Abstract
An association study conducted on 375 maize inbred lines indicates a strong relationship between Vgt1 polymorphisms and flowering time, extending former quantitative trait loci (QTL) mapping results. Analysis of allele frequencies in a landrace collection supports a key role of Vgt1 in maize altilatitudinal adaptation.
AMONG cereals, maize (Zea mays L.) is exceptional in its geographic distribution. Maize originated in tropical central Mexico and was spread during pre-Columbian times to cold temperate regions, with latitudes ranging from ∼40°S in Chile up to ∼45°N in Canada. This diffusion has been made possible via adaptation of flowering time to local environmental conditions, thus enabling the plant to reach the mature state within a shorter growing season. Linkage analyses have shown that flowering time variation involves numerous chromosomal regions, some of them displaying a major effect and often in diverse populations (Chardon et al. 2004). One of these major quantitative trait loci (QTL), namely Vegetative to generative transition 1 (Vgt1), located in bin 8.05, has been recently cloned using a map-based approach and assigned to a ∼2-kb noncoding region that regulates an APETALA2-like gene (ZmRap2.7) located ∼70 kb downstream (Salvi et al. 2007). The same study showed that three polymorphisms within Vgt1 were strongly associated with flowering time across a panel of 95 inbred lines. Reproducibility of association studies has proven to be a challenging issue in human genetics (Newton-Cheh and Hirschhorn 2005; Gorroochurn et al. 2007) but is not yet well-documented in plants. One exception is the relationship between the Dwarf8 gene and flowering time in maize addressed by Andersen et al. (2005) and Camus-Kulandaivelu et al. (2006), following the pioneering study by Thornsberry et al. (2001). These investigations showed that for traits and polymorphisms among highly differentiated genetic groups, sampling and evaluation of population structure are critical issues and can lead to contrasting results.
Because Vgt1 has been positionally cloned, it offers a unique opportunity to assess association mapping reliability in maize. The objectives of this study were therefore (i) to evaluate the reproducibility of the association between flowering time variation and molecular polymorphisms in the Vgt1 region using a large set of inbred lines specifically selected to include early materials and (ii) to investigate to what extent Vgt1 could have contributed to maize adaptation to temperate climates and possibly other adaptative constraints, by investigating a large and geographically dispersed set of open-pollinated varieties from America and Europe.
Association mapping and linkage disequilibrium in the Vgt1 region:
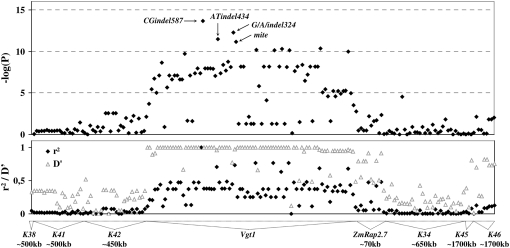
We identified 269 polymorphisms across 10 amplicons encompassing a region of 2.2 Mbp around the Vgt1 locus. The most significant associations with flowering-time variation evaluated under long days were obtained in Vgt1 itself: among the 81 tests performed using a mixed model, 67 gave a P-value < 1.10−2 (Figure 1). Three of the four most strongly associated polymorphisms corresponded to the ones highlighted by Salvi et al. (2007), including a miniature inverted-repeat transposable element (MITE) insertion. Additionally, a previously uncharacterized 2-bp indel, termed CGindel587, displayed the lowest P-value (P = 2.10−14) and explained 4% of the phenotypic variation not accounted for by population structure with the mixed model. This percentage increased to 17% when a general linear model (GLM) was applied. To further investigate which of these polymorphisms could underlie the Vgt1 effect, we performed association tests with CGindel587, ATindel434, G/A/indel324, or mite as a cofactor. We observed that CGindel587 remained significant (P < 1.10−3) when one of the other polymorphisms was included in the model, whereas the reverse was not true. Consistently, a haplotype-based analysis combining mite and CGindel587 led to a P-value of 4.10−13, higher than that of CGindel587 alone. Finally, one can note that including Dwarf8 in the model did not affect the significance levels and that no interaction was found between Dwarf8 and Vgt1.
Figure 1.—
Association of DNA polymorphisms with days to pollen shed (DPS) and extent of linkage disequilibrium in the vicinity of the Vgt1 region. Statistical significance is expressed as −log(P); LD is given as both r2 and D′ with CGindel587. Approximate positions of amplicons are given relative to Vgt1 according to the physical map (http://www.maizesequence.org). ATindel434, G/A/indel324, and mite correspond to the polymorphisms highlighted by Salvi et al. (2007). CGindel587 has been termed following the same nomenclature on the basis of the clustal alignment of N28 and its early derivative C22-4 (Salvi et al. 2007). The mapping population was composed of 375 inbred lines representative of the American and European diversity, with a wide range of flowering times, as previously described by Camus-Kulandaivelu et al. (2006). Association mapping analyses were performed using TASSEL version 2.0.1 (Bradbury et al. 2007). Polymorphisms with low allele frequency (<5%) were removed, leaving 181 of the 269 polymorphisms for analysis (see supplemental Materials and Methods for further details). Data presented here were obtained using a mixed linear model (MLM) approach accounting for both population structure and relatedness among individuals (YU et al. 2006). Estimation of population structure and Loiselle kinship coefficient (Loiselle et al. 1995) was performed using Structure (Pritchard et al. 2000) and SPAGeDi (Hardy and Vekemans 2002), respectively, using 55 tri- to hexa-nucleotide SSRs.
Outside Vgt1 itself, among the 11 polymorphisms considered in ZmRap2.7, 4 were significantly associated with flowering time at the P = 0.05 threshold (among which 2 were significant at the P = 0.01 threshold). Here again, the P-values were no longer significant when CGindel587 was included in the analysis as a cofactor.
Finally, nine significant associations (P < 0.01) were also found in amplicons located several hundreds of kilobases upstream or downstream of the Vgt1/ZmRap2.7 region; seven of them remained significant at a P = 0.05 threshold when CGindel587 was included in the analysis. Considering the false discovery rate calculated with Q-value software (Storey 2002) at a 5% level, five of these polymorphisms appeared as true positives, although it cannot be precluded that these significant tests may be due to a residual effect of population structure not fully accounted for by the mixed model we used, as discussed by Yu et al. (2006). The level of linkage disequilibrium (LD) was high (r2 > 0.7) over a distance of 1 kb at the Vgt1 locus, and r2 values >0.4 were observed between ZmRap2.7 polymorphisms and Vgt1 (Figure 1 and supplemental Figure 1). A modeling of LD, following the approach of Veyrieras et al. (2007), enabled us to distinguish between four ancestral haplotypes at Vgt1, which have undergone limited recombination (supplemental Figure 2). The most frequent haplotype combines all the early alleles at loci that display the strongest associations with earliness, including the MITE (absent in other ancestral haplotypes) and CGindel587. Another ancestral haplotype is characterized by the late allele at ATindel434, G/A/indel324, and mite and displays segregation at CGindel587, with 24 and 17 individuals bearing the early and late allele, respectively.
Congruence with QTL mapping results:
Flowering time has been the focus of several linkage studies or is often scored as a secondary trait. We investigated the congruence between QTL and association mapping at the Vgt1 locus on the basis of eight studies found in the literature (Table 1). Considering the significance levels reported above, we focused on the genotype at mite and CGindel587.
TABLE 1.
Comparison of QTL mapping studies with genotypes observed in parental lines
| Studya | Cross | Traitd | QTL at Vgt1 locus | R2(%)e | LODe | Additive effect (days)e/late parent | mitef | CGindel587f |
|---|---|---|---|---|---|---|---|---|
| Vladutu et al. (1999)b | N28 × E20 | DPS | Detected | NA | ∼15 | 2.82/N28 | +/− | +/− |
| Barriere et al. (2005) | F838 × F286 | SD | Detected | 28.8 | 17.8 | 2.3/F838 | +/− | +/− |
| Beavis et al. (1994) | B73 × Mo17 | DPS | Detected | 31 | 11.2 | NA/B73 | +/− | +/− |
| Blanc et al. (2006) | DE × F283 × F9005 × F810 | SD | Detected | 5 | NA | 0.4/F9005 | −/−/−/− | −/−/−/− |
| Austin and Lee (1996) | H99 × Mo17 | SD/DPS | Not detected | — | — | — | −/− | −/− |
| Poupard et al. (2001) | F2 × MBS847 | SD | Not detected | — | — | — | −/+ | −/− |
| Mechin et al. (2001) | F2 × MBS847 | SD | Not detected | — | — | — | −/+ | −/− |
| Bouchez et al. (2002)c | F2 × MBS847c | SD | Not detected | — | — | — | −/+ | −/− |
Other studies reported a QTL for days to flowering in the Vgt1 region (e.g., Abler et al. 1991; Koester et al. 1993; Tuberosa et al. 1997; Jiang et al. 1999) but, due to incomplete information, they cannot be included in our comparison.
Mean value of two environments.
In Bouchez et al. (2002) the iodent line (Io) was MBS847 (A. Charcosset, personal communication).
Days to pollen shed (DPS); silking date (SD).
NA, not available.
+, late allele; −, early allele, in parental line1/parental line2/…
Vgt1 was identified by Vladutu et al. (1999) in a cross between N28 and its early near-isogenic line, E20 (with Gaspé Flint allele at Vgt1). This study highlighted a large effect of Vgt1 (LOD ≈ 17). Two other studies revealed a QTL of large effect at Vgt1 in two different crosses: F838 × F286 (Barriere et al. 2005) and B73 × Mo17 (Beavis et al. 1994). The presence of a QTL at Vgt1 in these three crosses is consistent with the genotype observed both at mite and CGindel587, the early parents bearing the early alleles at these loci. Two studies conducted by Blanc et al. (2006) and Austin and Lee (1996) involved parental lines that were identical at both mite and CGindel587, which is consistent with no QTL of major effect being detected at Vgt1. Finally, no QTL has been reported in bin 8.05 in three distinct mapping populations involving F2 and MBS847 (Mechin et al. 2001; Poupard et al. 2001; Bouchez et al. 2002). These inbred lines differ at mite but share the same allele at CGindel587.
Relationship between allele frequencies and geographical origin:
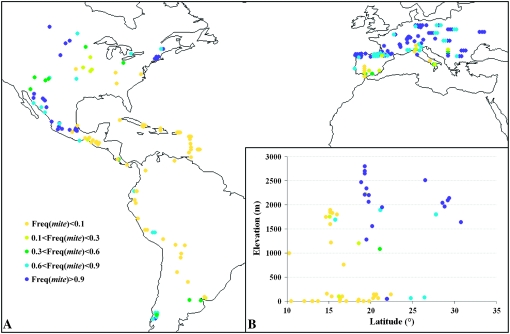
Data obtained in the inbred line panel suggested that Vgt1 could have been subjected to differential selection, with mite and CGindel587 allele frequencies varying from 0.3 in the late tropical group to 0.87 in the European and Northern Flint groups and with an intermediate frequency of 0.45 in Stiff Stalk and Corn Belt Dent groups. For ease of genotyping and considering its high LD with CGindel587 when a broad genetic diversity is addressed, we used mite as a proxy for CGindel587 and analyzed its frequency in a 256-landrace collection (Figure 2). This collection exhibits a latitudinal cline for flowering time (supplemental Figure 3) with days to pollen shed (DPS) under long days, ranging from 612 growing degree days (GDD) for the PPS488 population (latitude 42.9°N) to 1567 GDD for ECUA881 (latitude 3°S).
Figure 2.—
Geographical distribution of mite frequency (A) for 256 European and American landraces and (B) for a subset of 77 landraces from Central America and the Caribbean considering both latitude and elevation. The genotypic analysis of landraces was focused on mite due to its strong association with flowering time and its high level of LD with CGindel587. Moreover it could be easily genotyped by size analysis on a standard agarose gel. Genotyping was performed on a bulk of 15 plants per population as previously described by Dubreuil et al. (2006). Additional information is given in supplemental Materials and Methods.
MITE insertion frequency was generally high (>0.75) in European populations, except those originating from southern Spain and Italy. High frequencies were also observed in Northern Flint and northern Mexican materials. Populations originating from Corn Belt displayed variable frequencies (0–0.8). Caribbean populations exhibited low (<0.25) or null frequency. Populations originating from South America displayed a low frequency except those from southern Chile. We also observed a relationship between elevation and mite frequency (correlation coefficient = 0.71, P < 0.0001) for populations from Central America and the Caribbean. The frequency of the early allele reached a maximum in landraces collected in elevated sites and declined for landraces originating from lowlands (Figure 2).
Discussion:
Our study confirms the association between nucleotidic variation at Vgt1 and flowering time initially found by Salvi et al. (2007). Association tests appear extremely robust for this locus, as illustrated by very high levels of significance observed (down to P = 2.10−14 for CGindel587) while population structure, strongly related to both polymorphisms and flowering time [50% of the phenotypic variation according to Camus-Kulandaivelu et al. (2006)], was accounted for. P-values lower by several orders of magnitude than those observed by Salvi et al. (2007) suggest a higher power of the present experiment, due to a larger population size and a more diversified material, including early-flowering lines from Northern Flint and European genetic groups. Furthermore, population structure seems to “absorb” the effect of Vgt1 in the present panel: CGindel587 explained 17 and 4% of the phenotypic variation not related to population structure with the GLM and mixed model, respectively, vs. 32% for the polymorphisms reported by Salvi et al. (2007). Despite difficulties inherent to the estimation of polymorphism effects in such situations, our results suggest that the Vgt1 early allele shortens the plant cycle by ∼100 GDD, i.e., ∼7 days under northern France conditions. We can also notice that significance levels at loci other than CGindel587 were considerably reduced when including CGindel587 as a cofactor. This indicates that associations found at other positions, including those located in the ZmRap2.7 gene, are likely the result of linkage disequilibrium with CGindel587. It can be noted that −log(P) for these polymorphisms better correlates with r2 statistics with CGindel587 than with D′ (0.9 and 0.74, respectively). The region indeed shows a high level of LD, with r2 values up to 0.4 between Vgt1 and ZmRap2.7, located 70 kb apart. Although infrequent in maize, such a level of LD has also been reported by Palaisa et al. (2004) in the region surrounding the Y1 locus. This suggests that LD-based genomewide scans at the density of one marker per 10 kb may be relevant in maize to identify regions of the genome with strong contributions to the traits of interest.
Modeling of haplotype structure suggests that four ancestral haplotypes contributed to the variation observed at Vgt1. The two ancestral haplotypes bearing the early allele at CGindel587 display a high similarity in this subregion, suggesting a common origin and the possibility of a recombination event(s) assembling the early allele of CGindel587 with late alleles at the other strongly associated loci.
The effect of CGindel587 on early flowering is supported by both a highest significance level and a better consistency with results from QTL analyses. Indeed, our study also illustrates that comparison with QTL studies could be a beneficial way to validate or to pinpoint the weakness of association mapping results. No QTL for flowering time was observed in F2 × MBS847 crosses, although the MITE is inserted in F2 but not in MBS847. As a result, this mite transposon is not expected to be directly responsible for the Vgt1 effect. The results are more congruent with genotype observed at CGindel587 since the two lines share the same allele. The ultimate discrimination of the “causal polymorphism” will require further analyses but CGindel587 appears as a good candidate. It can be noted that this 2-bp indel is neither located in a conserved noncoding sequence (CNS) according to Salvi et al. (2007) nor in a known regulatory domain. The underlying molecular mechanisms by which it could impact on flowering time thus needs to be elucidated.
When considering this polymorphism, it is worth noting that the high LD of CGindel587 with mite makes mite a reliable proxy to estimate early allele frequency in the inbred line and population panels. These investigations show that early allele frequency at Vgt1 was highly correlated with geographical origins. Early allele of Vgt1 displayed a higher frequency in the Northern Flint and European groups. Frequencies were intermediate in the Corn Belt Dent genetic pool and lower in the late tropical pool. This further supports association results, showing that variation at Vgt1 was selected in adaptation of maize to cool temperate climates. As compared to the Dwarf8 early allele (Camus-Kulandaivelu et al. 2006), the Vgt1 early allele displays a higher frequency in tropical materials. This suggests that it was preexisting in this genetic pool, as corroborated by the detection of the MITE among a limited set of teosintes (data not shown).
Furthermore, the frequency of Vgt1 alleles among tropical populations varies with the altitude of the collection site, the early allele being rare at low elevations. This, in addition to an excess of haplotypes and a positive and significant Tajima's D at the Vgt1 locus in tropical inbred lines (results not shown) supports the hypothesis of diversifying selection following domestication of maize, with early and late materials adapted to high and low cultivation systems, respectively. Indeed, late materials display an advantage in warmer conditions of lowlands by preventing flowering at a stage where the plant would not have accumulated enough resources to ensure reproductive success. So, in addition to having played a key role in adaptation to a cool temperate climate, Vgt1 may also have been involved in the differentiation of maize varieties according to elevation in tropical Central America.
Acknowledgments
We are grateful to F. Dumas and V. Combes for their contribution to molecular analyses. We also acknowledge D. Brunel and colleagues at Centre National de Génotypage (France) for access to facilities and technical assistance for sequencing. We thank C. Mir, L. Moreau, M. Tenaillon, T. Zerjal, and two anonymous reviewers for helpful discussion and comments. PROMAIS contributed to analyses of open-pollinated varieties. This work was supported by Génoplante Consortium (France), the Bundesministerium für Bildung und Forschung (Bonn, Germany), and KWS SAAT AG (Einbeck, Germany) in the frame of the Genomanalyse im Biologischen System Pflanze Consortium.
References
- Abler, B. S. B., M. D. Edwards and C. W. Stuber, 1991. Isoenzymatic identification of quantitative trait loci in crosses of elite maize inbreds. Crop Sci. 31 267–274. [Google Scholar]
- Andersen, J. R., T. Schrag, A. E. Melchinger, I. Zein and T. Lubberstedt, 2005. Validation of Dwarf8 polymorphisms associated with flowering time in elite European inbred lines of maize (Zea mays L.). Theor. Appl. Genet. 111 206–217. [DOI] [PubMed] [Google Scholar]
- Austin, D. F., and M. Lee, 1996. Genetic resolution and verification of quantitative trait loci for flowering and plant height with recombinant inbred lines of maize. Genome 39 957–968. [DOI] [PubMed] [Google Scholar]
- Barriere, Y., G. Aurel, M. Briand, D. Denoue and A. Gueu, 2005. QTL mapping for cell wall constituents and cell wall digestibility in maize recombinant inbred line progeny F838 × F286 harvested at an early forage stage of maturity. Institut National de la Recherche Agronomique Technical Report. INRA, Paris.
- Beavis, W. D., O. S. Smith, D. Grant and R. Fincher, 1994. Identification of quantitative trait loci using a small sample of topcrossed and F4 progeny from maize. Crop Sci. 34 882–896. [Google Scholar]
- Blanc, G., A. Charcosset, B. Mangin, A. Gallais and L. Moreau, 2006. Connected populations for detecting quantitative trait loci and testing for epistasis: an application in maize. Theor. Appl. Genet. 113 206–224. [DOI] [PubMed] [Google Scholar]
- Bouchez, A., F. Hospital, M. Causse, A. Gallais and A. Charcosset, 2002. Marker-assisted introgression of favorable alleles at quantitative trait loci between maize elite lines. Genetics 162 1945–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury, P. J., Z. Zhang, D. E. Kroon, T. M. Casstevens, Y. Ram-Doss et al., 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23 2633–2635. [DOI] [PubMed] [Google Scholar]
- Camus-Kulandaivelu, L., J. B. Veyrieras, D. Madur, V. Combes, M. Fourmann et al., 2006. Maize adaptation to temperate climate: relationship between population structure and polymorphism in the Dwarf8 gene. Genetics 172 2449–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardon, F., B. Virlon, L. Moreau, M. Falque, J. Joets et al., 2004. Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome. Genetics 168 2169–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil, P., M. Warburton, M. Chastanet, D. Hoisington and A. Charcosset, 2006. More on the introduction of temperate maize into Europe: large-scale bulk SSR genotyping and new historical elements. Maydica 51 281–291. [Google Scholar]
- Gorroochurn, P., S. E. Hodge, G. A. Heiman, M. Durner and D. A. Greenberg, 2007. Non-replication of association studies: “Pseudo-failures” to replicate? Genet. Med. 9 325–331. [DOI] [PubMed] [Google Scholar]
- Hardy, O. J., and X. Vekemans, 2002. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2 618–620. [Google Scholar]
- Jiang, C., G. O. Edmeades, I. Armstead, H. R. Lafitte, M. D. Hayward et al., 1999. Genetic analysis of adaptation differences between highland and lowland tropical maize using molecular markers. Theor. Appl. Genet. 99 1106–1119. [Google Scholar]
- Koester, R. P., P. H. Sisco and C. W. Stuber, 1993. Identification of quantitative trait loci controlling days to flowering and plant height in two near isogenic lines of maize. Crop Sci. 33 1209–1216. [Google Scholar]
- Loiselle, B. A., V. L. Sork, J. Nason and C. Graham, 1995. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am. J. Bot. 82 1420–1425. [Google Scholar]
- Mechin, V., O. Argillier, Y. Hebert, E. Guingo, L. Moreau et al., 2001. Genetic analysis and QTL mapping of cell wall digestibility and lignification in silage maize. Crop Sci. 41 690–697. [Google Scholar]
- Newton-Cheh, C., and J. N. Hirschhorn, 2005. Genetic association studies of complex traits: design and analysis issues. Mutat. Res. 573 54–69. [DOI] [PubMed] [Google Scholar]
- Palaisa, K., M. Morgante, S. Tingey and A. Rafalski, 2004. Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proc. Natl. Acad. Sci. USA 101 9885–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupard, B., L. Moreau and A. Charcosset, 2001. Analyse de l'épistasie entre QTL pour trois caractères agronomiques chez le maïs. Institut National de la Recherche Agronomique Technical Report. INRA, Paris.
- Pritchard, J. K., M. Stephens and P. Donnely, 2000. Inference of population structure using multilocus genotype data. Genetics 155 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi, S., G. Sponza, M. Morgante, D. Tomes, X. Niu et al., 2007. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA 104 11376–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, J. D., 2002. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B Stat. Methodol. 64 479–498. [Google Scholar]
- Thornsberry, J. M., M. M. Goodman, J. Doebley, S. Kresovich, D. Nielsen et al., 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28 286–289. [DOI] [PubMed] [Google Scholar]
- Tuberosa, R., S. Salvi and R. L. Phillips, 1997. Bulked segregant analysis confirms the importance of the region near umc89a for days to pollen shed in maize. Maize Genet. Coop. News Lett. 72 71–72. [Google Scholar]
- Veyrieras, J. B., L. Camus-Kulandaivelu, B. Gouesnard, D. Manicacci and A. Charcosset, 2007. Bridging genomics and genetic diversity: linkage disequilibrium structure and association mapping in maize and other cereals. Crop Sci. 47(S3): S60–S71. [Google Scholar]
- Vladutu, C., J. McLaughlin and R. L. Phillips, 1999. Fine mapping and characterization of linked quantitative trait loci involved in the transition of the maize apical meristem from vegetative to generative structures. Genetics 153 993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. M., G. Pressoir, W. H. Briggs, I. Vroh Bi, M. Yamasaki et al., 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38 203–208. [DOI] [PubMed] [Google Scholar]