. 1998; New et al. 1998). It associates with RPA and stimulates its replacement by Rad51, thus avoiding competition between the two proteins for binding of single-stranded DNA (Sugiyama and Kowalczykowski 2002). Two paralogues of Rad51, forming the Rad55/Rad57 heterodimer, fulfill a similar recombination mediator activity and stabilize the Rad51 nucleofilament (Sung 1997b). While it has been reported in mouse and chicken that Rad51 deletion results in cell death and an early arrest of embryonic development (Tsuzuki et al. 1996; Sonoda et al. 1998), paradoxically, Rad52−/− mice are viable and the cells exhibit only moderate sensitivity to DNA-damaging agents, as well as only a slight impairment in HR (Rijkers et al. 1998). A commonly accepted assumption proposes functional redundancy of Rad52 with other proteins in vertebrates (Yamaguchi-Iwai et al. 1998). However, the possibility of a more fundamental difference of the roles of Rad52 in different organisms cannot be excluded.
Previous results have suggested that most recombination pathways described in S. cerevisiae are conserved in S. pombe (Osman and Subramani 1998). In the fission yeast, two homologs of Rad52 have been identified: Rad22 and Rti1 (also known as Rad22B; see van den Bosch et al. 2001). These two proteins show sequence similarity (supplemental Figure A1), and a former study proposed that Rad22 is not required for all recombination pathways because of overlapping functions with Rti1 (van den Bosch et al. 2002). More recent work, however, has led to the conclusion that Rad22 is as critical for vegetative cell survival as its S. cerevisiae counterpart (Doe et al. 2004). This discrepancy has been attributed to the fact that rad22 mutant strains frequently acquire suppressor mutations. The suppressor gene was identified as the F-box helicase Fbh1 (Osman et al. 2005), which has also been found in vertebrates, but not in budding yeast (J. Kim et al. 2002; Kohzaki et al. 2007).
Rad52 and most of the members of the RAD52 epistasis group in S. cerevisiae have also been reported to be involved in the processing of collapsed and stalled replication forks caused by DNA lesions, such as single-stranded gaps (Merrill and Holm 1998; Kuzminov 2001). Another type of lesion, thought to initiate mating-type switching in S. pombe, results from a DNA modification at the mat1 locus, which then leads to recombination of the mat1 cassette with an intact donor cassette at mat2 or mat3 (Arcangioli 1998; Dalgaard and Klar 1999; Egel 2005). A rad22 mutant allele has been shown to cause deletions in the mating-type region (Ostermann et al. 1993). It is therefore likely that recombination events involved in mating-type switching depend on Rad22.
Since previous accounts of the meiotic defects of S. pombe rad22 and rti1 mutants were compromised by the fbh1 suppressor mutations mentioned above, we have assessed the roles of S. pombe Rad52 homologs in meiotic recombination in a controlled genetic background. Here, we demonstrate that both proteins are expressed during meiosis and partially colocalize during prophase by immunostaining of spread meiotic nuclei. Results on partial colocalization with Rad51 (also called Rhp51; see Jang et al. 1994) foci and linear elements (Rec10 and Hop1 proteins; see Lorenz et al. 2004) are also presented. Genetic analysis shows that, unlike in S. cerevisiae, most CO and noncrossover (NCO) events between homologs (interchromosomal recombination) do not depend on Rad22 and Rti1. On the other hand, intrachromosomal recombination (i.e., recombination between sister chromatids or between loci within a chromatid) measured in a heteroallelic duplication construct with the ade6 gene (Schuchert and Kohli 1988) is concluded to be strongly dependent on Rad22 and Rti1.
MATERIALS AND METHODS
Strains and media:
The general genetic methods and media have been described in Gutz et al. (1974) and Moreno et al. (1991). The S. pombe strains used in this work are listed in supplemental Table A1. Cells were grown at 30° and the crosses were carried out at 25°. Vegetative cells were cultured in rich liquid (YEL) and on solid (YEA) media. Crosses were performed on malt extract agar (MEA). For antibiotic resistance or auxotrophy determination, rich YEA or GMA and MMA minimal media were used. Nutritional supplements were added at 100 mg/liter. Liquid synthetic minimal media PM and PM-N (lacking NH4Cl) were used for meiotic time-course experiments. For media preparation, see Watanabe et al. (1988), Parisi et al. (1999), and Forsburg and Rhind (2006).
C-terminal tagging of the rti1 gene:
The PCR-targeting method described by Bähler et al. (1998) was used to fuse a 3HA tag to the C-terminal end of the rti1 gene. Briefly, 90-bp-long PCR primers were generated, consisting of 20 bp homologous to the kanMX6 module in pFA6a plasmids and 70 bp homologous to the targeted locus (the 3′-end of the rti1 sequence and the flanking region directly downstream, respectively, at positions 1449–1519 and 1533–1603, relative to the position of the start codon). The sequences were 5′-TGATCCTCAG-TCGGCAATGAGGTCGCGAGAAAACTACGATGCTACGGTGGATAAGAAAGCCAA-AAAAGGAGCTGAAGCTTCGTACGCTGCA-3′ and 5′-TAAACAAATCATTAGTCATAAAACAGAAAATACTTGGTAAAAAACAAGTTGCCAATCATCACATTTTGCCATAT-CATCGATGAATTCGA-GC-3′ for the upstream and downstream primers, respectively. The primers were designed to delete a region of 14 bp, including the stop codon of the rti1 gene, to avoid truncations of the fusion protein. PCR reactions were subsequently performed using these two primers and the pFA6a-3HA-kanMX6 plasmid as a template. The products contained the epitope tag and the kanMX6 G418-resistance cassette, flanked by two 70-bp regions homologous to the C-terminal end of the rti1 gene. They were used to transform the h− wild-type strain 972, and the transformants that had integrated the kanMX6 cassette were selected by plating on YEA plates supplemented with 100 mg/liter G418. The integration was confirmed by PCR and by sequencing of the junction region between the 3′-end of the rti1 gene and the 3HA cassette. The functionality of the tagged protein was tested by measuring spore viability and interchromosomal recombination frequencies at conventional test loci in wild-type and rad22Δ backgrounds.
Meiotic time courses and chromatin spreading:
The methods used for synchronous meiosis induction in h+/h− diploids, chromatin spreading, and immunostaining were the same as previously described (Bähler et al. 1993; Molnar et al. 2003; Lorenz et al. 2004). Briefly, 200 ml of PM diploid cultures were grown at 30° to a density of 107–108 cells/ml. The cells were then harvested, washed in H2O, and resuspended in 200 ml PM-N to induce meiosis. Samples of 5 ml were taken at hourly intervals for chromatin spreading, together with 2-ml samples that were used to check meiotic progression by DAPI staining of the nuclei and FACS analysis.
Chromatin spreading of the nuclei was performed as follows: cells of sporulating cultures were harvested and incubated at 30° for 25 min in 1 ml of spheroplasting enzyme solution consisting of 0.65 m KCl containing 1.5 mg/ml of lysing enzymes from Trichoderma harzianum (L-2265, Sigma, St. Louis), 0.2 mg/ml of zymolyase 100T (Seikagaku, Tokyo), 10 mm DTT, and a 25-fold-diluted stock solution of complete protease inhibitor cocktail (Roche). Twenty microliters of the spheroplast suspension was mixed on a clean microscope slide with 40 μl of a fixative solution (4% paraformaldehyde, 3.4% sucrose) and 80 μl of Lipsol detergent (LIP). Opening of the cells and subsequent spreading of the nuclear chromatin content was terminated after 30–45 sec by addition of 80 μl of the fixative solution. The slides were then air dried and stored at −20° for further use.
For immunostaining, the slides were washed three times in PBS + 0.05% Triton X-100. Excess liquid was drained and the slides were incubated overnight at room temperature with 20–25 μl of the primary antibody at the adequate dilution. The following primary antibodies were used at the indicated dilutions: mouse monoclonal antirecombinant Rad51 antibody at 1:50 (clone 51RAD01, NeoMarkers), guinea pig polyclonal anti-S. pombe-Hop1 antibody at 1:50 (described in Lorenz et al. 2004), rabbit polyclonal anti-Rec10 antibody at 1:200 (described in Lorenz et al. 2004), mouse monoclonal anti-HA antibody at 1:100 (clone 12CA5, Roche Applied Science), rabbit polyclonal anti-Rad22 and anti-Rti1 at 1:100 (described in van den Bosch et al. 2002). After incubation, the slides were washed as indicated above and incubated in the dark for at least 4 hr at room temperature with the appropriate Cy3-, FITC-, and Alexa-conjugated secondary antibodies. After a final washing step, the slides were mounted with ∼8 μl of Vectashield mounting medium (Vector Labs) containing 4′,6-diamino-2-phenylindole (DAPI).
Immunofluorescence was detected with the same microscope setup as for whole-cell immunostaining experiments and black-and-white pictures were taken using a Nikon DXM1200 digital camera. False color assignment and merging of the pictures was done using Photoshop CS2 (Adobe) and ImageJ v. 1.35s (NIH) software. For each time point, 50 spread nuclei were scored.
DAPI staining and FACS analysis:
To determine meiotic stages, cells were washed with water, and aliquots were mixed with an equal amount of DAPI (2.5 μg/ml). Cells showing one nucleus, two or more nuclei, or a horsetail nucleus were scored. At least 200 cells were examined at every time point.
For FACS analysis, samples were washed twice in 50 mm Na–citrate (pH 7.0). For time points 0, 1, and 2 hr the cell titer was determined by counting. Cells were diluted to 4 × 106 cells/ml. Twenty-five microliters of RNAse A (10 mg/ml) was added to 1-ml samples, which were incubated at 37° for 1 hr. The cells were then transferred to 5-ml Falcon tubes and stained with a 1-ml propidium iodide solution (2.5 μg/ml in 50 mm Na–citrate). Samples were stored on ice in the dark for 3 days to allow diffusion of propidium iodide. Before subjecting them to FACS (FACS Scan, Beckton-Dickinson), samples were sonicated at 10% intensity with a Branson sonifier for 8 × 0.5 sec with 0.5-sec breaks after each cycle. Data were analyzed with the CellQuest program (BD Biosciences).
Determination of spore viability and meiotic recombination frequencies:
The parental strains for each cross were grown to an OD600 of ∼0.5 (∼2–6 × 106 cells/ml, mid-logarithmic phase). For each parent, equal amounts of cells (∼4 × 107 cells) were then washed in H2O, mixed in 0.85% NaCl, plated onto MEA, and incubated at 25° for 3 days. Material from the MEA plates was then incubated overnight at 30° in a solution of 1:500 snail digestive enzymes (Helix pomatia gastric juice, Biosepra) and 100 mg/liter of lysing enzymes (lysing enzymes from T. harzianum, Sigma) to digest cells. The spores were harvested, counted, and then used to determine spore viability and recombination frequencies.
Spore viability was determined by plating 104–105 spores on YEA plates. The plates were incubated at 30° for 24–30 hr, and the number of germinating spores vs. dead spores was counted by microscopic scoring. Spores forming microcolonies of at least four cells were counted as living. A minimum of 400 spores or microcolonies were counted for each cross.
For intergenic recombination analysis, an appropriate dilution of spores was germinated on YEA plates at 30°. Spore colonies were randomly picked, grown on YEA master plates, and replica plated to supplemented GMA plates for genotype determination. The recombination frequency (RF) was calculated as the ratio of recombinant colonies vs. total colonies scored. Genetic distances in centimorgans (d) were calculated according to the formula d = −50 × ln(1−2 × RF) (Munz 1994).
For determination of intragenic recombination frequencies, appropriate dilutions of spores were plated on selective (GMA) and nonselective (YEA) plates and incubated at 30°. Prototrophic recombinants were counted as growing colonies on the GMA plates. Prototroph frequencies were expressed as the number of prototrophs per 106 viable spores (ppm). Viable spore titer was determined by plating on YEA.
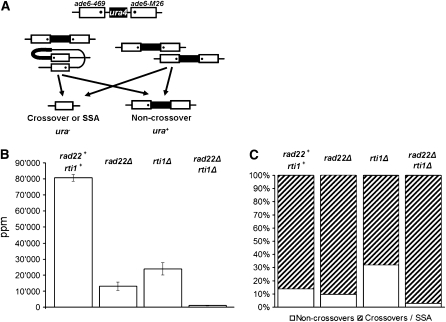
For determination of intrachromosomal recombination frequencies, appropriate strains carrying the heteroallelic ade6 duplication PS1 (Schuchert and Kohli 1988) (ade6-469 and ade6-M26, respectively) were crossed with strains carrying full deletions of the ade6 and ura4 genes (ade6-D20 and ura4-D18 alleles, respectively). Appropriate dilutions of spores were plated on selective (supplemented GMA lacking adenine) and nonselective (YEA) media and incubated at 30°. Recombinants prototrophic for adenine were counted and their number per 106 viable spores (ppm) was calculated. To determine the ratio between prototrophs still carrying the ura4+ marker and those that had lost it (Figure 8, A and C), 130 colonies were replicated on selective media (supplemented GMA lacking uracil) for each cross. Uracil auxotrophic and prototrophic recombinants were counted, and their relative percentages calculated.
Figure 8.—
Intrachromosomal recombination in single and double rad22Δ and rti1Δ mutants (A) Diagram of the construct used in the PS1 assay (Schuchert and Kohli 1988). Two repeats of the ade6 gene are separated by a copy of the ura4+ gene. One of the ade6 alleles carries the 469 mutation, the other the M26 mutation. Strains carrying the construct in single and double deletion backgrounds were constructed and then crossed with strains carrying a full deletion of the ade6 gene. In the progeny of such crosses, adenine prototrophic spores arise from unequal sister chromatid exchange (right) or from intrachromatid recombination events (left) of the duplication. (B) Prototrophic recombinant frequencies in the different genetic backgrounds. The bars represent the standard errors of the means. For primary data, see supplemental Table A6. (C) ade+ prototrophs from the four crosses were assayed for retention (open) vs. loss (shaded) of the ura4+ marker.
RESULTS
Vegetative growth phenotypes of strains carrying rad22 and/or rti1 deletions:
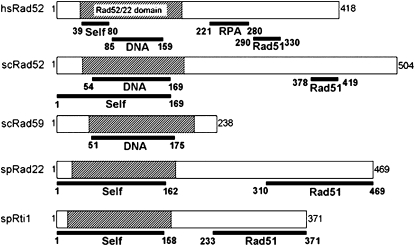
The two S. pombe proteins Rad22 and Rti1 show extensive similarity to each other and to Rad52 proteins from other organisms at both the nucleotide and the amino acid sequence levels. The two fission yeast proteins share 39% similarity and are identical for 30% of their amino acids. Rad22 shows 35% similarity and 24% identity with Rad52 of S. cerevisiae, while Rti1 shows 32 and 19%, respectively (supplemental Figure A1). Both fission yeast proteins display significant homology to Rad59, but unlike Rad59, both carry a C-terminal domain for interaction with Rad51 (Bai and Symington 1996; van den Bosch et al. 2002; Feng et al. 2007). Conservation is strongest in the N-terminal region, which contains the Rad52/22 homology domain common to all members of the RAD52 superfamily (Iyer et al. 2002), whereas the C-terminal ends of the proteins show more variability (Figure 1 and supplemental Figure A1). The N-terminal ends of Rad52 homologs are required for self-interaction, DNA binding, and annealing and thus play a key role in the function of these proteins (Asleson et al. 1999). The C-terminal regions are required for interaction with Rad51 and are more species specific (Milne and Weaver 1993; Kagawa et al. 2002; W. J. Kim et al. 2002) (Figure 1).
Figure 1.—
Protein domain alignment of Homo sapiens, S. cerevisiae, and S. pombe Rad52 homologs (van den Bosch et al. 2002; Bi et al. 2004). The full-length amino-acid sequences of the different genes are shown in supplemental Figure A1. Here the open reading frames are represented as open boxes, approximately to scale. The Rad52/22 homology domain required for protein multimer formation and interaction with DNA is indicated by hatching (Boundy-Mills and Livingston 1993; Asleson et al. 1999; Iyer et al. 2002; Singleton et al. 2002). The domains for interaction with RPA (Park et al. 1996; Hays et al. 1998; Gasior et al. 2001) and Rad51 (Milne and Weaver 1993; Song and Sung 2000) are located toward the C terminus. The end points of the various interaction domains are given below the boxes.
The mitotic generation times of rad22 and rad22 rti1 deletion mutants were found to be approximately twofold longer than in wild type. A large part of these mutant cells showed elongation and aberrant septation. None of these phenotypes were found in rti1Δ strains (data not shown; see van den Bosch et al. 2001; Osman et al. 2005). These observations suggest that the lack of Rad22 significantly slows cell cycle progression before the onset of M phase. Previous studies led to the same conclusion and showed that progression at late S to G2 phase was slowed or arrested (Suto et al. 1999).
The strains initially used for the study of genotoxin sensitivity were derived from previously published strains (van den Bosch et al. 2001) (supplemental Table A1) and showed low sensitivity to camptothecin, methyl methanesulfonate, and hydroxyurea (supplemental Figure A2) due to the accumulation of suppressor mutations. Suppressor mutations have previously been assigned to the fbh1 gene (Osman et al. 2005). After backcrossing with a wild-type strain, rti1 and unsuppressed rad22, as well as unsuppressed rad22 rti1 deletion mutants, were isolated. The unsuppressed rad22 and rad22 rti1 mutants displayed pronounced hypersensitivity to low doses of the above-mentioned DNA-damaging agents (supplemental Figure A2). However, the sensitivity of rti1Δ cells remained comparable to wild-type and suppressed strains. As expected, the deletion of fbh1 in a rad22Δ unsuppressed background restored damage resistance to wild-type levels. Interestingly, the fbh1Δ mutant also showed sensitivity to DNA-damaging agents, but only to higher doses (data not shown; see Morishita et al. 2005; Osman et al. 2005). These previous studies demonstrated that deletion of fbh1 impaired growth and viability, probably due to chromosome segregation defects. rad22 mutant strains gave rise to genotoxin-resistant derivatives after only a few rounds of subculturing. Careful checking of rad22 deletion strains was required to avoid suppressor accumulation.
Rad22 is expressed during mitotic growth and localizes to the nucleus:
The expression of Rad22 and Rti1 was analyzed during vegetative growth. The Rad22 protein was detected by Western blot analysis with polyclonal direct antibodies (van den Bosch et al. 2002) in wild-type cells and in rti1Δ mutants (data not shown), but no band was observed in rad22Δ and rad22Δ rti1Δ mutants, confirming the specificity of the antibody. In accordance with a previous report (van den Bosch et al. 2002), Western blot analysis did not detect Rti1 in vegetative cells (data not shown).
Next, the subcellular localization of the two proteins was analyzed by whole-cell immunostaining. In a wild-type strain, nuclear localization of Rad22 was observed (supplemental Figure A3A). A faint punctate pattern was also observed in the cytoplasm. In rad22Δ or rad22Δ rti1Δ mutants, these cytoplasmic structures were detected again, but no signal was in the nucleus. Therefore, the latter are considered to be unspecific background. When the Rti1-specific antibody was applied, no nuclear staining was detected, but punctate cytoplasmic staining was again observed (supplemental Figure A3B). In this case, it may also reflect unspecific binding of the primary or secondary antibodies to cytoplasmic structures. As an additional piece of evidence for the lack of Rti1 in vegetative nuclei, it should be mentioned that Rti1 foci were detected in spread meiotic nuclei (see below), while HA-tagged Rti1 was still not observed in spread nuclei 1 hr after the induction of meiosis (data not shown).
Taken together, these results indicate that Rad22 is expressed in vegetative cells and localizes to the nucleus, while Rti1 is not or is only weakly expressed.
Rad22 and Rti1 form nuclear foci during meiosis and partially colocalize:
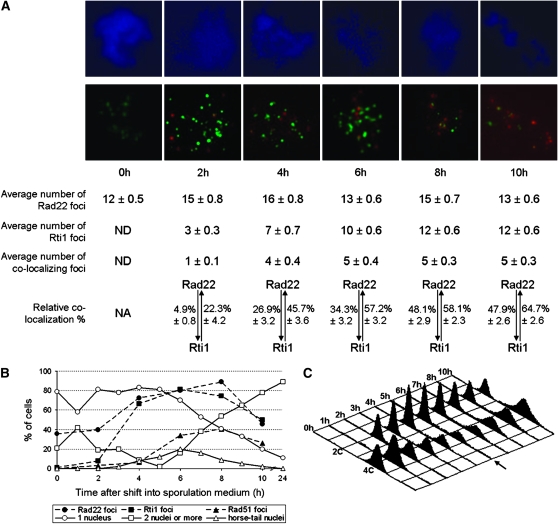
Chromatin spreads were prepared from diploid strains expressing wild-type Rad22 protein and an HA-tagged version of the Rti1 protein upon induction of meiosis. Distinct nuclear foci were observed after immunostaining of Rad22 and Rti1-HA (Figure 2A). To assess the meiotic stages of the cells that were positive for Rad22 and Rti1 immunostaining, parallel cell samples were stained with DAPI (Figure 2B). The lowest percentages of cells exhibiting two or more nuclei were counted at the time points 2–6 hr, corresponding to the stages of G1 after the last mitotic division and initiation of S phase. FACS analysis (Figure 2C) showed that the bulk of DNA synthesis occurred after 5 hr, the time point with the lowest 4C peak. Spread nuclei displaying foci of Rad22 and Rti1 were most frequent from 4 to 8 hr after the switch to sporulation medium (Figure 2B). This corresponds with meiotic prophase I, as indicated by the abundance of horsetail nuclei at these time points even though Rad22 and Rti1 foci already accumulate during meiotic S phase. No Rti1 foci were detected in spread meiotic chromatin at 0 hr and only very few at 2 hr, while Rad22 foci were detected in ∼40% of cells at 0 and 2 hr. Since a final mitotic division occurs ∼1 hr after meiosis induction (Doll et al. 2005), this observation confirms that Rad22, but not Rti1, is expressed during the mitotic cell cycle (see above). The number of Rad22 foci per nucleus was rather stable throughout the time course from 0 to 10 hr. It fluctuated between 12 and 16 on average (Figure 2A). For Rti1, the average number increased from zero foci at 0 hr to 12 foci at 8 and 10 hr (Figure 2A). When spread meiotic nuclei of an rti1Δ diploid were stained, no Rti1 foci were observed (data not shown). Thus, the Rad22 and Rti1 proteins are expressed during meiotic S phase and prophase I and localize in nuclear foci. Unfortunately, diploid strains homozygous for the rad22 deletion were not sufficiently stable for the performance of meiotic time-course experiments and visualization of foci.
Figure 2.—
Comparison of the localizations of Rad22 and Rti1 on immunostained meiotic chromatin spreads. (A) Pictures of spread nuclei representative of the different time points are shown. The chromatin was stained in blue by DAPI (top). Overlaid Rad22 and Rti1 foci appear in green and red, respectively (bottom). Fifty nuclei showing foci were analyzed per time point and the average number of Rad22, Rti1, and Rad22-Rti1 colocalization foci are given below the photos. No Rti1 foci were detected at 0 hr (ND). The respective colocalization percentages are also given, with the percentage of Rad22 foci that colocalized with Rti1 at the left of the double arrow, and the percentage of Rti1 foci that colocalized with Rad22 at the right. (B) Meiosis progression was assayed by DAPI staining. Cell samples were collected at the indicated times after induction of meiosis and stained with DAPI. The cells were classified with respect to the number and shape of their nuclei (one nucleus, open circles; two or more nuclei, open squares; horsetail nuclei, open triangles). The peak of cells with two nuclei at 1 hr is due to the last mitotic division before meiosis. At 5 hr, only 2% of the cells had more than two nuclei, indicating that the meiotic divisions occurred largely after this time point. The dashed lines indicate the percentage of nuclei containing at least one focus of Rad22 (solid circles), Rti1 (solid squares), and Rad51 (solid triangles). (C) The meiotic time courses performed for the immunostaining experiments showed the expected FACS profile (Doll et al. 2005). The cells started with a 4C DNA content (G2). After the last mitosis, they entered G1 (2C DNA content). The 4C peak increased again, when bulk DNA synthesis occurred in S phase. The arrow at 5 hr indicates the lowest number of cells with 4C DNA content.
The distribution pattern for Rad22 and Rti1 signals indicated partial colocalization, as determined from partial or total overlay of foci. The ratio of colocalizing foci increased early in the time course (Figure 2A): on average, only one colocalization focus per nucleus was observed 2 hr after meiotic induction, but four to five colocalization foci were found at 4 hr and later. A precise analysis of this colocalization pattern revealed that 2 hr after the shift into sporulation medium, on average only 4.9% of Rad22 foci also contained Rti1 (Figure 2A). This percentage increased to reach a peak of 48% at 8 hr. On the other hand, 22% of Rti1 foci were already colocalizing with Rad22 at 2 hr after meiotic induction, and this ratio increased to a maximum of 65% at 10 hr.
Rad22 and Rti1 colocalize with Rad51:
To check whether Rad22 and Rti1 proteins colocalize with Rad51 in S. pombe, as Rad52 does in S. cerevisiae (Shinohara et al. 1992; Milne and Weaver 1993; Gasior et al. 1998), an antibody directed against Rad51 was used in combination with the Rad22- and Rti1-specific antibodies described above (Figure 3). No Rad51 foci were observed in spread meiotic chromatin at meiosis induction (t = 0 hr), and only very few were observed during the last mitotic division and prior to prophase I (t = 2 and 4 hr) (Figure 2B). The majority of the foci were detected at 6–10 hr, during meiotic S phase and prophase I, with a maximum of 40% of the spread nuclei showing Rad51 at t = 8 hr. On average, 10–20 foci per Rad51-positive nucleus were observed from 6 to 10 hr after meiosis induction.
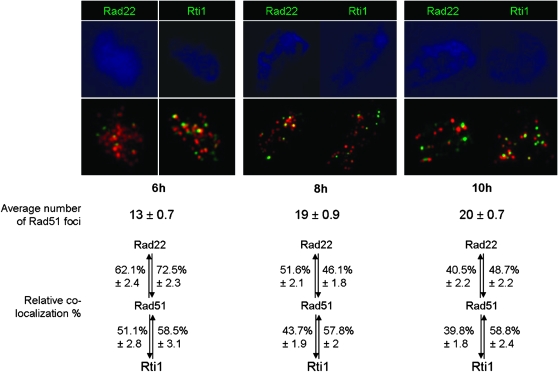
Figure 3.—
Colocalization of Rad51 with Rad22 and Rti1. The photos show representative spread nuclei, where the chromatin was stained in blue by DAPI and Rad51 foci appear in red. For each time point, Rad22 foci appear in green in the left photo and Rti1 foci in green in the right photo. Fifty nuclei were analyzed per time point, and the average number of Rad51 foci per nucleus is given below the photos. No Rad51 foci were detected at 0 hr, and only a few at 2 and 4 hr. The colocalization percentages of Rad51 with Rad22 and Rti1 are also given, with the percentage of Rad51 foci that colocalized with Rad22 at the left of the top double arrows, the percentage of Rad22 foci that colocalized with Rad51 at the right of the top double arrows, the percentage of Rad51 foci that colocalized with Rti1 at the left of the bottom double arrows, and the percentage of Rti1 foci that colocalized with Rad51 at the right of the bottom double arrows.
The foci of Rad51 colocalized to a fair extent with those of Rad22 and Rti1. The frequency of Rad51 colocalization with Rad22 was highest at 6 hr (72.5%) and then decreased to 45–50% at 8 and 10 hr. At the same time, the amount of Rti1 foci colocalizing with Rad51 remained stable at ∼58%.
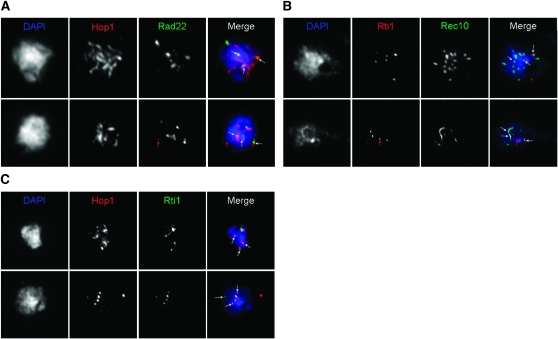
Rad22 and Rti1 localize to the linear elements:
S. pombe Rec10, a distant homolog of S. cerevisiae Red1, has been shown to be a component of linear elements, filamentous structures that are likely to be related to the axial/lateral elements of the synaptonemal complex in other eukaryotes (Molnar et al. 2003; Lorenz et al. 2004; Loidl 2006). Hop1 has been shown to associate with Rec10 and thus to be a marker for linear elements as well (Lorenz et al. 2004). Since several recombination proteins (including Rad51) localize to linear elements (Lorenz et al. 2004, 2006), the localizations of the Rad22 and Rti1 proteins was compared with those of Rec10 and Hop1.
The Rec10 and Hop1 immunosignals observed at 5 and 7 hr after the shift to sporulation medium (Figure 4) were similar to those observed in previous studies (Lorenz et al. 2004, 2006), revealing the most prevalent classes of linear elements: mostly interconnected networks and then threads and dots. The Hop1 staining (Figure 4, A and C) was more discontinuous, in comparison to Rec10 staining (Figure 4B). Hop1 staining was more often found in spread nuclei with long linear elements. In every case, virtually all the Rad22 and Rti1 foci colocalized with Rec10 or Hop1 staining (indicated by white arrows in the final picture of each row in Figure 4). However, not all of the linear elements were decorated with Rad22 or Rti1 foci, and the ratio of Rad22- and Rti1-containing linear elements vs. free linear elements was estimated to be ∼1:1 for both proteins. In rare cases, Rad22 or Rti1 foci were not localized to linear elements (indicated by red arrows).
Figure 4.—
Spread meiotic nuclei showing localization of Rad22 and Rti1 to linear elements. Chromatin was stained by DAPI (blue in the final photo of each row). (A) Localization of Hop1 (red) and Rad22 (green) in a representative “network” (7 hr, top) and a “thread” (7 hr, bottom) nucleus. (B) Localization of Rec10 (green) and Rti1 (red) in a “dot” (6 hr, top) and a “thread” (6 hr, bottom) nucleus. (C) Localization of Hop1 (red) and Rti1 (green) in a “thread” (5 hr, top) and a “dot” (5 hr, bottom) nucleus. For definition of the different types of spread nuclei, see text. Most of the Rad22 and Rti1 foci colocalized with linear elements (indicated by white arrows). Red arrows indicate Rad22 and Rti1 foci away from linear elements.
Deletion of rad22 and rti1 affects mating and meiosis and causes spore death:
As mentioned in the Introduction, previous studies have shown that rad22 mutants tend to rapidly acquire suppressor mutations in the fbh1 gene. Therefore, all the parental strains from the crosses described below were derived from newly constructed rad22 and rti1 deletion mutants (strains MCW1285 and MCW1686; see Osman et al. 2005 and supplemental Table A1) and carried an smt-0 deletion. The smt-0 deletion at the mat1 gene makes cells unable to perform mating-type switching by prevention of mat1 DNA breaks, which initiate the switching process (Klar 1990 and see below). The genotoxin sensitivity of the parents was then carefully checked during construction of the strains and before each experiment to ensure that suppression acquired during vegetative growth remained as low as possible.
The aberrant mitotic growth phenotypes and DNA-damage sensitivity of rad22Δ and rad22Δ rti1Δ strains were similar to those of rad51Δ (Grishchuk et al. 2004) and rad50Δ strains (Hartsuiker et al. 2001). Since mating efficiency was also reduced in rad51Δ (Grishchuk et al. 2004) and rad50Δ (Hartsuiker et al. 2001) strains, the mating efficiency of cells carrying single and double deletions of rad22 and rti1 was measured (supplemental Table A2). In a rad22Δ strain, mating efficiency was reduced ∼4-fold compared to wild type, while the rti1 mutants exhibited wild-type mating efficiency. The rad22 rti1 double mutant showed a stronger (10-fold) reduction in mating efficiency than the rad22 single mutant. Thus, although Rti1 seems to have a limited role in mitotic cells (supplemental Figure A2; van den Bosch et al. 2001, 2002), it has partially overlapping functions with Rad22 during the mating of cells.
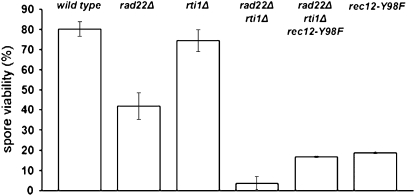
The viability of spores obtained from crosses homozygous for rad22Δ, rti1Δ, and the double mutant was measured using the microscopy method described in materials and methods (Figure 5). These measurements revealed a 2-fold reduction of spore viability in the rad22Δ mutant. The viability of rti1Δ spores remained at wild-type level. A striking 20-fold decrease of viability was observed in the rad22Δ rti1Δ mutant. These results indicate a crucial role for Rad22 and Rti1 in the production of viable spores.
Figure 5.—
Reduced spore viability in crosses homozygous for the rad22 deletion. The progeny of crosses involving wild-type parents (1-11 and 3-99), rad22Δ parents (GO123 and GO126), rti1Δ parents (GO179 and GO181), rad22Δ rti1Δ parents (GO228 and GO243), rad22Δ rti1Δ rec12-Y98F parents (GO247 and GO248), and finally rec12-Y98F parents (KLY144 and GO244) were tested. For details, see supplemental Table A3. The error bars represent the standard error of the mean. A 2-fold reduction of spore viability was observed for rad22Δ, while the rad22Δ rti1Δ double mutant displayed a 20-fold reduction.
In S. pombe, Rec12 is the homolog of the Spo11 protein required for formation of meiotic DSBs and recombination (Keeney et al. 1997; Cervantes et al. 2000). Therefore, the rec12-Y98F catalytic site mutation (described in Sharif et al. 2002) was crossed into a rad22Δ rti1Δ double-mutant background to test whether spore lethality depends on DSB formation. A fourfold increase of spore viability was observed in the triple mutant, to a level similar to that of rec12-Y98F mutants (Sharif et al. 2002). The very low spore viability in the rad22Δ rti1Δ double mutant thus depends on DSB formation and indicates that spore lethality at least partially arises from missing or aberrant processing of DSBs.
Deletion of rad22 and rti1 only slightly impairs meiotic recombination between homologous chromosomes:
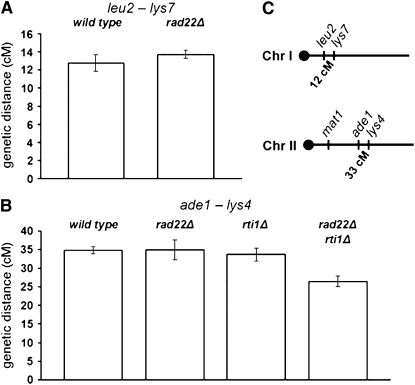
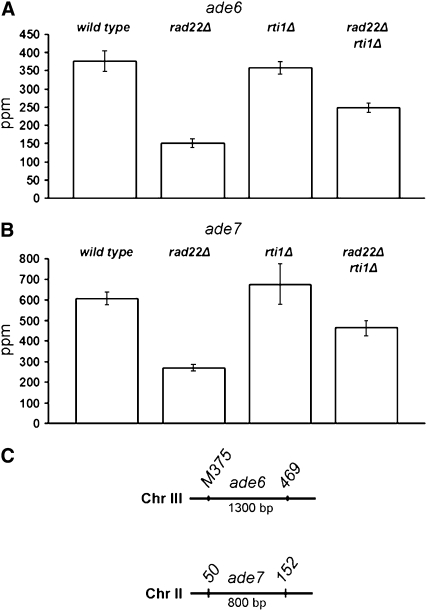
To clarify the contribution of Rad22 and Rti1 to meiotic recombination, auxotrophy markers were crossed into rad22 and rti1 deletion backgrounds, and the segregation of these markers was analyzed in the progeny by random spore analysis. Intergenic recombination in the leu2–lys7 (chromosome I) and ade1–lys4 (chromosome II) intervals was measured (Figure 6 and supplemental Table A4), as well as intragenic recombination at the ade6 locus between the ade6-M375 and ade6-469 alleles and at the ade7 locus between the ade7-50 and ade7-152 alleles (Figure 7 and supplemental Table A5).
Figure 6.—
Intergenic recombination in single and double rad22Δ and rti1Δ mutants. The genetic distance in centimorgans was calculated as indicated in materials and methods. The error bars represent the standard error of the mean. For primary data, see supplemental Table A4. (A) Genetic distances measured at the leu2–lys7 interval. (B) Genetic distances measured at the ade1–lys4 interval. (C) Approximate location of the intervals on chromosomes I and II.
Figure 7.—
Intragenic recombination in single and double rad22Δ and rti1Δ mutants. The number of prototrophic recombinants per 106 viable spores (ppm) was determined as indicated in materials and methods. The error bars represent the standard error of the mean. For primary data, see supplemental Table A5. (A) Conversion frequencies in crosses of the ade6-M375 and ade6-469 alleles. (B) Conversion frequencies were also measured between the ade7-50 and ade7-152 alleles. (C) Location of the ade6 and ade7 alleles in the respective genes on chromosomes II and III.
Surprisingly, the rad22 deletion showed no effect on intergenic recombination in the two intervals analyzed (Figure 6). This indicates that Rad22 plays no significant role in CO formation. In addition, no effect of the rti1 deletion was observed. Finally, deletion of rad22 and rti1 led to only a 1.3-fold reduction in the ade1–lys4 interval. Taken together, these results indicate that Rad22 and Rti1 play only a minor role in CO formation between homologous chromosomes.
With respect to intragenic recombination, the rad22 deletion exhibited a twofold reduction of intragenic recombination at the two loci analyzed (Figure 7). This result suggests that Rad22 also plays only a minor enhancing role in meiotic gene conversion. The rti1 deletion showed no effect. Deletion of both rad22 and rti1 genes led to a small reduction of intragenic recombination similar to that observed in the rad22 single mutant.
Reduction of intrachromosomal recombination in rad22 and rti1 mutants:
Since the reduction of recombination between homologs was small after deletion of rad22 and rti1, we asked whether Rad22 and Rti1 may be involved in other meiotic recombination processes, and assayed the effect of the deletions on intrachromosomal recombination. For that purpose, the heteroallelic duplication of the ade6 gene (Schuchert and Kohli 1988) was crossed into the rad22 deletion mutants (Figure 8A). Briefly, the construct consists of two repeats of the ade6 gene separated by a wild-type copy of the ura4 gene and is heterozygous over a full deletion of the ade6 gene. In a rad22+ background, an average recombination frequency of 80,800 ppm (Figure 8B) was observed. The parts-per-million values measured in rad22Δ and rti1Δ backgrounds were 6-fold and 3-fold lower, respectively. A synergistic 100-fold reduction was observed in the double mutant (Figure 8B; for detailed data, see supplemental Table A6).
The assay system used does not allow a distinction between unequal sister chromatid exchange and intrachromatid recombination, but NCOs can be distinguished from COs and single-strand annealing (SSA) events (Figure 8A and see Schuchert and Kohli 1988). A minority of ade+ recombinants (14%) were uracil-prototrophic NCOs in the rad22+ rti1+ cross (Figure 8C). An increase of NCOs was observed in the rti1Δ cross, while the rad22Δ cross was not significantly different from the wild-type cross. Deletion of both genes led to a fivefold decrease of NCOs.
Crosses heterozygous for the rad22 deletion lead to mating-type-specific death of the rad22 deletion progeny:
During the construction of the strains needed for the genetic experiments, some combinations of mating types and rad22 or rad22 rti1 double deletions were extremely difficult to obtain when only one of the parents was carrying the rad22 deletion. The rad22 gene is located on the left arm of chromosome I, while the mat genes are on the right arm of chromosome II, so this phenomenon could not be explained by genetic linkage. For further investigation, the mating-type frequencies of rad22Δ colonies resulting from heterozygous crosses were analyzed (Table 1).
TABLE 1.
Mating-type segregation of rad22Δ progeny in crosses heterozygous for rad22 and fbh1 deletions
| Genotype of the parents and strain designationsa | No. of rad22Δ colonies of each mating typeb
|
Total no. of colonies analyzed | |||
|---|---|---|---|---|---|
| Cross | h+ | h− | h90 | ||
| 1 | h+N rad22Δ (MCW1285) | 60 | 4 | 0 | 64 |
| h−Ssmt+ (EPY5) | |||||
| 2 | h−rad22Δc | 6 | 89 | 0 | 95 |
| h+N smt+ (EPY17) | |||||
| 3 | h+N rad22Δ (MCW1285) | 16 | 48 | 0 | 64 |
| h−smt-0 (EPY51) | |||||
| 4 | h+N fbh1Δ rad22Δ (MCW1553) | 25 | 38 | 1 | 64 |
| h−Ssmt+ (EPY5) | |||||
| 5 | h+N fbh1Δ rad22Δ (MCW1553) | 30 | 32 | 2 | 64 |
| h−smt-0 (EPY51) | |||||
All rad22Δ strains were not characterized with respect to the smt locus.
Since the deletion of rad22 is followed by the insertion of a ura4+ marker (complete genotypes are indicated in supplemental Table A1), progeny spores were germinated on uracil-containing minimal medium to select for rad22Δ colonies.
Strain obtained from cross 1.
The rad22Δ progeny of such crosses largely lacks the mating type of the rad22+ parent (Table 1, crosses 1 and 2). One of the rare rad22Δ colonies showing the underrepresented mating type (h− in cross 1) was then crossed with a wild-type strain. The resulting rad22Δ colonies again mostly showed the mating type of the rad22Δ parent, in this case h− (cross 2). To assess whether the observed mating-type discrepancy was related to initiation of mating-type switching, an h− rad22+ parent carrying a deletion of the smt region (smt-0) at mat1 was crossed with a rad22Δ h+ smt+ strain (Table 1, cross 3). From this cross, both mating types were obtained rather frequently. When similar experiments were carried out with a strain carrying a rad22 deletion and the suppressor fbh1Δ, the mating-type ratio of the rad22Δ progeny was normal, irrespective of the presence of the smt-0 deletion in the rad22+ parent (Table 1, crosses 4 and 5).
To verify the observed discrepancies of the frequency of mating types in crosses 1 and 3 of Table 1, 40 tetrads from each cross were dissected. In cross 1, homozygous for smt+, none of the rad22Δ colonies were h−, the mating type of the rad22+ parent. The mating-type ratio among the rad22+ progeny colonies did not deviate from 1:1. Only 5% of the tetrads had four viable spores; the remaining tetrads formed only two or three colonies. This was due to lethality of the h− rad22Δ and also to lethality of some of the h+ rad22Δ spores. The observation of the partial lethality of h+ rad22Δ spores indicates that the statistically significant but weaker bias observed in the random spore experiment (cross 3 in Table 1) most likely was not a real deviation from a 1:1 ratio, but rather an effect of this lethality. Forty-five percent of tetrads of cross 3 formed four colonies, and 17 h− rad22Δ segregants were recovered vs. 10 h+ rad22Δ segregants. The ratio of mating types among the rad22+ progeny was 1 to 1.
In summary, when the rad22 deletion is recombined into spores with a smt+ mating type derived from the rad22+ parent, the viability is strongly reduced. Microcolonies of two to four cells resulted from rad22Δ spores with the “lethal” smt+ mating type. Since the Rad22 protein is present throughout meiosis of crosses heterozygous for rad22Δ, it is proposed that a specific lesion causing lethality needs to be repaired at spore germination when Rad22 is missing in rad22Δ spores. The lethal lesion may form during DNA replication in the germinating spore.
DISCUSSION
Rad22 is important for the survival of vegetative cells and for successful meiosis, while Rti1 plays only a role in meiosis. Immunofluorescence microscopy revealed that the rise of Rti1 foci (Figure 2) coincides with the peak of mRNA concentration (Mata et al. 2002). The upregulation of rti1 expression in later prophase is paralleled by increased colocalization of Rti1 with Rad22. The substantial number of Rad22 and Rti1 foci not showing colocalization may be due to transient association, to independent functions at different sites, or to incomplete immunodetection. The early presence of Rad22 foci in meiosis is compatible with a role of Rad22 in meiotic S phase (see below).
At the time points with maximal numbers of Rti1 foci, the number of Rad51 foci exceeded that of Rad22 and Rti1 foci by about one-third (Figures 2 and 3). The observed number of foci colocalizing suggests that they may have contained all three proteins at a certain point or that a majority of Rad51 foci may have contained only one of the Rad52 homologs. Interaction between Rad22 and Rti1 has been reported (van den Bosch et al. 2002) and indicates that some Rad51-dependent repair mechanisms are enhanced by the presence of both Rad52 homologs.
The timing of appearance of Rad22 and Rti1 foci and their colocalization with linear elements is consistent with a role in DSB repair (Figure 4), similar to what was reported for Rad51 (Lorenz et al. 2006). However, sometimes Rad22 and Rti1 foci were not associated with linear elements (Figure 4). As previously suggested, this might be caused by linear elements disintegrating at a certain step or by late recombination intermediates being no longer associated with linear elements but occurring in the chromatin loops (Lorenz et al. 2006).
Gamete viability is the final readout for successful completion of all meiotic processes. Compared to wild type, deletion of rad22 and rti1 reduced spore viabilities to 44 and 88%, respectively. The double mutant retained only 4% viability, indicating that the Rad22 and Rti1 proteins partially substitute for each other (Figure 5).
Crossovers between homologous chromosomes are required for formation of intact gametes in many organisms. In S. cerevisiae, Rad52 is essential for formation of CO and NCO (Klein 1988). Thus, it was very surprising to find that Rad22 and Rti1 in S. pombe are largely dispensable for intergenic and intragenic recombination in meiosis (Figures 6 and 7). Our data agree with results obtained by another group at the ura4-aim2–his3-aim interval (Osman et al. 2003) for intergenic recombination and at ade6 for intragenic recombination (M. Whitby, personal communication). What other function of Rad22 and Rti1 then ensures high spore viability?
With help of an ade6 duplication construct (Schuchert and Kohli 1988; see Figure 8), intrachromosomal recombination, including unequal pairing of sister chromatids and intrachromatid events, was assessed. The occurrence of recombination between homologs was excluded by crossing the construct with an ade6 deletion strain. Significant reductions of prototrophic recombinants were detected in rad22Δ and rti1Δ crosses (Figure 8). The double mutant showed an even greater reduction, indicating that Rad22 and Rti1 partially substitute for each other. We suggest that Rad22 and Rti1 have a vital function in meiotic DNA repair involving sister chromatids.
Since we reckoned that spore lethality in the rad22 and rti1 mutants might be caused by a defect in DSB processing, experiments were performed with the rec12-Y98F mutation that abolishes meiotic DSB formation (Sharif et al. 2002). In this mutant, spore viability amounted to 22% (Figure 5). This still high spore viability in the absence of CO has been attributed to random segregation and backup systems for segregation of achiasmate chromosomes (Molnar et al. 2001; Sharif et al. 2002; Davis and Smith 2003). Introduction of rec12-Y98F into the rad22Δ rti1Δ double mutant restored spore viability to a level only 5% lower than that of the rec12-Y98F single mutant (Figure 5), strongly suggesting that Rad22 and Rti1 act downstream of DSB formation. This is consistent with the interpretation that functions of Rad22 and Rti1 not involved in the repair of Rec12-dependent DNA DSBs do not contribute much to overall spore viability. Such additional functions may involve repair of stalled replication forks in meiotic S phase (Figure 9), consistent with the role of Rad22 in vegetative S phase (Noguchi et al. 2004; Coulon et al. 2006).
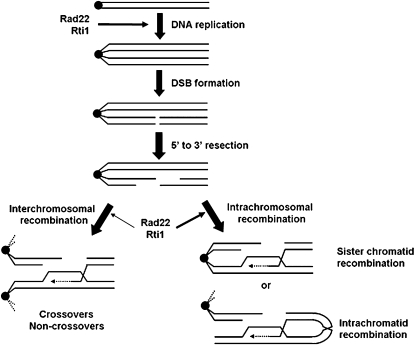
Figure 9.—
Hypothesis on the roles of Rad22 and Rti1 in S. pombe meiosis. The little understood early functions in meiotic S phase are proposed to contribute much less to spore viability than the roles in intrachromosomal repair of meiotic double-strand breaks. In contrast to S. cerevisiae Rad52, the role of Rad22 and Rti1 in meiotic recombination between homologous chromosomes is negligible. This parallels the observation of a preponderance of Holliday junctions between sister chromatids (Cromie et al. 2006).
The results described above led to the hypothesis that Rad22 and Rti1 contribute to meiotic DSB processing that largely does not result in CO formation, but in the repair of DSBs by interaction between sister chromatids (Figure 9). The identification of intrachromosomal recombination as the major meiotic function of Rad22 and Rti1 is in line with the observation that meiotic Holliday junctions in S. pombe involve sister chromatids four times more often than homologs (Cromie et al. 2006). A recent study has shown that truncated RAD52 polypeptides mimicking murine splicing variants are able to favor sister chromatid recombination in S. cerevisiae (Thorpe et al. 2006). In addition, we estimate that in a cell the overall number of HR events including repair involving sister chromatids largely exceeds the average observed number of 20 Rad51 foci (Figure 3). Forty-five CO/cell occur on average in S. pombe meiosis (Munz 1994), and an unknown number of intersister events essential for spore viability has to be added on the basis of this work. Therefore, it is reasonable to assume that Rad51 foci are transient or that several repair events occur in the same focus. Alternatively, some of the repair events may be catalyzed by Rad22, Rti1, or Rad51 proteins at low local concentrations not detectable as foci.
Rad22 is required for mating-type switching in mitotic cells (Ostermann et al. 1993). In crosses heterozygous for the rad22 deletion, most of the rad22Δ spores inheriting the mating type from the rad22+ parent were unable to form colonies (Table 1). We speculated that the few survivors with the underrepresented mating type might have resulted from mutation of the smt site. Tetrad analysis revealed that the bias was actually abolished in the cross with the rad22+ smt-0 strain. In addition, it was demonstrated that the lethality of the rad22Δ spores with the “wrong” mating type is Fbh1 dependent (Table 1). We propose (supplemental Figure A4) that a lethal lesion occurs at the smt+ site next to mat1 and that at spore germination Rad22 performs an essential repair function, as it does during mating-type switching in vegetative cells. Promotion of 3′-end invasion into one of the storage cassettes mat2 or mat3 on the same chromosome or on the homolog would be consistent with the biochemical function of Rad52 proteins (Sung 1997a; Benson et al. 1998; New et al. 1998; Sugiyama and Kowalczykowski 2002). Further work is required to elucidate this phenomenon.
In conclusion, this study has demonstrated that Rad22 and Rti1, the two S. pombe homologs of Rad52, have little effect on meiotic recombination between homologous chromosomes, but are important for meiotic DSB repair by intrachromosomal recombination. This function substantially contributes to spore viability. The situation in S. pombe meiosis indicates that considerable diversity exists within eukaryotes with respect to the functions of the Rad52 enzymes.
Acknowledgments
We thank Albert Pastink and Femke A. T. de Vries (University of Leiden) for the kind gift of Rad22- and Rti1-specific antibodies. We are also indebted to Matthew C. Whitby, Fekret Osman, and Weili Sun (University of Oxford) for exchange of results before publication and to Gerald Smith (Hutchinson Cancer Research Center, Seattle) and Scott Keeney (Sloane Kettering Institute, New York) for challenging discussions. Thanks also go to Jacob Dalgaard (Marie Curie Research Institute, Oxted, UK) for strains and advice with respect to mating-type switching and to Maria Siomos (Gregor Mendel Institute, Vienna) for critical reading of the manuscript. This work was supported by grants from the Swiss National Science Foundation and by grant P18186 from the Austrian Science Fund.
References
- Arcangioli, B., 1998. A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J. 17 4503–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asleson, E. N., R. J. Okagaki and D. M. Livingston, 1999. A core activity associated with the N terminus of the yeast RAD52 protein is revealed by RAD51 overexpression suppression of C-terminal rad52 truncation alleles. Genetics 153 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler, J., T. Wyler, J. Loidl and J. Kohli, 1993. Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J. Cell Biol. 121 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie, III et al., 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14 943–951. [DOI] [PubMed] [Google Scholar]
- Bai, Y., and L. S. Symington, 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10 2025–2037. [DOI] [PubMed] [Google Scholar]
- Benson, F. E., P. Baumann and S. C. West, 1998. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391 401–404. [DOI] [PubMed] [Google Scholar]
- Bi, B., N. Rybalchenko, E. I. Golub and C. M. Radding, 2004. Human and yeast Rad52 proteins promote DNA strand exchange. Proc. Natl. Acad. Sci. USA 101 9568–9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, D. K., D. Park, L. Xu and N. Kleckner, 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69 439–456. [DOI] [PubMed] [Google Scholar]
- Boundy-Mills, K. L., and D. M. Livingston, 1993. A Saccharomyces cerevisiae RAD52 allele expressing a C-terminal truncation protein: activities and intragenic complementation of missense mutations. Genetics 133 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes, M. D., J. A. Farah and G. R. Smith, 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5 883–888. [DOI] [PubMed] [Google Scholar]
- Coulon, S., E. Noguchi, C. Noguchi, L. L. Du, T. M. Nakamura et al., 2006. Rad22Rad52-dependent repair of ribosomal DNA repeats cleaved by Slx1-Slx4 endonuclease. Mol. Biol. Cell 17 2081–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie, G. A., R. W. Hyppa, A. F. Taylor, K. Zakharyevich, N. Hunter et al., 2006. Single Holliday junctions are intermediates of meiotic recombination. Cell 127 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard, J. Z., and A. J. Klar, 1999. Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature 400 181–184. [DOI] [PubMed] [Google Scholar]
- Davis, L., and G. R. Smith, 2003. Nonrandom homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics 163 857–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe, C. L., F. Osman, J. Dixon and M. C. Whitby, 2004. DNA repair by a Rad22-Mus81-dependent pathway that is independent of Rhp51. Nucleic Acids Res. 32 5570–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll, E., M. Molnar, Y. Hiraoka and J. Kohli, 2005. Characterization of rec15, an early meiotic recombination gene in Schizosaccharomyces pombe. Curr. Genet. 48 323–333. [DOI] [PubMed] [Google Scholar]
- Egel, R., 2005. Fission yeast mating-type switching: programmed damage and repair. DNA Repair 4 525–536. [DOI] [PubMed] [Google Scholar]
- Feng, Q., L. During, A. A. de Mayolo, G. Lettier, M. Lisby et al., 2007. Rad52 and Rad59 exhibit both overlapping and distinct functions. DNA Repair 6 27–37. [DOI] [PubMed] [Google Scholar]
- Forsburg, S. L., and N. Rhind, 2006. Basic methods for fission yeast. Yeast 23 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior, S. L., A. K. Wong, Y. Kora, A. Shinohara and D. K. Bishop, 1998. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 12 2208–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior, S. L., H. Olivares, U. Ear, D. M. Hari, R. Weichselbaum et al., 2001. Assembly of RecA-like recombinases: distinct roles for mediator proteins in mitosis and meiosis. Proc. Natl. Acad. Sci. USA 98 8411–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk, A. L., and J. Kohli, 2003. Five RecA-like proteins of Schizosaccharomyces pombe are involved in meiotic recombination. Genetics 165 1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk, A. L., R. Kraehenbuehl, M. Molnar, O. Fleck and J. Kohli, 2004. Genetic and cytological characterization of the RecA-homologous proteins Rad51 and Dmc1 of Schizosaccharomyces pombe. Curr. Genet. 44 317–328. [DOI] [PubMed] [Google Scholar]
- Gutz, H., H. Heslot, U. Leupold and N. Loprieno, 1974. Schizosaccharomyces pombe, pp. 395–446 in Handbook of Genetics. Plenum Press, New York.
- Hartsuiker, E., E. Vaessen, A. M. Carr and J. Kohli, 2001. Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J. 20 6660–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays, S. L., A. A. Firmenich, P. Massey, R. Banerjee and P. Berg, 1998. Studies of the interaction between Rad52 protein and the yeast single-stranded DNA binding protein RPA. Mol. Cell. Biol. 18 4400–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, L. M., E. V. Koonin and L. Aravind, 2002. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics 3 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, Y. K., Y. H. Jin, E. M. Kim, F. Fabre, S. H. Hong et al., 1994. Cloning and sequence analysis of rhp51+, a Schizosaccharomyces pombe homolog of the Saccharomyces cerevisiae RAD51 gene. Gene 142 207–211. [DOI] [PubMed] [Google Scholar]
- Kagawa, W., H. Kurumizaka, R. Ishitani, S. Fukai, O. Nureki et al., 2002. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol. Cell 10 359–371. [DOI] [PubMed] [Google Scholar]
- Keeney, S., C. N. Giroux and N. Kleckner, 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 375–384. [DOI] [PubMed] [Google Scholar]
- Kim, J., J. H. Kim, S. H. Lee, D. H. Kim, H. Y. Kang et al., 2002. The novel human DNA helicase hFBH1 is an F-box protein. J. Biol. Chem. 277 24530–24537. [DOI] [PubMed] [Google Scholar]
- Kim, W. J., E. J. Park, H. Lee, R. H. Seong and S. D. Park, 2002. Physical interaction between recombinational proteins Rhp51 and Rad22 in Schizosaccharomyces pombe. J. Biol. Chem. 277 30264–30270. [DOI] [PubMed] [Google Scholar]
- Klar, A. J., 1990. The developmental fate of fission yeast cells is determined by the pattern of inheritance of parental and grandparental DNA strands. EMBO J. 9 1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, H. L., 1988. Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics 120 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohzaki, M., A. Hatanaka, E. Sonoda, M. Yamazoe, K. Kikuchi et al., 2007. Cooperative roles of vertebrate Fbh1 and Blm DNA helicases in avoidance of crossovers during recombination initiated by replication fork collapse. Mol. Cell. Biol. 27 2812–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov, A., 2001. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. USA 98 8461–8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl, J., 2006. S. pombe linear elements: the modest cousins of synaptonemal complexes. Chromosoma 115 260–271. [DOI] [PubMed] [Google Scholar]
- Lorenz, A., J. L. Wells, D. W. Pryce, M. Novatchkova, F. Eisenhaber et al., 2004. S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J. Cell Sci. 117 3343–3351. [DOI] [PubMed] [Google Scholar]
- Lorenz, A., A. Estreicher, J. Kohli and J. Loidl, 2006. Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma 115 330–340. [DOI] [PubMed] [Google Scholar]
- Malkova, A., E. L. Ivanov and J. E. Haber, 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93 7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata, J., R. Lyne, G. Burns and J. Bahler, 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32 143–147. [DOI] [PubMed] [Google Scholar]
- Merrill, B. J., and C. Holm, 1998. The RAD52 recombinational repair pathway is essential in pol30 (PCNA) mutants that accumulate small single-stranded DNA fragments during DNA synthesis. Genetics 148 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne, G. T., and D. T. Weaver, 1993. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev. 7 1755–1765. [DOI] [PubMed] [Google Scholar]
- Molnar, M., J. Bahler, J. Kohli and Y. Hiraoka, 2001. Live observation of fission yeast meiosis in recombination-deficient mutants: a study on achiasmate chromosome segregation. J. Cell Sci. 114 2843–2853. [DOI] [PubMed] [Google Scholar]
- Molnar, M., E. Doll, A. Yamamoto, Y. Hiraoka and J. Kohli, 2003. Linear element formation and their role in meiotic sister chromatid cohesion and chromosome pairing. J. Cell Sci. 116 1719–1731. [DOI] [PubMed] [Google Scholar]
- Moreno, S., A. J. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe, pp. 795–823 in Methods in Enzymology. Vol. 194, edited by C. Guthrie and G. Fink. Academic Press, San Diego. [DOI] [PubMed]
- Morishita, T., F. Furukawa, C. Sakaguchi, T. Toda, A. M. Carr et al., 2005. Role of the Schizosaccharomyces pombe F-Box DNA helicase in processing recombination intermediates. Mol. Cell. Biol. 25 8074–8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz, P., 1994. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics 137 701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muris, D. F., K. Vreeken, H. Schmidt, K. Ostermann, B. Clever et al., 1997. Homologous recombination in the fission yeast Schizosaccharomyces pombe: different requirements for the rhp51+, rhp54+ and rad22+ genes. Curr. Genet. 31 248–254. [DOI] [PubMed] [Google Scholar]
- New, J. H., T. Sugiyama, E. Zaitseva and S. C. Kowalczykowski, 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391 407–410. [DOI] [PubMed] [Google Scholar]
- Noguchi, E., C. Noguchi, W. H. McDonald, J. R. Yates, III and P. Russell, 2004. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol. Cell. Biol. 24 8342–8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman, F., and S. Subramani, 1998. Double-strand break-induced recombination in eukaryotes. Prog. Nucleic Acid Res. Mol. Biol. 58 263–299. [DOI] [PubMed] [Google Scholar]
- Osman, F., J. Dixon, C. L. Doe and M. C. Whitby, 2003. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12 761–774. [DOI] [PubMed] [Google Scholar]
- Osman, F., J. Dixon, A. R. Barr and M. C. Whitby, 2005. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol. Cell. Biol. 25 8084–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermann, K., A. Lorentz and H. Schmidt, 1993. The fission yeast rad22 gene, having a function in mating-type switching and repair of DNA damages, encodes a protein homolog to Rad52 of Saccharomyces cerevisiae. Nucleic Acids Res. 21 5940–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, S., M. J. McKay, M. Molnar, M. A. Thompson, P. J. van der Spek et al., 1999. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol. Cell. Biol. 19 3515–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M. S., D. L. Ludwig, E. Stigger and S. H. Lee, 1996. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J. Biol. Chem. 271 18996–19000. [DOI] [PubMed] [Google Scholar]
- Rijkers, T., J. Van Den Ouweland, B. Morolli, A. G. Rolink, W. M. Baarends et al., 1998. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 18 6423–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, G. S., 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11 2600–2621. [DOI] [PubMed] [Google Scholar]
- Rothstein, R., B. Michel and S. Gangloff, 2000. Replication fork pausing and recombination or “gimme a break.” Genes Dev. 14 1–10. [PubMed] [Google Scholar]
- Sauvageau, S., A. Z. Stasiak, I. Banville, M. Ploquin, A. Stasiak et al., 2005. Fission yeast rad51 and dmc1, two efficient DNA recombinases forming helical nucleoprotein filaments. Mol. Cell. Biol. 25 4377–4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchert, P., and J. Kohli, 1988. The Ade6-M26 mutation of Schizosaccharomyces pombe increases the frequency of crossing over. Genetics 119 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif, W. D., G. G. Glick, M. K. Davidson and W. P. Wahls, 2002. Distinct functions of S. pombe Rec12 (Spo11) protein and Rec12-dependent crossover recombination (chiasmata) in meiosis I; and a requirement for Rec12 in meiosis II. Cell Chromosome 1 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara, A., H. Ogawa and T. Ogawa, 1992. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69 457–470. [DOI] [PubMed] [Google Scholar]
- Singleton, M. R., L. M. Wentzell, Y. Liu, S. C. West and D. B. Wigley, 2002. Structure of the single-strand annealing domain of human RAD52 protein. Proc. Natl. Acad. Sci. USA 99 13492–13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, B., and P. Sung, 2000. Functional interactions among yeast Rad51 recombinase, Rad52 mediator, and replication protein A in DNA strand exchange. J. Biol. Chem. 275 15895–15904. [DOI] [PubMed] [Google Scholar]
- Sonoda, E., M. S. Sasaki, J. M. Buerstedde, O. Bezzubova, A. Shinohara et al., 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama, T., and S. C. Kowalczykowski, 2002. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 277 31663–31672. [DOI] [PubMed] [Google Scholar]
- Sung, P., 1997. a Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272 28194–28197. [DOI] [PubMed] [Google Scholar]
- Sung, P., 1997. b Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11 1111–1121. [DOI] [PubMed] [Google Scholar]
- Sung, P., and D. L. Robberson, 1995. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82 453–461. [DOI] [PubMed] [Google Scholar]
- Sung, P., L. Krejci, S. Van Komen and M. G. Sehorn, 2003. Rad51 recombinase and recombination mediators. J. Biol. Chem. 278 42729–42732. [DOI] [PubMed] [Google Scholar]
- Suto, K., A. Nagata, H. Murakami and H. Okayama, 1999. A double-strand break repair component is essential for S phase completion in fission yeast cell cycling. Mol. Biol. Cell 10 3331–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington, L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe, P. H., V. A. Marrero, M. H. Savitzky, I. Sunjevaric, T. C. Freeman et al., 2006. Cells expressing murine RAD52 splice variants favor sister chromatid repair. Mol. Cell. Biol 26 3752–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki, T., Y. Fujii, K. Sakumi, Y. Tominaga, K. Nakao et al., 1996. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA 93 6236–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch, M., K. Vreeken, J. B. Zonneveld, J. A. Brandsma, M. Lombaerts et al., 2001. Characterization of RAD52 homologs in the fission yeast Schizosaccharomyces pombe. Mutat. Res. 461 311–323. [DOI] [PubMed] [Google Scholar]
- van den Bosch, M., J. B. Zonneveld, K. Vreeken, F. A. de Vries, P. H. Lohman et al., 2002. Differential expression and requirements for Schizosaccharomyces pombe RAD52 homologs in DNA repair and recombination. Nucleic Acids Res. 30 1316–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., Y. Lino, K. Furuhata, C. Shimoda and M. Yamamoto, 1988. The S.pombe mei2 gene encoding a crucial molecule for commitment to meiosis is under the regulation of cAMP. EMBO J. 7 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai, Y., E. Sonoda, J. M. Buerstedde, O. Bezzubova, C. Morrison et al., 1998. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell. Biol 18 6430–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]