Abstract
Single-chain variable fragments (scFvs) of anti-Lewisy hu3S193 humanized antibody were constructed by joining the VH and VL domains with either +2 residues, +1 residue, or by directly linking the domains. In addition two constructs were synthesized in which one or two C-terminal residues of the VH domain were removed (−1 residue, −2 residue) and then joined directly to the VL domain. An scFv construct in the reverse orientation with the VL joined directly to the VH domain was also synthesized. Upon transformation into Escherichia coli all scFv constructs expressed active protein. Binding activity, multimeric status, and multivalent properties were assessed by flow cytometry, size exclusion chromatography, and biosensor analysis. The results for hu3S193 scFvs are consistent with the paradigm that scFvs with a linker of +3 residues or more associate to form a non-covalent dimer, and those with a shorter linker or directly linked associate predominantly to form a non-covalent trimer and tetramer that are in equilibrium. While the association of V domains to form either a dimer or trimer/tetramer is governed by the length of the linker, the stability of the trimer/tetramer in the equilibrium mixture is dependent on the affinity of the interaction of the individual V domains to associate to form the larger Fv module.
Keywords: scFv multimers, diabody, triabody, trimer, tetrabody, anti-Lewisy antibody, hu3S193
Single-chain variable fragments (scFvs) were designed to link the variable heavy-chain domain (VH) and variable light-chain domain (VL) via a flexible peptide linker to stabilize and enhance their interaction with each other to form an antigen-binding site (Huston et al. 1991; Malby et al. 1993). Peptide linkers >12 amino-acid residues provide sufficient spatial flexibility for the VH and VL domains to associate predominantly as scFv monomers with only small amounts of higher molecular mass multimers evident (Griffiths et al. 1993; Kortt et al. 1994; Whitlow et al. 1994). Decreasing the linker length to <10 residues prevents the V domain from interacting with its attached partner and promotes the interaction of complementary VH/VL pairs from two scFv molecules to associate to form noncovalent multivalent multimers (Holliger et al. 1993; Iliades et al. 1997; Kortt et al. 1997; Pei et al. 1997; Atwell et al. 1999; Le Gall et al. 1999; Dolezal et al. 2000). The formation of scFv dimers, trimers, and tetramers by noncovalent association of tethered V domains is dependent on the length of the linker joining the domains and the orientation of the V domains in the scFv construct (Kortt et al. 2001). In general, with linkers of 3 to 10 residues, scFv dimers (diabodies) are predominantly formed whereas trimers (triabodies) and tetramers (tetrabodies) are formed with linkers of 2 residues or less.
The use of scFvs as immunodiagnostic and immunotherapeutic reagents is gaining momentum (Wu et al. 1996; Adams et al. 1998a; Pavlinkova et al. 1999; Yazaki et al. 2001). ScFv multimers such as diabodies (55 kD) and minibodies (80 kD) are significantly larger than scFv monomers and have in vivo application advantages of slower clearance from the circulation, good tumor accumulation, and show an increase in binding avidity (Wu et al. 1996; Adams et al. 1998b, Adams and Schier 1999). Recent studies using radiolabeled diabodies in vivo showed these molecules have a significant advantage over monomeric scFvs as indicated by tumor:blood ratios and better performance than disulphide-linked scFv monomers and F(ab′)2 (Adams et al. 1998b, 1999).
The maximum peak tumor uptake of diabodies (~55 kD) is within the range of 5–15% injected dose per gram (%ID/g), which is unlikely to deliver sufficient of a radiolabeled β-emitter for therapeutic efficacy. Diabodies will therefore require tagging with a toxic agent that is effective at low doses (Bera et al. 1999) or alternatively labeled with a strong α-emitter (Adams et al. 2000; McDevitt et al. 2001) to achieve effective tumor killing. scFv Trimers (~80 kD) are likely to provide higher %ID/g tumor uptake because of the increased avidity providing increased accumulation of scFv at the tumor site and therefore are more likely to provide therapeutic levels of reagent for tumor killing. Hu et al. (1996) have shown that anti-CEA T84.66 scFv minibody (80 kD) exhibited pharmacokinetic properties more therapeutically effective than the smaller diabodies.
We have previously shown that an scFv of anti-Lewisy hu3S193 humanized monoclonal antibody (hu3S193; Scott et al. 2000), with a five residue linker formed a bivalent diabody (Power et al. 2001). Radiolabeling of this diabody with 111Indium via the bifunctional metal chelator, CHX-A″-DTPA (Tahtis et al. 2001), demonstrated a blood clearance rate similar to other diabodies (Adams et al. 1998a; Wu et al. 1999). Biodistribution and tumor localization of hu3S193 diabody was compared to labeled parent F(ab′)2 with a tumor:blood ratio of 20:1 for the diabody and 5:1 for the F(ab′)2 at 8 h post injection (Tahtis et al. 2001). To investigate the properties and efficacy of a hu3S193 scFv trimer as an imaging and therapeutic reagent, we undertook the construction of a series of scFvs with different linker lengths to define the optimum conditions for the formation of a trimer. Here we characterize the hu3S193 scFv multimers formed with linkers of +2 residues to (−2) residues and their solution properties.
Results
Expression and purification of hu3S193 scFv multimers
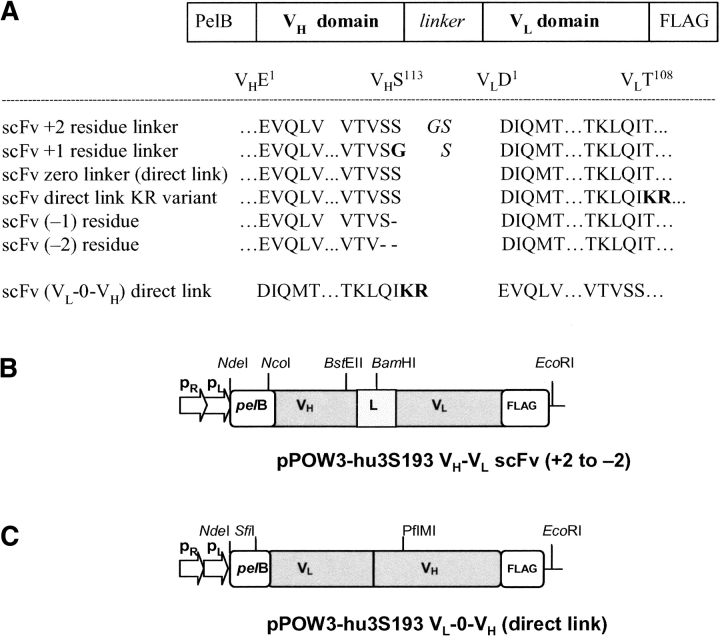
The pPOW3 plasmid vectors expressing the seven hu3S193 scFv constructs are shown in Figure 1 ▶. To investigate the effect of linker length upon the multimer produced and to define the switch between dimer and trimer for the hu3S193 scFv in the VH-VL orientation, the following constructs were made where the V domains were joined with either +2 residues or +1 residues, directly linking the scFv domains, and a VH domain lacking one of the C-terminal serine residues to produce scFv (−1), and scFv (−2) lacking both C-terminal Ser residues (Fig. 1A,B ▶). Three scFv direct-linked constructs were made. In the first construct (Fig. 1A ▶), the VH-0-VL were directly linked. In the second construct, VH-0-VL, where the VL domain C-terminal threonine was replaced with basic residues KR to give VH-VLKR variant. In the construction of the third construct, VL-0-VH were directly linked in the reverse orientation with the VL domain first, (containing the C-terminal KR residues) linked directly to the N-terminal glutamic acid residue of the VH domain to give VL-0-VH KR variant (Fig. 1C ▶). The rationale for these KR variants arose from the observation that the sequence alignment of the light-chain Vκ of hu3S193 with other Vκ sequences showed that the C-terminal residue was a threonine rather than a lysine or arginine as observed in many Vκ sequences (Johnson and Wu 2001). The C-terminal threonine in the parent hu3S193 Vκ was replaced by residues KR, to introduce a basic C terminus as in other VL domains, and to enhance the solubility of the protein and to provide an additional lysine residue for possible attachment of a metal ion chelator for radiolabeling studies (Fig. 1C ▶). All constructs contain a C terminal FLAG affinity purification tail (DYKDDDDK).
Figure 1.
Schematic diagram of the seven different hu3S193 scFv expression units. (A) Sequences of the hu3S193 scFv linker regions, with definition of the constructs demonstrating various linker lengths joining the VH and VL domains. The amino-acid sequence of the VH domain C terminus, N terminus of the VL domain and the linker residues used in each construct including the reverse orientation (VL-VH) construct are shown. (B) VH-VL and (C) VL -VH constructs in the heat-inducible pPOW3 vector, showing unique restriction sites used during the gene construction.
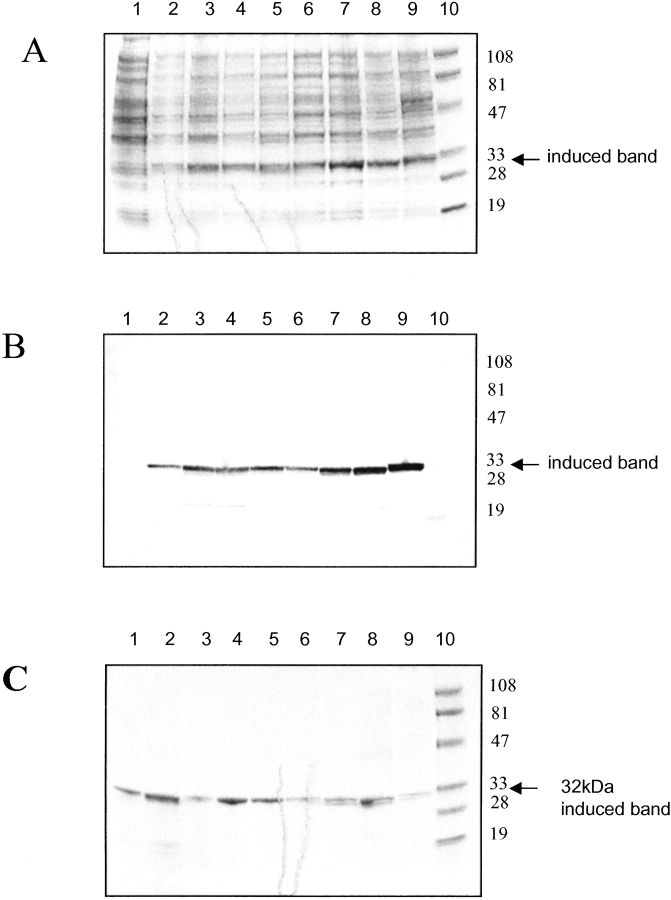
Each scFv construct was expressed from pPOW vector in Escherichia coli strain TOP 10 and produced similar amounts of protein as indicated by SDS-PAGE analysis (Fig. 2A ▶) and Western transfer of duplicate gel (Fig. 2B ▶). The induced protein is clearly visible as a major 32-kD band. Minor levels of cleaved scFv, as indicated by the faint 19-kD band, are also present in some of the crude extracts.
Figure 2.
Reduced SDS-PAGE (12% polyacrylamide gels) of bacterial cultures of hu3S193 scFvs expressed using the heat-inducible pPOW3 expression vector in Escherichia coli Topp10 cells. (A) Coomassie Blue R-250 stained gel; (B) Western transfer of a duplicate gel probed with anti-FLAG FLYTAG antibody; (C) affinity-purified hu3S193 scFv multimer series (Coomassie stained). Each lane was loaded with a standardized amount of cell culture for each hu3S193 scFv multimer construct. The samples were taken at 2 h postinduction, unless otherwise indicated. Gels A and B loaded: lane 1, hu3S193 +5 (preinduction control); lane 2, hu3S193 +5; lane 3, hu3S193 +2; lane 4, hu3S193 +1; lane 5, hu3S193 direct link; lane 6, hu3S193 direct link KR variant; lane 7, hu3S193 (−1); lane 8, hu3S193 (−2); lane 9, hu3S193 reverse VL-0-VH; lane 10, low-range, molecular-weight markers (kD) 102, 81, 47, 32.7, 30, 24. Gel C contained the same samples after affinity purification, with lane 1, soluble hu3S193 +5; lane 2, hu3S193 +5 urea solubilized; all other lanes loaded as for Gels A and B.
Each hu3S193 scFv construct was routinely expressed in a 10-L fermenter batch run as previously described (Bayly et al. 2002). Approximately 75% of the expressed hu3S193 scFv was initially insoluble and was presumably associated with the cell membrane fraction. Thus, each scFv was extracted from the bacterial cell pellet as a soluble fraction or a urea-solubilized fraction and then isolated in a single affinity chromatography step using a batch adsorption method (Bayly et al. 2002). SDS-PAGE analysis of the affinity purified scFvs showed a single protein band migrating with an apparent molecular mass of ~32 kD, as expected for an scFv under denaturing conditions (Fig. 2C ▶).
Characterization of hu3S193 scFv multimers by size exclusion chromatography
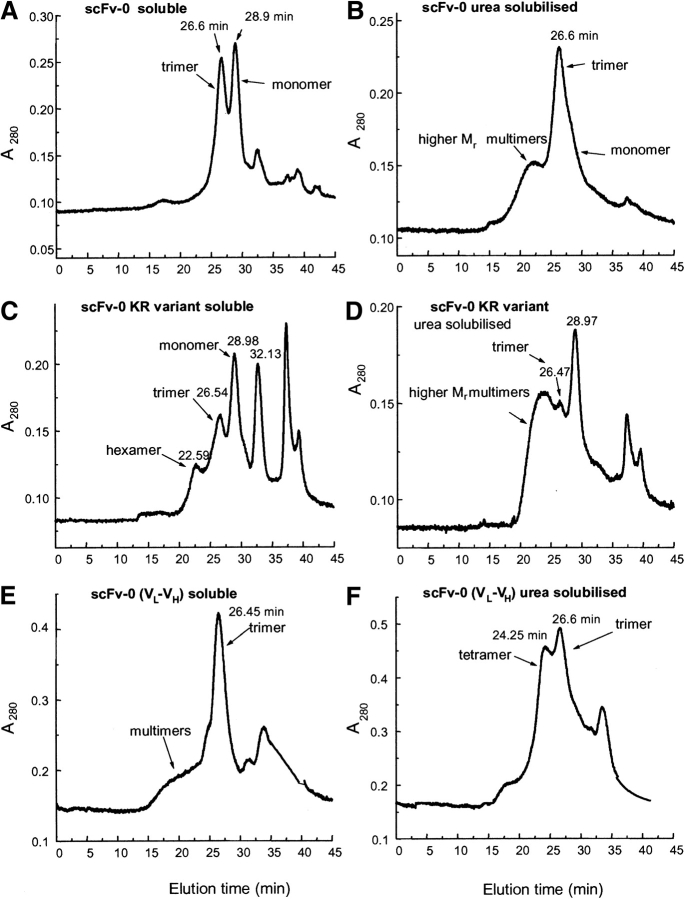
The multimeric state of affinity purified hu3S193 scFvs with the different linker lengths was analyzed by gel filtration on a calibrated Superose 12 column. The hu3S193 scFv +15 monomer (VH -VL) with Mr 26.8 kD and hu3S193 scFv +5 dimer with Mr 53.4 kD have been previously characterized (Power et al. 2001). The hu3S193 scFvs with +2 and +1 residue linkers both produced a major peak eluting with an apparent molecular mass of 54 kD (Fig. 3 ▶) for the soluble and urea-solubilized scFvs preparations as previously found for the hu3S193 scFv +5 diabody molecule (Power et al. 2001). Both fractions of scFv +2 and +1 contained a small amount of higher molecular mass multimers, and some preparation runs also contained a lower molecular mass peak of ~15 kD (Fig. 3B ▶). Peak purified scFv +2 dimer from the urea-soluble fraction was stable at 4°C for >4 wk (Fig. 3C ▶). The ~15 kD peak (Fig. 3B ▶) isolated by gel filtration gave a single protein band of Mr ~18 kD on SDS-PAGE that did not react with the anti-FLAG antibody on Western blot analysis. N-terminal sequence analysis revealed the sequence of the VH domain indicating that this peak was a fragment of the linked VH-VL domains arising from proteolytic cleavage within the VL domain.
Figure 3.
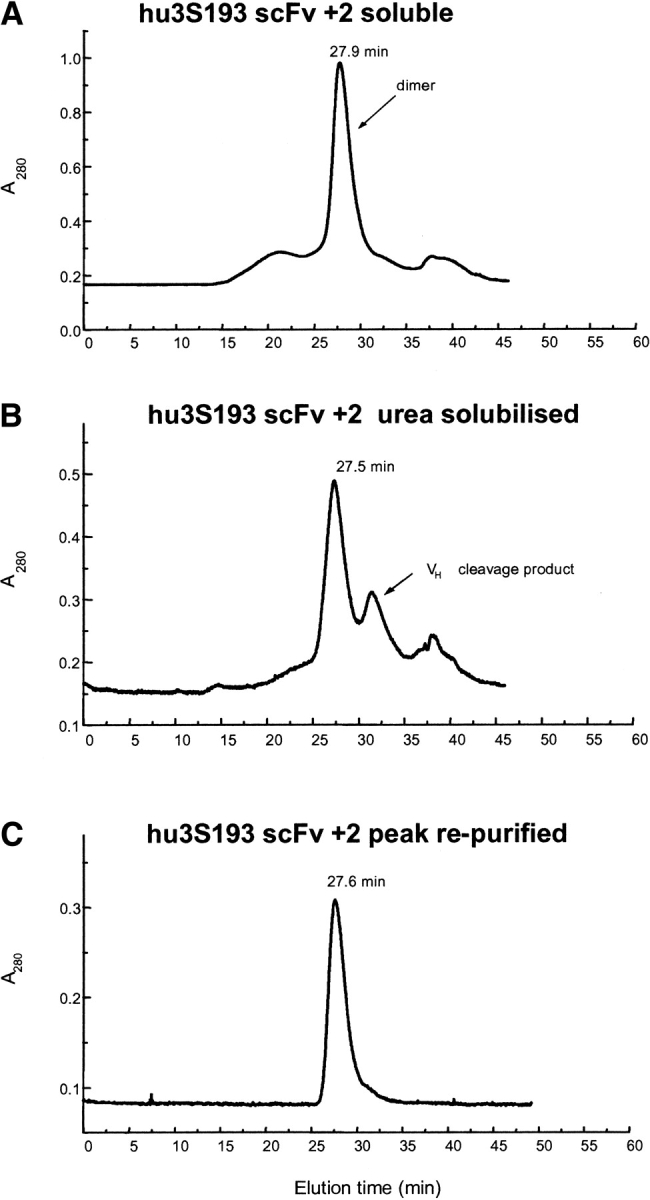
Size exclusion chromatography profiles of affinity-purified hu3S193 scFv +2 on a calibrated Superose 12 HR10/30 column equilibrated in PBS, pH 7.4 and run at a flow rate of 0.5 mL/min. (A) hu3S193 +2 isolated from soluble protein fraction, predominantly dimer (54 kD); (B) urea-solubilized fraction of the cell pellet, also predominantly dimer and the VL cleavage fragment; and (C) rechromatography of the hu3S193 +2 dimer peak from panel B.
Affinity purified scFv (VH-0-VL) from the soluble fraction yielded two peaks (Fig. 4A ▶) with elution times consistent with trimeric (Mr ~81 kD) and monomeric (Mr ~26.5 kD) protein. However, the monomer peak from this (VH-0-VL) preparation elutes just prior to the time observed for the scFv +15 monomer peak (previously characterized), suggesting that the scFv monomer and trimer are in equilibrium, and their elution profile reflects that it may be a reaction boundary, which is likewise true for all of the elution profiles in Figures 3 ▶–5 ▶. Isolation of both the scFv (VH-0-VL) trimer and monomer peaks by gel filtration, concentration, and subsequent rechromatography showed that both species rapidly reequilibrated to the trimer/monomer mixture.
Figure 4.
Size exclusion chromatography profiles of affinity purified hu3S193 direct linked (scFv-0) proteins isolated from either the soluble protein fraction or the urea-solubilized fraction. (A) hu3S193 VH-0-VL soluble, (B) hu3S193 VH-0-VL urea solubilized, (C) VH-0-VL KR variant soluble, (D) VH-0-VL KR variant urea solubilized, (E) reverse VL-0-VH soluble, and (F) reverse VL-0-VH urea-solubilized fraction.
Figure 5.
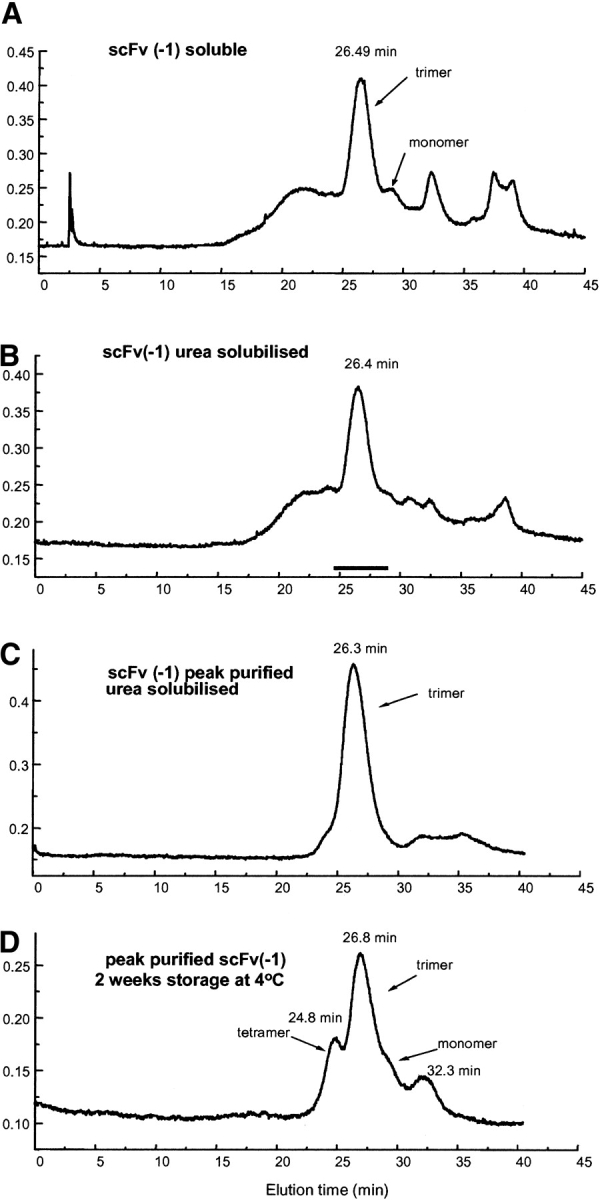
Size exclusion chromatography profiles of affinity purified hu3S193 scFv (−1) isolated from (A) soluble protein fraction, predominantly as trimer (Mr ~81 kD), (B) urea-solubilized fraction, (C) rechromatography of the hu3S193 (−1) trimer peak from panel B, and (D) rechromatography of the hu3S193 (−1) trimer peak from panel B after 2 wk storage at 4°C showing formation of tetramer, monomer, and the VL cleavage fragment.
Affinity-purified scFv (VH-0-VL) from the urea-solubilized fraction contained predominantly a trimer, with a small amount of monomer observed in the trailing edge of the trimer profile (Fig. 4B ▶) as well as higher-molecular-mass multimers.
The soluble fraction of the scFv (VH-0-VL) variant with KR residues at the C terminus of the VL domain yielded a series of peaks consistent with a mixture of scFv monomer, trimer, and higher Mr multimers (possibly a hexamer). In addition, the scFv-VL domain cleavage fragment (Rt = 32.13 min) was found in appreciable amounts in soluble scFv (VH-0-VL) preparations (Fig. 4C ▶), which seems to increase with the age of the sample. The urea-solubilized hu3S193 scFv (VH-0-VL) KR variant yielded a similar elution profile, but the trimer and higher-sized multimers were poorly resolved (Fig. 4D ▶).
In contrast, reverse VL-0-VH on gel filtration showed that the affinity-isolated soluble scFv fraction contained a mixture of predominantly trimer (Fig. 4E ▶), whereas the urea-solubilized scFv fraction contained a mixture of trimer and tetramer (Fig. 4F ▶).
The two scFv constructs in which the C-terminal serine, scFv (−1), and the C-terminal and penultimate serine, scFv (−2), of the VH domain were removed and directly linked to the VL domain, expressed active scFv products that were isolated by affinity chromatography. Soluble and urea solubilized hu3S193 scFv (−1) yielded a major peak, eluting with an apparent molecular mass of a trimer (~81 kD) with only a trace of the monomer peak that was observed for the hu3S193 scFv (VH-0-VL) construct. Both affinity purified scFvs from the soluble and urea-solubilized fractions contained multimers larger than the trimer, but these were not resolved into discrete multimer peaks on the Superose 12 column (Fig. 5 ▶). Peak-purified hu3S193 scFv (−1) trimer from the urea-soluble fraction eluted largely as a symmetrical peak on rechromatography and was stable at 4°C for ~1 wk (Fig. 5C ▶). After 2 wk at 4°C, tetramer, monomer, and the VL cleavage fragment appeared indicating that the trimer is in equilibrium with these other multimeric forms and additionally, some proteolytic cleavage occurred on storage (Fig. 5D ▶). Gel filtration of hu3S193 scFv (−2), soluble and urea solubilized preparations, yielded a mixture of multimers that was not resolved into discrete peaks (data not shown), and further characterization was not pursued.
Formation of hu3S193 scFv-Fab complexes to determine valency of multimers
Two anti-hu3S193 antibodies that compete with the binding of the Ley tetrasaccharide antigen to antibody hu3S193 were used to investigate the valency of the scFv multimers by measuring the sizes of the complexes formed. The two antibodies were cleaved with papain as described previously (Gruen et al. 1993). Antibody F2–3A produced a Fab fragment as expected, but antibody F3–27 yielded an F(ab′)2 fragment that was not reduced to Fab′ with the standard mercaptoethylamine reduction protocol.
The anti-idiotype F2–3A Fab formed a stable complex with hu3S193 Fab resulting in an elution time consistent with the formation of a 1:1 complex with a molecular mass of ~100 kD. The interaction between F2–3A Fab and hu3S193 IgG resulted in a complex with a Mr of ~250 kD consistent with two Fabs binding to each arm of the IgG. Similarly, the hu3S193 scFv +15 monomer formed a stable complex with F2–3A Fab with the Mr of ~77 kD expected for a 1:1 complex. Surprisingly, the hu3S193 scFv +2 dimer and hu3S193 scFv (−1) trimer yielded complexes eluting with molecular masses of ~104 kD and ~131 kD, respectively, consistent with the binding of only one Fab per mole of dimer and trimer (Fig. 6 ▶). Similar results were obtained with scFvs isolated from either the soluble or urea-solubilized factions. These results suggested that (i) only one arm of the diabody and triabody was active, or (ii) the binding of one F2–3A precludes the binding of subsequent Fabs. However, BIAcore binding data clearly show that both hu3S193 scFv dimer and trimer exhibit multivalent binding to immobilized F2–3A antibody consistent with two and three active antigen binding sites per molecule expected for correctly assembled diabodies and triabodies.
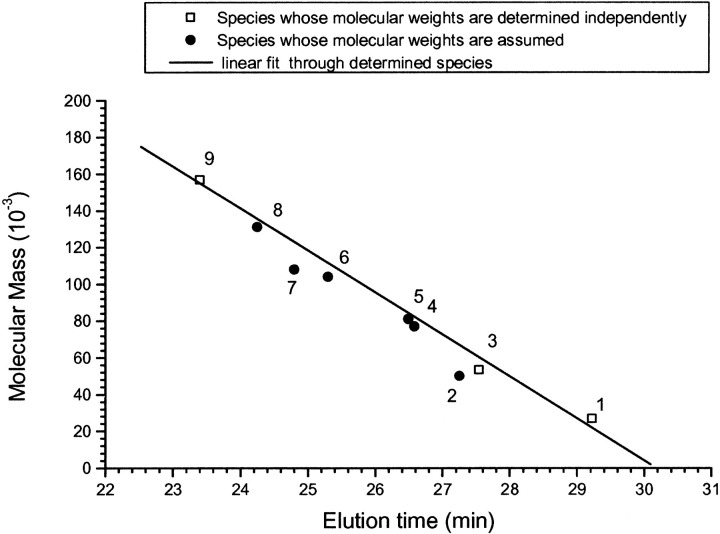
Figure 6.
Relationship between molecular mass and elution time on a Superose 12 column for the family of hu3S193 scFv multimers, anti-idiotype hu3S193 antibodies F2–3A and F3–27 fragments, and the hu3S193 scFv-anti-idiotype antibody complexes formed. Samples were run on calibrated Superose 12 HR10/30 column equilibrated in PBS, pH 7.4 and run at a flow rate of 0.5 mL/min. (1) hu3S193 +15 monomer (26.8 kD), (2) anti-idiotype F2–3A Fab (50 kD), (3) hu3S193 +2 dimer (53.4 kD), (4) hu3S193 +15 monomer/F2–3A Fab complex (77 kD), (5) hu3S193 (−1) trimer (81 kD), (6) hu3S193 +5 dimer/F2–3A Fab (1:1) complex (104 kD), (7) hu3S193 (−1) tetramer (108 kD, see Fig. 5D ▶), (8) hu3S193 (−1) trimer/F2–3A Fab 1:1 complex (131 kD), (9) hu3S193 IgG (157 kD). The solid line of linear fit through the independently determined data points is plotted with R-square value = 0.991.
Size exclusion data for complex formation with anti-idiotype F3–27 F(ab′)2 and hu3S193 scFv (−1) trimer showed that complexes with two and three Fabs per mole of trimer are formed (1:2 complex ~280 kD and 1:3 complex ~380 kD). The interaction between hu3S193 IgG and F3–27 F(ab′)2 resulted in complexes consistent with one and two F(ab′)2 molecules binding per mole of IgG (1:1 complex ~250 kD and 1:2 complex ~350 kD) (data not plotted).
A plot of the calculated molecular masses of the hu3S193 scFv multimers, hu3S193 IgG, and various complexes with anti-idiotype antibody fragments against elution times showed a linear relationship for this family of proteins (Fig. 6 ▶). This linear relationship provides confidence in the molecular masses assigned to the hu3S193 scFv multimers based on their elution times on Superose 12.
Binding activity of hu3S193 scFv multimers
Flow cytometry binding analysis
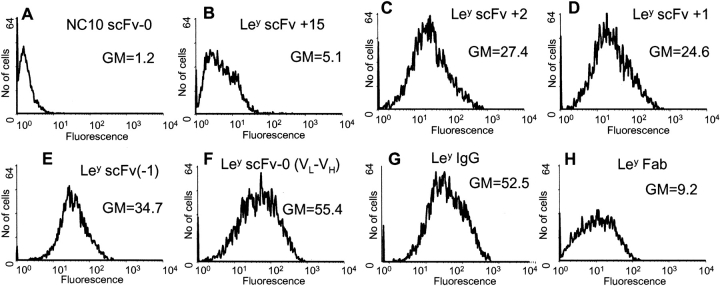
The binding activity of peak purified hu3S193 scFv multimers to native Ley tetrasaccharide antigen expressed on a cell surface was determined by flow cytometry binding analysis using the breast cancer cell line MCF-7 (Fig. 7 ▶). To overcome detection problems as a result of epitope accessibility and recognition by a secondary-labeled antibody, the hu3S193 scFv multimers were labeled with Alexa Fluor 488 fluorescent dye for direct detection. The labeling procedure incorporated between two and four dye molecules per mole of hu3S193 scFv multimer, and the labeled scFv multimers retained between 30–70% binding activity compared to unlabeled hu3S193 scFv. Directly labeled hu3S193 scFv monomer, dimer, trimer, tetramer, and labeled Fab specifically bound to the native antigen on the MCF-7 cells (Fig. 7 ▶), with good yields of mean fluorescent intensity for all species. No interaction was observed with the antigen-negative SK-MEL-28 cells and irrelevant labeled NC10 scFv-0 trimer on MCF-7 cells used as a control.
Figure 7.
Flow cytometry analysis of hu3S193 multimers, Fab and IgG binding to Ley positive cell line, MCF-7. SK-MEL28 cells were used as an antigen negative cell line (results not shown). Cells were incubated in 100 μL of growth medium containing 5 μg hu3S193 scFv or antibody. The hu3S193 scFv multimers, Fab and IgG, were labeled with Alexa Fluor 488 and fluorescence was measured directly with a 488 argon laser. Labeled NC10 scFv-0 was used as a negative control (A). The geometric mean fluorescence (GM) is shown on each panel.
BIAcore binding analysis
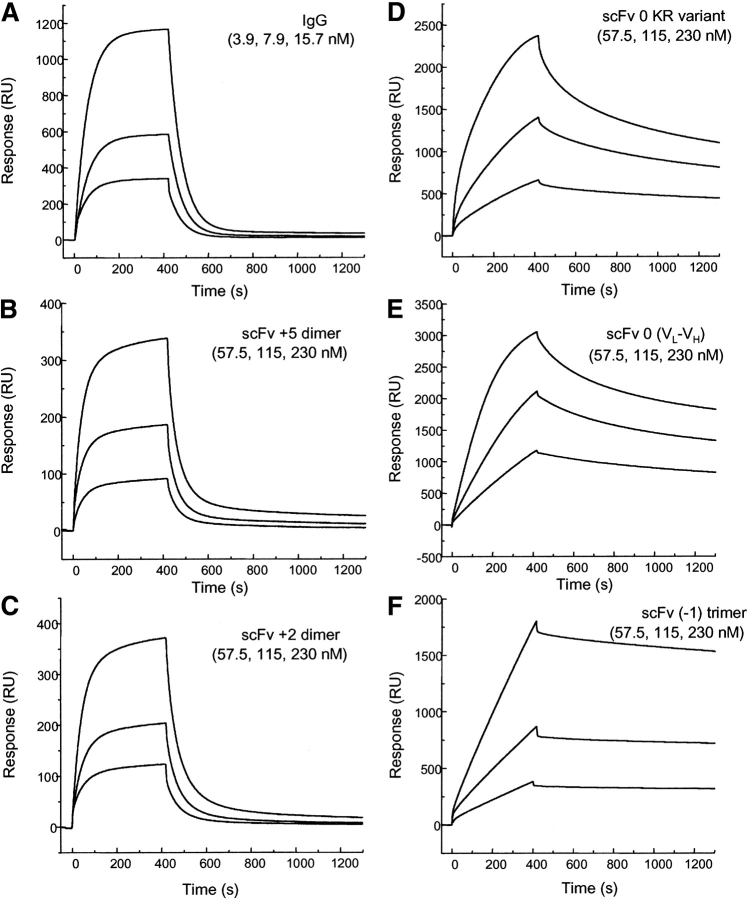
The binding interactions of the hu3S193 scFv multimers with immobilized Ley tetrasaccharide-BSA and anti-hu3S193 anti-idiotype antibody F2–3A were measured by BIAcore biosensor. Hu3S193 scFv +2 and hu3S193 scFv +1 dimers isolated as soluble protein or refolded after solubilization with 4 M urea showed identical binding kinetics with immobilized Ley tetrasaccharide-BSA as found for the hu3S193 scFv +5 dimer and parent IgG (Power et al. 2001) consistent with the 1:1 Langmuir binding model (Fig. 8 ▶). The dissociation rate constants for hu3S193 scFv +1 and hu3S193 scFv +2 dimers (kd = 8.0 × 10−3 s−1) are essentially the same as those for the hu3S193 scFv +5 dimer (kd = 7.8 × 10−3 s−1) and parent IgG (kd = 8.4 × 10−3 s−1) (Power et al. 2001).
Figure 8.
Sensorgrams illustrating the binding hu3S193 scFv multimers to immobilized synthetic Ley tetrasaccharide-BSA complex (2100 RU) at a constant flow rate of 5 μL/min with an injection volume of 35 μL. The surface was regenerated with 10 μL of 100 mM HCl after each cycle. (A) hu3S193 IgG (3.9, 7.9, 15.7nM), (B) hu3S193 +5 dimer (57.5, 115, 230nM), (C) hu3S193 +2 dimer (57.5, 115, 230nM), (D) VH-0-VL KR variant trimer (57.5, 115, 230nM), (E) reverse VL-0-VH trimer/tetramer (57.5, 115, 230nM), (F) hu3S193 (−1) trimer (57.5, 115, 230 nM). Monomeric forms (hu3S193 +15 and Fab) not shown, and dimeric forms (IgG and hu3S193 +5 and hu3S193 +2) exhibit monovalent binding with the surface whereas hu3S193 trimer and tetramer show multivalent binding as indicated by the dissociation rates.
The binding of direct-linked hu3S193 (VH-0-VL and scFv-0 [KR variant]) trimers and the direct-linked reverse orientation (VL-0-VH) (a trimer/tetramer equilibrium) to immobilized Ley tetrasaccharide-BSA showed a significantly slower dissociation rate consistent with multivalent binding (Fig. 8 ▶). These results show that (i) the hu3S193 trimer and tetramer contain at least two active antigen-binding regions per molecule, and (ii) the spatial arrangement of the antigen-binding regions in these hu3S193 scFv multimers is such that the binding regions can bind to two or more Ley tetrasaccharide antigen molecules coupled to BSA and immobilized on the sensor surface. This is in contrast to the bivalent dimers (hu3S193 scFv +5, +2, +1) and parent IgG, which do not interact with the synthetic Ley tetrasaccharide-BSA complex on the sensor surface in a bivalent manner, as previously reported in Power et al. (2001).
A comparison of the binding interaction of hu3S193 Fab and IgG with immobilized hu3S193 anti-idiotype antibody F2–3A showed that the dissociation rate for the IgG (~10−5 s−1) was significantly slower than for the monomeric Fab fragment consistent with an increase in avidity as a result of the bivalent binding by the parent hu3S193 IgG.
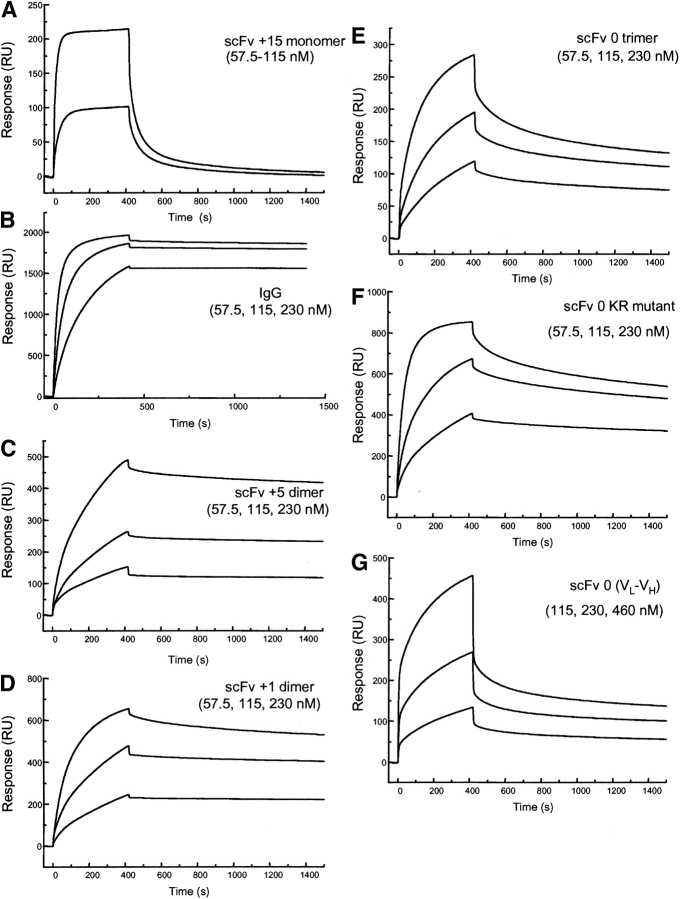
The binding of hu3S193 scFv +5, +2, and +1 dimers, direct-linked (VH-0-VL) trimers and reverse (VL-0-VH) trimer/tetramer showed dissociation rates slower than those for the hu3S193 scFv +15 monomer consistent with multivalent binding to immobilized hu3S193 anti-idiotype antibody F2–3A (Fig. 9 ▶). The binding data shows that the two antigen-binding regions of the hu3S193 scFv dimers are both active and can cross-link two antigen molecules when the steric orientation of the antigens on the sensor surface is favorable for such an interaction to occur. A comparison of the biosensor data for the two immobilized antigens, Ley tetrasaccharide-BSA and hu3S193 anti-idiotype antibody F2–3A indicates that binding data obtained with synthetic immobilized antigens may not necessarily reflect the valency of the molecule or its activity in solution toward native antigen.
Figure 9.
Sensorgrams illustrating the binding of hu3S193 multimers to immobilized anti-idiotype hu3S193 antibody F2–3A (2340 RU) at a constant flow rate of 5 μL/min with an injection volume of 35 μL. The surface was regenerated with 10 μL of 100mM HCl after each cycle. All samples are shown at 57.5, 115, and 230 nM concentration. (A) hu3S193 scFv +15 monomer, (B) hu3S193 IgG, (C) hu3S193 +5 dimer, (D) hu3S193 +2 dimer, (E) VH-0-VL trimer/monomer mixture, (F) VH-0-VL [KR variant] trimer, (G) reverse VL-0-VH trimer/tetramer mixture. The hu3S193 scFv dimer and trimer peaks were isolated by size exclusion chromatography prior to biosensor analysis to remove higher molecular mass species from each multimer in the analysis.
Discussion
The linking of the VH domain to the VL domain of anti-Lewisy hu3S193 antibody with a two residue, one residue, or directly linked successfully produced active scFv multimers with multivalent binding sites as determined by flow cytometry, biosensor binding analysis, and complex formation with anti-idiotype antibodies. Hu3S193 scFv constructs with +2 and +1 residue linkers formed a dimer, whereas the direct-linked constructs formed an equilibrium mixture of predominantly trimer and tetramer. Earlier studies (Atwell et al. 1999) on NC10 (antineuraminidase) scFvs (VH-0-VL) demonstrated that with linkers of +3 or more residues, scFv dimers were formed, while with +2 to zero residues, scFv trimers were formed. In comparing the effect of linker lengths on the transition from dimer to trimer/tetramer for these two scFvs (NC10 and hu3S193), it should be noted that NC10 lacks the C-terminal SerH113 residue that is found in hu3S193 scFv and hence the hu3S193 scFv constructs contain an additional residue that may act as a de facto linker residue. Thus, hu3S193 scFv +2 has three residues and hu3S193 scFv +1 has two residues that can be considered as linker residues according to the linker length definition used for the NC10 scFv based on the crystal structure of NC10 scFv (Kortt et al. 2001). Hu3S193 scFv +2 therefore forms a dimer as predicted from the result for NC10 that showed that linkers of +3 or longer formed dimers (Atwell et al. 1999). Similarly, based on the results for NC10 scFv +2, one would have expected hu3S193 scFv +1 to form a trimer, however, surprisingly it assembles into a dimer. The results reported for hu3S193 scFv provide further evidence that the preferable multimer form of scFvs in which the V domains are linked with +3 residues is a dimer. The result for hu3S193 scFv +1, however, shows that the transition from dimer to trimer depends not only on the linker length but also on each specific VH and VL domain and their noncovalent association to form an active Fv module.
The directly linked hu3S193 scFv (VH-0-VL) and (VH-0-VL-KR) variant constructs form predominantly a trimer (Mr 81 kD) that is in a rapid equilibrium with a monomeric form (Mr 27 kD); in addition, higher molecular mass multimers are also found (Fig. 4B ▶). Rechromatography of trimer and monomer peak fractions isolated by size exclusion chromatography showed that the trimer-monomer equilibrium was rapidly reestablished. The gel filtration data shows that in solution, direct linked (VH-0-VL) occurs as a monomer that is presumably inactive in binding antigen as the linked V domains cannot associate to a Fv module, which reversibly associates with other V domains (monomer) to form active trimers and higher Mr multimers. In contrast, NC10 scFv-0 (VH-0-VL) yielded a stable trimer with no evidence of a monomeric VH-0-VL.
Linking the hu3S193 V domains directly in the reverse orientation VL-0-VH, using the VL KR variant domain, yielded an equilibrium mixture of trimer and tetramer as reported previously for NC10 scFv-0 (VL-0-VH) (Dolezal et al. 2000). The hu3S193 scFv (−1) construct predominantly formed a trimer as well as a mixture of poorly resolved higher Mr multimers (Fig. 3C ▶). Peak purified hu3S193 scFv (−1) trimer (Fig. 3C ▶) on storage at 4°C showed conversion to an equilibrium mixture of trimer and tetramer with a trace of monomer evident indicating that the multimeric forms of this construct were not structurally stable. Reengineering the VH-VL interface in these short-linker scFv species could make them more stable and is something to consider for future work.
This study provides further support for the paradigm that scFvs in which the V domains linked with +3 residues or more associate to form noncovalent dimers and those linked with a shorter linker or directly linked, form a trimer/tetramer mixture. The exception is hu3S193 scFv +1, which formed a dimer. While hu3S193 scFvs with +2 to +5 residue linkers formed stable dimers, scFvs with directly linked V domains did not form a single stable multimer but rather associated to form an equilibrium mixture of predominantly trimer and tetramer. Studies on the effect of linker length on the assembly of NC10 multimers showed that linking VH-0-VL directly yielded a stable trimer (Kortt et al. 1997) while in the reverse orientation directly linked VL-0-VH domains yielded an equilibrium mixture of trimer and tetramer (Dolezal et al. 2000). Thus, while the size of noncovalent multimer (dimer or trimer/tetramer) formed is governed mainly by the linker length, the stability of the trimer/tetramer in the equilibrium is dependent upon the affinity of the interaction of the V domains to associate into the Fv module. The direct-linked hu3S193 VH-0-VL and VL-0-VH constructs generate trimeric and tetrameric units that provide increased avidity, which will make them interesting reagents for use in imaging studies depending on their clearance rate, or possibly for therapeutic use as they may be able to achieve a greater accumulation %ID/g at the tumor-targeting site (than the hu3S193 scFv dimers) a required property for therapeutic use. The most favorable constructs for further development are hu3S193 scFv(−1) VH VL for the trimer work and the direct-linked hu3S193 VL-0-VH scFv for tetrameric studies.
Materials and methods
Reagents
Restriction enzymes and DNA modifying enzymes were all purchased from New England Biolabs (Genesearch) unless otherwise stated. The cloning vector kit PCR- script amp (KS+) was purchased from Stratagene (Integrated Sciences). Oligonucleotide primers used in PCR amplification and for DNA sequence analysis were all synthesized on ABI Applied Biosystems 470 DNA synthesizer. The hybridoma cell line, KM5–1C7–8–5, producing an anti-FLAG antibody was kindly provided by Dr. N. Nicola (CRC for Cellular Growth Factors and WEHI). Secondary antibody goat antimouse IgG (H+L) HRP conjugate was purchased from Bio-Rad Laboratories, as were the prestained, low-range protein molecular-weight markers.
Construction of hu3S193 scFv genes with various linker lengths
Genes encoding the VH and VL domains genes were isolated by PCR from the hybridoma cell line expressing hu3S193 as previously described (Power et al. 2001). The scFv gene constructs with different linker lengths joining the VH and VL domains of hu3S193 ranging from +2 residues down to (−2) residues (residues removed from the C terminus of the VH domain) are shown in Figure 1 ▶. Sequences of the oligonucleotide primers used to assemble these constructs are listed in Table 1. A BamHI site was engineered into the (Gly4Ser)3 linker region of hu3S193 scFv +5 (Power et al. 2001) to assist in the construction of scFvs with various linker lengths. The VH domain was cloned as NcoI-BamHI and the VL domain as BamHI-EcoRI encoding either FLAG purification tag or a KR variant that replaces the VL C-terminal threonine with a lysine and arginine before the FLAG tag (Fig. 1 ▶).
Table 1.
Synthetic oligonucleotides used for isolation and construction of hus3S193 scFv multimer genes
| N4793 | CTG GCG GCC CAG CCG GCC ATG GCC GAG GTC CAA CTG GTG GAG AGC GGT (48 mer) |
| N4810 | GGG ACC CCG GTC ACC GTC TCC TCA GAC ATC CAG ATG ACC CAG AGC CCA AGC AGC CTG (57 mer) |
| N5456 | GTC ACC GTC TCC TCA G (16 mer) |
| N5457 | GA TCC TGA GGA GAC G (15 mer) |
| N5536 | GTC ACC GTC TCC G (13 mer) |
| N5535 | GA TTC GGA GAC G (12 mer) |
| N5911 | GGG ACC CCG GTC ACC GTC TCC GAC ATC CAG ATG ACC CAG AGC (42 mer) |
| N5912 | GCT CTG GGT CAT CTG GAT GTC GGA GAC GGT GAC CGG GGT CCC (42 mer) |
| N5196 | GGG ACC CCG GTC ACC GTC GAC ATC CAG ATG ACC CAG AGC CCA AGC AGC CTG (51 mer) |
| N5183 | CTC GCG GCC CAG CCG GCC ATG GCC GAG ATC CAG ATG ACC CAG AGC CCA AGC AGC CTG (57 mer) |
| N5181 | ACC GCT CTG CAC CAG TTG GAC CTG CCG TTT GAT TTG CTT GGT CCC TTG GCC GAA CGT GAA (63 mer) |
| N5182 | AA CGA GAA TTC TTA CTT GTC ATC GTC GTC CTT GTA ATC TGA GGA GAC GGT GAC CGG GGT CCC TTG GCC CCA GTC (74 mer) |
| N2175 | TGT GTG ATA CGA AAC GA General pPOW3 vector forward primer (17 mer) |
| N2357 | GCG CGT CGG GCT CTA GA General pPOW3 vector reverse primer (17 mer) |
| N4905 | TTT TGT GCA AGA GGC ACC (18 mer) |
| N4796 | TGT GAT TTG CAG CTT GGT CCC TTG GCC GAA (30 mer) |
| N5205 | GCT TGG GCT CTG GGT CAT CTG G (22 mer) |
Nucleotide sequence of the oligonucleotide primers used to attach the linkers between the VR and the VL domains to form the hu3S193 scFv dimer, trimer, and tetramer molecules.
The direct-linked (VH–0-VL) construct was made by utilizing the BstEII site near the C terminus of the VH domain. The hu3S193 scFv +5 residue construct was digested with BstEII and EcoRI to remove the C terminus of the VH domain, the GGGGS linker and the VL domain. A forward VL domain PCR primer N4810 was utilized that restores the C terminus of the VH domain and the N terminus of the VL domain to directly ligate the VH and VL domains. The +2 residue construct was engineered by ligating a dsDNA linker N5456 and N5457 (Table 1) to replace the region between the BstEII site and the BamHI site, using the +5 residue construct as a template. The +1 residue construct was made in a similar way utilizing the BstEII site and the BamHI sites and restored the residues spanning these two sites ligating a dsDNA linker N5536 and N5535 (Table 1). The C terminus of the VH domain was shortened by one residue (Ser) to make the (−1) construct using a pair of oligonucleotide primers N5911 and N5912 utilizing the QuikChange (Stratagene, Integrated Sciences) site-directed mutagenesis strategy. The VH domain was further shortened by another Ser residue to make the (−2) construct, using the same PCR strategy as for the direct linkage construct. The VH domain was digested with BstEII and the VL domain was PCR amplified with a new primer N5196, which removed the two C-terminal serine residues.
The hu3S193 scFv gene with the domains in the reverse orientation (VL-0-VH) with a direct linkage as shown in Figure 1 ▶, was constructed by utilizing the SfiI site for cloning into pPOW3 vector and the PflMI site at the N terminus of the VH domain for joining the two domains. The VL domain gene in the VL-VH construct was PCR amplified with primer N5183 containing the SfiI site and N5181, which contains the VL domain with the lysine and arginine residues at the C terminus and the first few residues of the N-terminal VH domain sequence and the PflMI site. The VH domain gene was made with the primers N4793 and N5182 (Table 1).
PCR amplification was used to synthesize the VH and VL gene fragments incorporating restriction endonuclease sites required for the cloning of the scFv fragments. PCR products were digested with restriction enzymes and fragment sizes were confirmed by electrophoresis on 8% polyacrylamide gels and the DNA fragments were excised and eluted from the gels. The scFv genes were cloned into pPOW3 bacterial expression vector for protein production (Power and Hudson 2000). Oligonucleotide primers that bind to the pPOW3 vector N2175 and N2357 were used for colony screening and dsDNA sequencing within the pPOW3 vector. Colony screening with these two oligonucleotides generated a band of 215 bp in addition to the expected size of the cloned gene. The scFv constructs contain a C-terminal extension FLAG octapeptide affinity tag (DYKDDDDK). The DNA constructs were confirmed by nucleotide sequencing using internal primers N4905 VH domain forward primer binding to the sequence FCARGT, N5205 VH domain reverse primer and N4796 VL domain reverse primer.
Hu3S193 scFv expression and purification
Cultures of E. coli TOP 10 (Invitrogen, Silenus Labs) transformed with pPOW3 vector encoding the appropriate scFv gene were grown and induced to express protein as described (Power and Hudson 2000). After initial, small-scale expression in 1-L shake flasks to establish that the correct product was synthesized, 10-L fermenter runs were performed as described previously (Bayly et al. 2002). ScFv protein was extracted from the bacterial cell pellet to yield soluble and urea solubilized protein fractions as described (Bayly et al. 2002). The scFvs were isolated from these fractions by affinity chromatography on anti-FLAG (KM5–1C7–8–5) antibody resin by a batch procedure as described previously (Bayly et al. 2002). The affinity-purified protein in 1 X TBS (Tris-buffered Saline) pH 8.0 was stored frozen at −20°C. The purity of the affinity-isolated scFvs was monitored by SDS-PAGE and Western blot analysis (Kortt et al. 1994).
Hu3S193 scFv multimer characterization by size exclusion chromatography
The multimeric state of each affinity purified hu3S193 scFv was evaluated by size exclusion chromatography on a calibrated Superose 12 HR 30/10 column (Amersham Pharmacia) in PBS pH 7.4, at a flow rate of 0.5mL/min. The apparent molecular mass of each hu3S193 scFv multimer was estimated from a calibration curve (Fig. 6 ▶) derived for this family of proteins, with the molecular mass of hu3S193 scFv +5 dimer whose molecular mass was determined by ultracentrifugation (Power et al. 2001) as an internal standard for the calibration. The concentrations of the scFvs were determined spectrophotometrically using an extinction coefficient for a 0.1% (w/v) solution at 280 nm of 1.68 calculated from the protein sequence (Gill and Hippel 1989).
Formation of hu3S193 scFv multimer-anti-idiotype Fab complexes
The anti-Ids were generated by Ludwig Institute of Cancer Research and were Protein A purified in the Biological Production Facility, LICR. Fab fragments of anti-idiotype hu3S193 antibodies F2–3A and F3–27 were prepared as described in Kortt et al. (1997). Purified hu3S193 scFv multimers were mixed with a small molar excess of the anti-idiotype Fab and the complexes formed were analyzed by size exclusion chromatography on Superose 12 as described above. The apparent molecular mass estimated for each complex was used to calculate the size and valency of each multimer.
Flow cytometry binding analysis
The binding of various hu3S193 scFv multimers to Ley tetrasaccharide antigen expressed on breast carcinoma cells was demonstrated by flow cytometry. The fluorescent dye Alexa Fluor™ 488 (Molecular Probes) was conjugated to each hu3S193 scFv multimer according to manufacturer’s directions. The breast carcinoma cell line, MCF-7 (ATCC HTB-22), which expresses high quantities of the Ley tetrasaccharide on its membrane surface was used for the binding studies. SK-MEL-28 melanoma (ATCC HTB-72) cells were used as a negative control line. The cells were maintained in RPMI 1640 (supplemented with 100 U/mL penicillin, 100μg/mL streptomycin and 10% heat-inactivated fetal bovine serum (FBS) (all from Invitrogen) at 37°C in 5% CO2 in humidified air.
For binding studies, cells were harvested at about 75% confluence, treated with trypsin-EDTA buffer (defined as trypsin at 0.25 mg/mL diluted 1 in 10 into PBS containing 0.02% EDTA) and washed in at least 10 volumes of growth medium. 1 × 105 cells in 100 μL of growth medium were incubated with 5 μg of hu3S193 scFv multimer-Alexa Fluor 488 conjugate for 30 min on ice. Trypsin does not cleave the Ley antigen from the surface of the cells (data not shown). NC10 scFv-0 (synthesized and purified from the same bacterial expression vector)-Alexa Fluor 488 conjugate was used as a negative control. After washing with cold wash buffer (2 mL PBS +2% FBS +0.1% sodium azide), the cells were resuspended in 200 μL wash buffer containing 2 μL propidium iodide (1 mg/mL) (Sigma) to detect dead cells. Measurements of relative fluorescence of stained viable cells were performed with a flow cytometer (Epics Elite, Beckman-Coulter, NSW) with a 488 nm argon laser. Data analysis was performed on WinMDI software (Scripps). Green fluorescence was estimated on propidium iodide negative cells gated on forward and side scatter.
Biosensor binding analysis
The binding properties of the hu3S193 scFv multimers to immobilized Ley tetrasaccharide antigen and to anti-hu3S193 anti-idiotype antibody F2–3A, were measured using a BIAcore biosensor (BIACORE AB).
Synthetic Ley tetrasaccharide ligand coupled to BSA (at a ratio of 30:1) (Alberta Research Council) was immobilized to a CM5 BIAsensor chip at a concentration of 155 μg/mL in 10 mM sodium acetate buffer pH 3.0, using standard amide coupling reactions (N-hydroxysuccinimide and N-ethyl-N′-dimethylaminopropyl-carbodimide). The immobilization was carried out at 25°C at a flow rate of 5 μL/min and on injecting 20 μL of the antigen ~2340 RU was coupled to the sensor chip surface. Anti-idioytpe antibody F2–3A was also immobilized on a CM-5 chip at at concentration of 20 μg/mL in 10 mM sodium acetate buffer, pH 4.5 using standard amine coupling chemistry as previously described (Kortt et al. 1999). Injection of 40 μL of IgG at a flow rate of 5 μL/min resulted in the immobilization of 2480 RU of F2–3A. The total concentration of immobilized protein may be lower than initially calculated, which would not alter the overall shape of the curve but only alter the scale of the RU units.
Binding experiments were performed in HBS buffer (10 mM HEPES, 0.15 M NaCl, 3.4 mM EDTA, 0.005% surfactant P20, pH 7.4) at a constant flow rate of 5 μL/min. Aliquots (30–50 μL) of each hu3S193 scFv multimer and hu3S193 Fab were injected over the sensor chip surfaces to measure the association and dissociation reactions. The three sensor surfaces were regenerated with a 10 μL aliquot of 100-mM HCl with negligible loss of binding activity.
The hu3S193 scFv multimers and hu3S193 IgG and Fab samples for binding analyses were peak purified by gel filtration on Superose 12 to remove any higher molecular mass oligomers that may have formed on storage following freezing and thawing. The binding data, when appropriate, was evaluated using the BIAevaluation 3.0.2 software as described previously (Kortt et al. 1999).
Acknowledgments
We thank Phil Strike for N-terminal protein sequencing, Nick Bartone for oligonucleotide synthesis, Kiki Tahtis and Con Panousis for helpful discussions, and Fiona Smyth for careful review of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
scFvs, single-chain variable fragments
VH, variable heavy-chain domain
VL, variable light-chain domain
Vκ, kappa light chain
Fv, variable fragment consisting of VH and VL domains
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0228503.
References
- Adams, G.P. and Schier, R. 1999. Generating improved single-chain Fv molecules for tumor targeting. J. Immunol. Methods 231 249–260. [DOI] [PubMed] [Google Scholar]
- Adams, G.P., Schier, R., McCall, A.M., Crawford, R.S., Wolf, E.J., Weiner, L.M., and Marks, J.D. 1998a. Prolonged in vivo tumour retention of a human diabody targeting the extracellular domain of human HER2/neu. Br. J. Cancer 77 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, G.P., Schier, R., Marshall, K., Wolf, E.J., McCall, A.M., Marks, J.D., and Weiner, L.M. 1998b. Increased affinity leads to improved selective tumor delivery of single-chain Fv antibodies. Cancer Res. 58 485–490. [PubMed] [Google Scholar]
- Adams, G.P., Shaller, C.C., Chappell, L.L., Wu, C., Horak, E.M., Simmons, H.H., Litwin, S., Marks, J.D., Weiner, L.M., and Brechbiel, M.W. 2000. Delivery of the α-emitting radioisotope bismuth-213 to solid tumors via single-chain Fv and diabody molecules. Nucl. Med. Biol. 27 339–346. [DOI] [PubMed] [Google Scholar]
- Atwell, J.L., Breheney, K.A., Lawrence, L.J., McCoy, A.J., Kortt, A.A., and Hudson, P.J. 1999. scFv multimers of the anti-neuraminidase antibody NC10: Length of the linker between VH and VL domains dictates precisely the transition between diabodies and triabodies. Protein Eng. 12 597–604. [DOI] [PubMed] [Google Scholar]
- Bayly, A.M., Kortt, A.A., Hudson, P.J., and Power, B.E. 2002. Large-scale bacterial fermentation and isolation of scFv multimers using a heat-inducible bacterial expression vector. J. Immunol. Methods 262 217–227. [DOI] [PubMed] [Google Scholar]
- Bera, T.K., Viner, J., Brinkmann, E., and Pastan, I. 1999. Pharmacokinetics and antitumor activity of a bivalent disulfide-stabilized Fv immunotoxin with improved antigen binding to erbB2. Cancer Res. 59 4018–4022. [PubMed] [Google Scholar]
- Dolezal, O., Pearce, L.A., Lawrence, L.J., McCoy, A.J., Hudson, P.J., and Kortt, A.A. 2000. ScFv multimers of the anti-neuraminidase antibody NC10: Shortening of the linker in single-chain Fv fragment assembled in V(L) to V(H) orientation drives the formation of dimers, trimers, tetramers and higher molecular mass multimers. Protein Eng. 13 565–574. [DOI] [PubMed] [Google Scholar]
- Gill, S.C. and von Hippel, P.H. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182 319–326. [DOI] [PubMed] [Google Scholar]
- Griffiths, A.D., Malmqvist, M., Marks, J.D., Bye, J.M., Embleton, M.J., McCafferty, J., Baier, M., Holliger, K.P., Gorick, B.D., Hughes-Jones, N.C., et al. 1993. Human anti-self antibodies with high specificity from phage display libraries. EMBO J. 12 725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruen, L.C., Kortt, A.A., and Nice, E. 1993. Determination of relative binding affinity of influenza virus N9 sialidases with the Fab fragment of monoclonal antibody NC41 using biosensor technology. Eur. J. Biochem. 217 319–325. [DOI] [PubMed] [Google Scholar]
- Holliger, P., Prospero, T., and Winter, G. 1993. “Diabodies”: Small bivalent and bispecific antibody fragments. Proc. Natl. Acad. Sci. 90 6444–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, S., Shively, L., Raubitschek, A., Sherman, M., Williams, L.E., Wong, J.Y., Shively, J.E., and Wu, A.M. 1996. Minibody: A novel engineered anti-carcinoembryonic antigen antibody fragment (single-chain Fv-CH3) which exhibits rapid, high-level targeting of xenografts. Cancer Res. 56 3055–3061. [PubMed] [Google Scholar]
- Huston, J.S., Mudgett-Hunter, M., Tai, M.S., McCartney, J., Warren, F., Haber, E., and Oppermann, H. 1991. Protein engineering of single-chain Fv analogs and fusion proteins. Methods Enzymol. 203 46–88. [DOI] [PubMed] [Google Scholar]
- Iliades, P., Kortt, A.A., and Hudson, P.J. 1997. Triabodies: Single chain Fv fragments without a linker form trivalent trimers. FEBS Lett. 409 437–441. [DOI] [PubMed] [Google Scholar]
- Johnson, G. and Wu, T.T. 2001. Kabat database and its applications: Future directions. Nucleic Acids Res. 29 205–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortt, A.A., Malby, R.L., Caldwell, J.B., Gruen, L.C., Ivancic, N., Lawrence, M.C., Howlett, G.J., Webster, R.G., Hudson, P.J., and Colman, P.M. 1994. Recombinant anti-sialidase single-chain variable fragment antibody. Characterization, formation of dimer and higher-molecular-mass multimers and the solution of the crystal structure of the single-chain variable fragment/sialidase complex. Eur. J. Biochem. 221 151–157. [DOI] [PubMed] [Google Scholar]
- Kortt, A.A., Lah, M., Oddie, G.W., Gruen, C.L., Burns, J.E., Pearce, L.A., Atwell, J.L., McCoy, A.J., Howlett, G.J., Metzger, D.W., et al. 1997. Single-chain Fv fragments of anti-neuraminidase antibody NC10 containing five- and ten-residue linkers form dimers and with zero- residue linker a trimer. Protein Eng. 10 423–433. [DOI] [PubMed] [Google Scholar]
- Kortt, A.A., Nice, E., and Gruen, L.C. 1999. Analysis of the binding of the Fab fragment of monoclonal antibody NC10 to influenza virus N9 neuraminidase from tern and whale using the BIAcore biosensor: Effect of immobilization level and flow rate on kinetic analysis. Anal. Biochem. 273 133–141. [DOI] [PubMed] [Google Scholar]
- Kortt, A.A., Dolezal, O., Power, B.E., and Hudson, P.J. 2001. Dimeric and trimeric antibodies: High avidity scFvs for cancer targeting. Biomolecular Engineering 18 95–108. [DOI] [PubMed] [Google Scholar]
- Le Gall, F., Kipriyanov, S.M., Moldenhauer, G., and Little, M. 1999. Di-, tri- and tetrameric single chain Fv antibody fragments against human CD19: Effect of valency on cell binding. FEBS Lett. 453 164–168. [DOI] [PubMed] [Google Scholar]
- Malby, R.L., Caldwell, J.B., Gruen, L.C., Harley, V.R., Ivancic, N., Kortt, A.A., Lilley, G.G., Power, B.E., Webster, R.G., Colman, P.M., et al. 1993. Recombinant anti-neuraminidase single chain antibody: Expression, characterization, and crystallization in complex with antigen. Proteins 16 57–63. [DOI] [PubMed] [Google Scholar]
- McDevitt, M.R., Ma, D., Lai, L.T., Simon, J., Borchardt, P., Frank, R.K., Wu, K., Pellegrini, V., Curcio, M.J., Miederer, M., et al. 2001. Tumor therapy with targeted atomic nanogenerators. Science 294 1537–1540. [DOI] [PubMed] [Google Scholar]
- Pavlinkova, G., Booth, B.J., Batra, S.K., and Colcher, D. 1999. Radioimmunotherapy of human colon cancer xenografts using a dimeric single-chain Fv antibody construct. Clin. Cancer Res. 5 2613–2619. [PubMed] [Google Scholar]
- Pei, X.Y., Holliger, P., Murzin, A.G., and Williams, R.L. 1997. The 2.0-Å resolution crystal structure of a trimeric antibody fragment with noncognate VH-VL domain pairs shows a rearrangement of VH CDR3. Proc. Natl. Acad. Sci. 94 9637–9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, B.E. and Hudson, P.J. 2000. Synthesis of high avidity antibody fragments (scFv multimers) for cancer imaging. J. Immunol. Methods 242 193–204. [DOI] [PubMed] [Google Scholar]
- Power, B.E., Caine, J.M., Burns, J.E., Shapira, D.R., Hattarki, M.K., Tahtis, K., Lee, F.-T., Smyth, F.E., Scott, A.M., Kortt, A.A., et al. 2001. Construction, expression and characterisation of a single-chain diabody derived from a humanised anti-Lewis Y cancer targeting antibody using a heat-inducible bacterial secretion vector. Cancer Immunol. Immunother. 50 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, A.M., Geleick, D., Rubira, M., Clarke, K., Nice, E.C., Smyth, F.E., Stockert, E., Richards, E.C., Carr, F.J., Harris, W.J., et al. 2000. Construction, production, and characterization of humanized anti-Lewis Y monoclonal antibody 3S193 for targeted immunotherapy of solid tumors. Cancer Res. 60 3254–3261. [PubMed] [Google Scholar]
- Tahtis, K., Lee, F.T., Smyth, F.E., Power, B.E., Renner, C., Brechbiel, M.W., Old, L.J., Hudson, P.J., and Scott, A.M. 2001. Biodistribution properties of (111)indium-labeled C-functionalized trans-cyclohexyl diethylenetriaminepentaacetic acid humanized 3S193 diabody and F(ab′)(2) constructs in a breast carcinoma xenograft model. Clin. Cancer Res. 7 1061–1072. [PubMed] [Google Scholar]
- Whitlow, M., Filpula, D., Rollence, M.L., Feng, S.L., and Wood, J.F. 1994. Multivalent Fvs: Characterization of single-chain Fv oligomers and preparation of a bispecific Fv. Protein Eng. 7 1017–1026. [DOI] [PubMed] [Google Scholar]
- Willuda, J., Kubetzko, S., Waibel, R., Schubiger, P.A., Zangemeister-Wittke, U., and Pluckthun, A. 2001. Tumor targeting of mono-, di-, and tetravalent anti-p185(HER-2) miniantibodies multimerized by self-associating peptides. J. Biol. Chem. 276 14385–14392. [DOI] [PubMed] [Google Scholar]
- Wu, A.M., Chen, W., Raubitschek, A., Williams, L.E., Neumaier, M., Fischer, R., Hu, S.Z., Odom-Maryon, T., Wong, J.Y., and Shively, J.E. 1996. Tumor localization of anti-CEA single-chain Fvs: Improved targeting by non-covalent dimers. Immunotechnology 2 21–36. [DOI] [PubMed] [Google Scholar]
- Wu, A.M., Williams, L.E., Zieran, L., Padma, A., Sherman, M., Bebb, G.G., Odom-Maryon, T., Wong, J.Y.C., Shively, J.E., and Raubitschek, A.A. 1999. Anti-carcinoembryonic antigen (CEA) diabody for rapid tumor targeting and imaging. Tumor Targeting 4 47–58. [Google Scholar]
- Yazaki, P.J., Wu, A.M., Tsai, S.W., Williams, L.E., Ikler, D.N., Wong, J.Y., Shively, J.E., and Raubitschek, A.A. 2001. Tumor targeting of radiometal labeled anti-CEA recombinant T84.66 diabody and T84.66 minibody: Comparison to radioiodinated fragments. Bioconjug. Chem. 12 220–228. [DOI] [PubMed] [Google Scholar]