Abstract
Nuclear magnetic resonance (NMR) methods were employed to study the interaction of the cytokine Interleukin-2 (IL-2) with the α-subunit of its receptor (IL-2Rα), and to help understand the behavior of small molecule inhibitors of this interaction. Heteronuclear 1H-15N HSQC experiments were used to identify the interaction surface of 15N-enriched Interleukin-2 (15N-IL-2) in complex with human IL-2Rα. In these experiments, chemical shift and line width changes in the heteronuclear single-quantum coherence (HSQC) spectra upon binding of 15N-IL-2 enabled classification of NH atoms as either near to, or far from, the IL-2Rα interaction surface. These data were complemented by hydrogen/deuterium (H/D) exchange measurements, which illustrated enhanced protection of slowly-exchanging IL-2 NH protons near the site of interaction with IL-2Rα. The interaction surface defined by NMR compared well with the IL-2Rα binding site identified previously using mutagenesis of human and murine IL-2. Two low molecular weight inhibitors of the IL-2/IL-2Rα interaction were studied: one (a cyclic peptide derivative) was found to mimic a part of the cytokine and bind to IL-2Rα; the other (an acylphenylalanine derivative) was found to bind to IL-2. For the interaction between IL-2 and the acylphenylalanine, chemical shift perturbations of 15N and 15NH backbone resonances were tracked as a function of ligand concentration. The perturbation pattern observed for this complex revealed that the acylphenylalanine is a competitive inhibitor—it binds to the same site on IL-2 that interacts with IL-2Rα.
Keywords: Nuclear magnetic resonance, Interleukin-2, Interleukin-2 receptor, protein–protein interactions, HSQC perturbation mapping, protein binding inhibitors
Interleukin-2 (IL-2) is a 15.5-kD cytokine that has a prevalent role in the growth, activation, and differentiation of T cells (Kaempfer 1994). The ability to disrupt the IL-2/Interleukin-2-receptor (IL-2/IL-2R) interaction is important in suppression of immune responses such as those associated with organ transplant rejection, IL-2R-expressing leukemia, and other autoimmune diseases (Waldmann 1993). IL-2 stimulates T-cell proliferation by binding to a heterotrimeric receptor complex consisting of α, β, and γ chains (IL-2Rα, IL-2Rβ, IL-2Rγ) on the T-cell surface. Monoclonal antibodies that recognize IL-2Rα and block IL-2 binding have proven clinically effective as immunosuppressive agents (Waldmann 1993; Hakimi et al. 1997). Small molecules capable of blocking the extracellular IL-2/IL-2Rα interaction would be valuable as potential second-generation versions of these immunosuppressive antibodies. The identification of such small molecule inhibitors could lead to a drug with more desirable cost and toxicity profiles, particularly if oral bioavailability could be achieved. A program to discover and optimize such inhibitors has been pursued, and has benefited from NMR characterization of the relevant binding interactions at a molecular level.
The three-dimensional structure of IL-2 is known from both X-ray (Brandhuber et al. 1987; McKay 1992; M. Hatada, S. Surgenor, D. Weber, W. Danho, and V. Madison, unpubl.) and NMR (Mott et al. 1995) studies (Fig. 1 ▶). The IL-2 fold is similar to other cytokines, such as IL-4 (Hage et al. 1999) and GM-CSF (Sprang and Bazan 1993), which form four-helix bundles with an up-up-down-down arrangement of the four main α-helices (referred to as helices A, B, C, and D). IL-2 has three structural elements that deviate from related four-helix bundles: an irregular one-turn helix in the loop between helices A and B (AB-loop); a kink in the middle of helix B (which causes this region to be considered as two helices, designated B and B′); and a two-stranded antiparallel β-structure formed by portions of the AB- and CD-loops. No experimental structure has been determined for the complex between IL-2 and IL-2Rα, although recombinant forms of each protein can be expressed in milligram quantities. Mutagenesis studies of human IL-2 have identified residues in the AB loop (K35, R38, F42, K43; Sauve et al. 1991) as being critical for binding to IL-2Rα. Subsequent mutagenesis studies of human (G. Ju, unpubl.) and murine (Zurawski et al. 1993) IL-2 have implicated additional residues—located in the B-helix, the BC loop, the CD loop, and the D-helix—as interacting with IL-2Rα.
Figure 1.
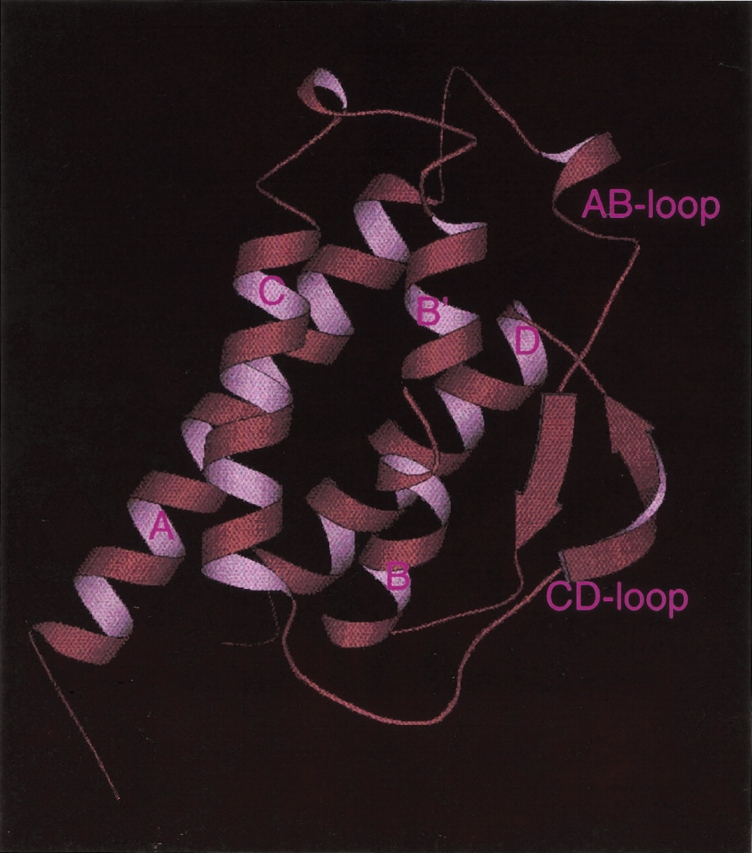
Molscript (Kraulis 1991) rendering of the X-ray crystal structure of human IL-2 (M. Hatada, S. Surgenor, D. Weber, W. Danho, and V. Madison, unpubl.). Labels correspond to secondary structural elements including the α-helices A, B, B′, C, and D; the AB-loop; and the CD-loop.
Here we describe structural information about the IL-2/IL-2Rα interaction obtained from NMR studies. Because IL-2 is smaller (15.5 kD) than IL-2Rα (55 kD), and therefore more amenable to NMR observations, we have focused on characterizing the changes in IL-2 signals due to complex formation. The site on IL-2 that interacts with IL-2Rα has been examined by two different NMR methods, which we refer to as: (1) HSQC perturbation mapping, and (2) Hydrogen/deuterium (H/D) exchange protection. We compare our results to those derived from site-directed mutagenesis mapping.
We also describe NMR studies of two types of small molecule inhibitors that were developed as part of a drug discovery effort. The first type is a constrained peptide, designed to mimic the binding region of IL-2 in the helical portion of the AB loop. We report the NMR structure of this peptide, free in solution, and compare it to the X-ray derived IL-2 substructure that it was designed to emulate. The second type of inhibitor is a small organic molecule (Tilley et al. 1997), also designed to mimic the receptor-binding segment of IL-2, but shown by NMR to inhibit via binding to IL-2 rather than to IL-2Rα.
Results
HSQC perturbation mapping of the IL-2 surface that interacts with IL-2R α
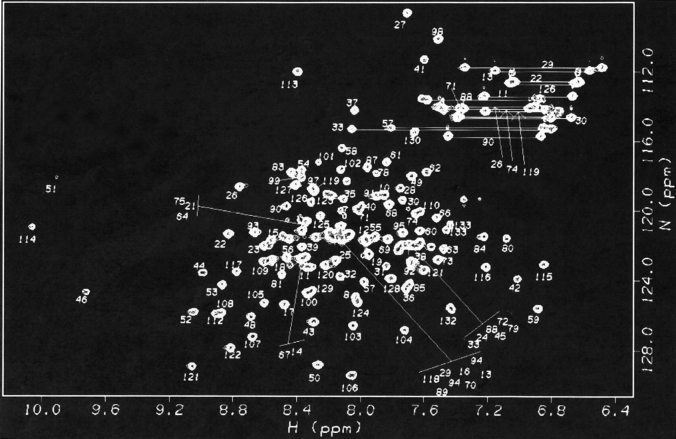
HSQC perturbation mapping (Yu et al. 1992; van Nuland et al. 1993; Emerson et al. 1995) involves comparison of spectral changes in 1H-15N HSQC spectra upon formation of a complex, in this case the complex between 15N-IL-2 and unlabeled IL-2Rα. Figure 2 ▶ shows the assigned 1H-15N HSQC spectrum of the 15.5 kD 15N-IL-2 protein. 1H-15N assignments have been reported previously (Mott et al. 1992). The IL-2 protein used in our NMR studies showed some minor differences in chemical shifts when compared with the previously reported NH assignments. One reason could be differences in expression and/or purification conditions. We chose to express IL-2 as a soluble protein in yeast, rather than expressing it in Escherichia coli with subsequent refolding of the insoluble protein. The soluble protein was purified on an affinity column containing immobilized IL-2Rα. The protein did not appear to be glycosylated, based on lack of heterogeneity on an SDS gel, and its reactivity with an antibody specific for the nonglycosylated form. NH assignments have been verified for our sample using 15N-edited 3D NOESY and TOCSY methods (Marion et al. 1989).
Figure 2.
500-MHz 1H-15N HSQC NMR spectrum of 15N-IL-2 at pH 4.6, 40°C. Numbers indicate the sequence specific resonance assignments. For crowded regions, the assignment labels are displaced yet retain the same pattern of relative positions as the resonances they represent. Crosspeaks connected by a horizontal line represent side-chain NH2 groups of Asn and Gln residues.
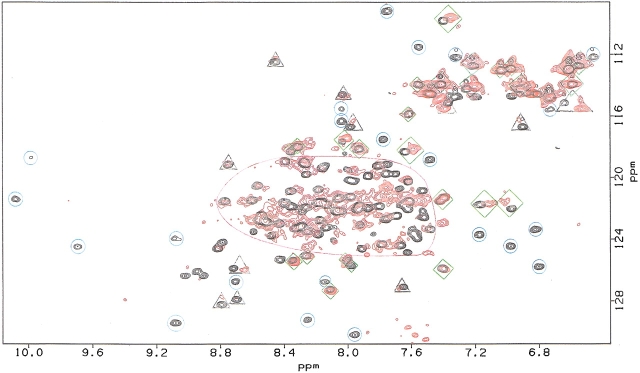
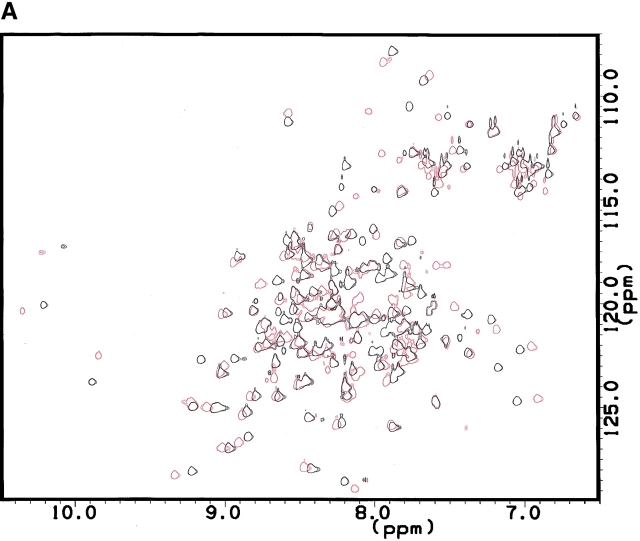
An overlay of the 1H-15N HSQC spectrum of free 15N-IL-2 (black) with that of 15N-IL-2 in complex with IL-2Rα (red) is shown in Figure 3 ▶. The latter spectrum is shown plotted at a much lower threshold than the spectrum of free IL-2. Although many of the 15N-IL-2 signals from the 60-kD complex remain observable, others are broadened beyond detection. Although assignment of the 15N-IL-2 resonances in the complex is impractical, differential broadening is readily evaluated by comparing the HSQC spectrum of IL-2 in the complex to that of IL-2 alone. These effects have been classified into four categories. Category one (blue circles in Fig. 3 ▶) contains signals that are well resolved in the free 15N-IL-2 spectrum, but have been broadened beyond detection in the spectrum of the complex. These resonances are likely to originate from atoms close to the interaction surface between 15N-IL-2 and IL-2Rα (Matsuo et al. 1999). In contrast, category two resonances (green diamonds in Fig. 3 ▶) are those that remain observable in the spectrum of the complex at an intensity at least 7% of that for uncomplexed IL-2. In addition, category two resonances exhibit 15N and 15NH chemical shifts within two line widths of the original values in the spectrum of free 15N-IL-2. These resonances most likely correspond to atoms far from the intermolecular interaction surface. Category three (gray triangles) contains resonances that exhibit an intermediate response. These signals remain above the threshold, but are either below the 7% intensity limit, or have chemical shift changes greater than two line widths from the free values. Finally, category four encompasses all remaining resonances, including those whose response is rendered uninterpretable due to spectral overlap (outlined in magenta).
Figure 3.
Overlay of 500-MHz 1H-15N HSQC NMR spectra of 15N-IL-2 in the presence (red, pH 6.1) and absence (black, pH 4.4) of IL-2Rα at 40°C. The red spectrum is displayed at a lower threshold to allow visualization of the relatively weaker signals of the 15N-IL-2/IL-2Rα complex. Blue circles, gray triangles, and green diamonds illustrate category 1, 2, and 3 resonances, respectively, as described in the text. The region encircled in magenta defines the area of the spectrum where resonance overlap restricted attempts to categorize the behavior of individual signals upon complex formation.
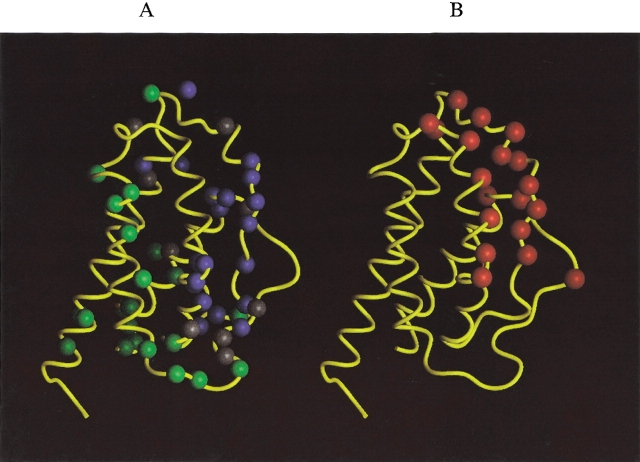
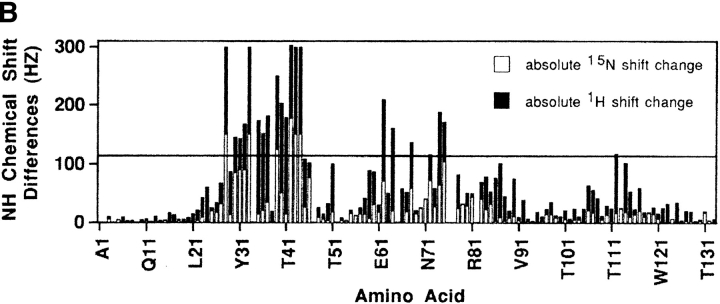
The classification results from the HSQC perturbation experiment have been mapped onto the crystal structure of IL-2 as shown in Figure 4A ▶. In this figure, nitrogen atoms corresponding to NH resonances from categories one, two, and three are shown as blue, green, and gray spheres, respectively. Atoms belonging to each category are clustered together, consistent with the expectation that category one atoms are located at the IL-2Rα binding site, category two atoms are away from this site, and category three atoms represent the fringe of the interaction surface. Comparison of Figure 4A and B ▶ shows that there is a strong correlation between the region containing the most highly perturbed atoms (blue spheres) and the analogous region of murine IL-2, which is implicated by mouse mutagenesis (red spheres) to interact with murine IL-2Rα.
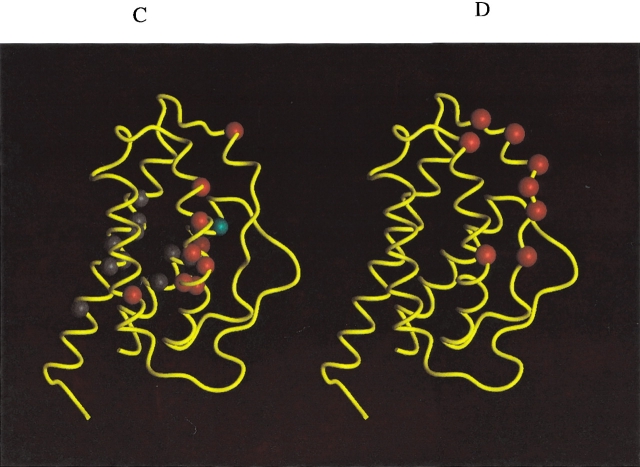
Figure 4.
Peptide backbone of IL-2 as determined by X-ray crystallography (M. Hatada, S. Surgenor, D. Weber, W. Danho, and V. Madison, unpubl.) with colored balls illustrating specific atom positions of interest. (A) Blue, gray, and green balls represent N atom positions corresponding to HSQC 15NH signal behavior defined as categories 1, 2, and 3, respectively (see text). (B) Red spheres represent alpha carbon atom (Cα) positions where mutagenesis studies have identified murine IL-2 side-chain interactions important in the recognition of murine IL-2Rα (Zurawski et al. 1993). The human IL-2 residues homologous to those found in the murine IL-2 mutagenesis study include: N30, N33, K35, R38, M39, L40, F42, K43, F44, Y45, E61, E62, P65, V69, L72, Q74, D109, T113, and V115. (C) H/D exchange results. Red spheres mark N atom positions where human IL-2/IL-2Rα complex formation showed significant reduction in NH exchange rate with solvent. Gray spheres showed no change in H/D exchange behavior upon complex formation. The cyan sphere is the F117 N atom position where NH exchange is more rapid upon IL-2/IL-2Rα complex formation. (D) Red spheres represent the Cα positions where human IL-2 mutagenesis studies identified residues important for complex formation with human IL-2Rα: K35, R38, F42, and K43 (Sauve et al. 1991) and T41, Y45, E62, and L72 (G. Ju, unpubl).
H/D exchange mapping of the IL-2 surface which interacts with IL-2R α
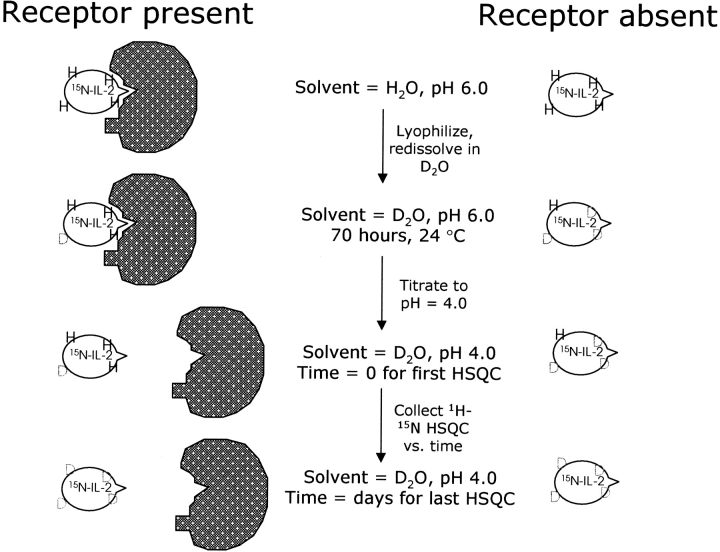
H/D exchange protection experiments have been performed to provide a complementary mapping of the IL-2/IL-2Rα interaction surface. The H/D exchange experiment is based on differential solvent exchange properties for IL-2 NH protons in the presence and absence of receptor. Figure 5 ▶ illustrates the experimental strategy wherein the 15N-IL-2/IL-2Rα complex is lyophilized out of H2O, then dissolved in D2O and incubated as a complex for 70 h at pH 6.0. At the end of the high pH incubation period, the pH is dropped to 4.6 and 1H-15N HSQC spectra are collected as a function of time. The pH drop serves two purposes: (1) it causes dissociation of the 15N-IL-2/IL-2Rα complex, returning the HSQC spectrum to the same chemical shift and line width characteristics as seen for the uncomplexed spectrum at pH 4.6; (2) it helps to reduce any further exchange of dissociated 15N-IL-2 NH protons with solvent beyond that which occurred in the bound state. The NH signal intensities from the pH 4.6 spectra of the 15N-IL-2/IL-2Rα sample were compared to those from control HSQC spectra, where 15N-IL-2 was treated according to the same scheme but in the absence of receptor, as shown in the right half of Figure 5 ▶. A single, long incubation time in D2O of 70 h was used to ensure that any differential NH exchange effects would represent substantial differences in protection factors. Table 1 summarizes the results of the H/D protection measurements. NH signals whose exchange rates with solvent were dramatically reduced in the presence of receptor are defined as “protected” (N atoms shown as red spheres in Fig. 4C ▶). NH atoms of 15N-IL-2 whose exchange behavior remained the same in the presence of IL-2Rα are listed as “not affected” (gray spheres in Fig. 4C ▶). The NH of F117 (cyan sphere in Fig. 4C ▶) exhibited an increased exchange rate upon binding IL-2Rα, and is therefore listed as “antiprotected.” The pattern of protection from exchange, as shown in Figure 4C ▶, was found to represent a subset of the IL-2Rα interaction surface mapped by the HSQC method (Fig. 4A ▶). This is expected, because for some surface residues exchange is too fast to allow detection of residual NH magnetization within the time period needed to acquire the first HSQC spectrum. The involvement of such residues in the binding interaction cannot be assessed in this type of H/D exchange study. Additionally, while amide exchange rates could have been reduced by an overall “tightening” of the IL-2 structure upon binding to the receptor, the regiospecific pattern of protection argues against this explanation.
Figure 5.
Experimental strategy for the H/D exchange study.
Table 1.
Behavior of NH resonances in NMR experiments
| NMR experiment | NH behavior | Peptide NH atoms | Side-chain NH atoms |
| 1H-15N HSQC perturbations due to IL-2/IL-2Rα complex formation | Category 1: blue atoms in Figure 4A ▶ | G27, T41, T42, K43, F44, M46, K48, A50, T51, C58, L59, E61, E62, E106, E114, V115, E116 | N29 Nδ, N33 Nδ, W121 Nɛ |
| Category 2: gray atoms in Figure 4A ▶ | N26, T37, M104, C105, Y107, T113, I122 | N32 Nδ, Q57 Nɛ | |
| Category 3: green atoms in Figure 4A ▶ | K8, K49, K54, L80, R83, D84, S87, G98, E100, T101, F103, F124, I129, S130, T131, L132, T133 | Q11 Nɛ, Q13 Nɛ, Q22 Nɛ, Q74 Nɛ, N90 Nδ, Q121 Nɛ | |
| H/D Exchange perturbations due to IL-2/IL-2Rα complex formation | Protected: red atoms in Figure 4C ▶ | R38, L59, E60, E62, E63, L66, V69, V93, R120 | |
| Not affected: gray atoms in Figure 4C ▶ | L12, L17, L18, L19, L21, Q22, I24, L118, I122, C125 | W121 Nɛ | |
| Antiprotected: cyan atom in Figure 4C ▶ | F117 | ||
| 1H-15N HSQC perturbations due to IL-2/24 complex formation | Sum of the 1H and 15N chemical shifts is greater than 117 Hz (at 600 MHz 1H frequency): blue atoms in Figure 10 ▶ | I28, N30, Y31, K32, N33, K35, L36, T37, M39, L40, T41, F42, K43, F44, E62, L72, Q74, S75 | N33 Nδ |
A small molecule that inhibits the IL-2/IL-2R α interaction by binding to IL-2Rα
Previously reported antipeptide antibody mapping (Danho et al. 1987) and mutagenesis (Collins et al. 1988; Sauve et al. 1991) studies implicated a contiguous segment of the IL-2 protein as being involved in binding to the IL-2Rα. In the antibody mapping study, a series of linear peptides 12 to 19 residues in length, corresponding to overlapping fragments covering the entire IL-2 protein sequence, were used to generate the corresponding antipeptide antibodies. The antisera to peptides 27–41 and 23–41 significantly inhibited IL-2 binding to IL-2Rα (Danho et al. 1987). Focused mutagenesis studies of human IL-2 (Fig. 4D ▶) showed that the IL-2 residues important for binding to human IL-2Rα included K35, R38, F42, and K43 (Sauve et al. 1991), in agreement with the antipeptide antibody results. These residues occur in the helical portion of the AB loop according to the IL-2 crystal structure. Linear peptides encompassing these residues were not able to inhibit the IL-2/IL-2Rα interaction. One of the likely reasons for the lack of inhibitory activity of these peptides is their excessive flexibility.
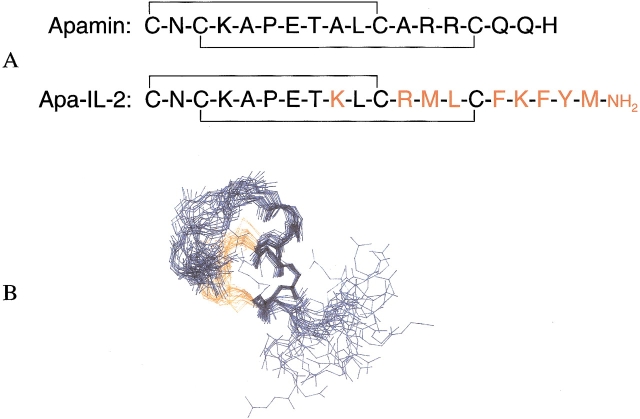
A constrained peptide (1 in Table 2) was designed in an attempt to hold the four amino acids found critical for binding more rigidly, in their native orientation. The scaffold used to achieve this rigidification was based on the bee venom peptide apamin, whose helical structure is stabilized by two disulfide bonds (Wemmer and Kallenbach 1983; Pease and Wemmer 1988). The IL-2-apamin hybrid (apa-IL-2, 1) was constructed by replacing apamin residues, that are noncritical for maintaining the secondary structure, with IL-2 residues, in a pattern designed to maintain their native spacing in IL-2 (Fig. 6A ▶). Apa-IL-2 was found to be an inhibitor of the IL-2/IL-2Rα interaction in a solid-phase binding assay. The IC50 value was measured as 230 ± 90 μM (Danho et al. 1996). More potent analogs in the 20 to 70 μM range were subsequently developed by replacing certain residues and by incorporating nonnatural amino acid substitutions. For example, replacement of the apa-IL-2 residue designed to mimic F42 with 2,6-dichlorobenzyl tyrosine increased the potency of the constrained peptide to 70 ± 40 μM (compare 1 and 2 in Table 2). Table 2 illustrates structure–activity relationships for a series of apa-IL-2 peptides with other selected mutations. The trends in the SAR of the peptide were found to match those obtained by mutagenesis of the corresponding residues in IL-2. The most potent inhibitor (7, 20 ± 10 μM) contained a double mutation with A30 and 2,6-dichlorobenzyl tyrosine42.
Table 2.
Apa-IL-2 structure–activity relationships
| IL-2 sequence | G27 | I28 | N29 | N30 | Y31 | K32 | N33 | P34 | K35 | L36 | T37 | R38 | M39 | L40 | T41 | F42 | K43 | F44 | Y45 | M46 |
| Apa-IL-2 sequence | C1 | N2 | C3 | K4 | A5 | P6 | E7 | T8 | K9 | L10 | C11 | R12 | M13 | L14 | C15 | F16 | K17 | F18 | Y19 | M20 |
| Compound number | Amino acid substitutions to Apa-IL-2 sequence | IC50 (μM) | ||||||||||||||||||
| 1 | None | 230 ± 90 | ||||||||||||||||||
| 2 | 2,6-Dichlorobenzyl-Tyrosine16 | 70 ± 40 | ||||||||||||||||||
| 3 | Alanine9; 2,6-Dichlorobenzyl-Tyrosine16 | inactive | ||||||||||||||||||
| 4 | Alanine12; 2,6-Dichlorobenzyl-Tyrosine16 | inactive | ||||||||||||||||||
| 5 | Alanine16 | inactive | ||||||||||||||||||
| 6 | 2,6-Dichlorobenzyl-Tyrosine;16 Alanine17 | inactive | ||||||||||||||||||
| 7 | Alanine4; 2,6-Dichlorobenzyl-Tyrosine16 | 20 ± 10 | ||||||||||||||||||
Figure 6.
(A) Primary sequence comparison of Apamin versus apa-IL-2. (B) Superposition of 30 solution structures determined by NMR for apa-IL-2 (backbone only is shown).
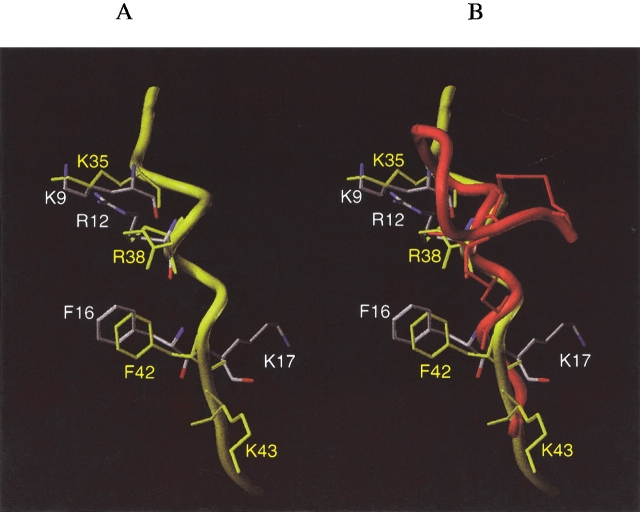
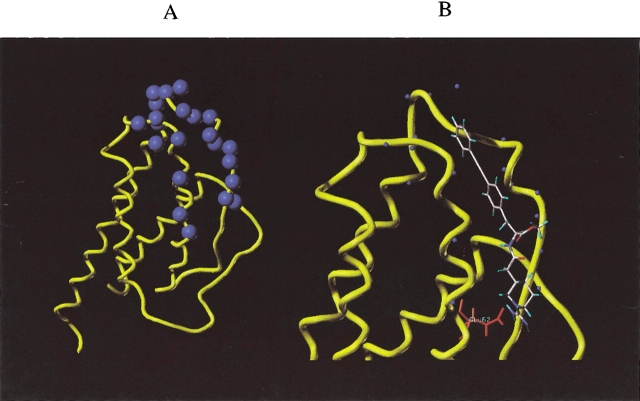
Figure 6B ▶ shows the backbone solution structure of apa-IL-2 determined by NMR as described previously (Fry et al. 1992; Greeley et al. 1992). The peptide backbone is held in an α-helical conformation by two disulfide bonds, as intended by the design, with a structure very similar to that of apamin (Wemmer and Kallenbach 1983). The residues K9, R12, F16, and K17 were designed to mimic IL-2 residues K35, R38, F42, and K43, of the AB-loop. Figure 7A ▶ shows a comparison of the positioning of these four side chains, using a representative solution structure of apa-IL-2 (in red, white, and blue), and the X-ray structure of IL-2 (shown in yellow). The alignment was chosen to maximize overlap of the four key side chains. For clarity, the apamin backbone is not shown in Figure 7A ▶. It is clear from this comparison that the positioning was successful for the residues representing K35, R38, and F42, but the C-terminal K17 does not overlap well with K43 of IL-2. Furthermore, the apamin backbone appears to obstruct access of these side chains to the IL-2Rα binding site, as shown in Figure 7B ▶, where the apa-IL-2 backbone is depicted in red. This partial steric blocking of key side-chain interactions by the apamin backbone is likely to be an additional limiting factor (in addition to mismatch of the K17/K43 position) for attaining high affinity for IL-2Rα.
Figure 7.
Overlay of the NMR structure of apa-IL-2 (colored by atom type) and the X-ray structure of the portion of IL-2 it was designed to mimic (colored yellow). In (A), only the side chains of apa-IL-2 are shown; in (B), both the side chains and the backbone (colored red) of apa-IL-2 are shown.
A small molecule that inhibits the IL-2/IL-2R α interaction by binding to IL-2
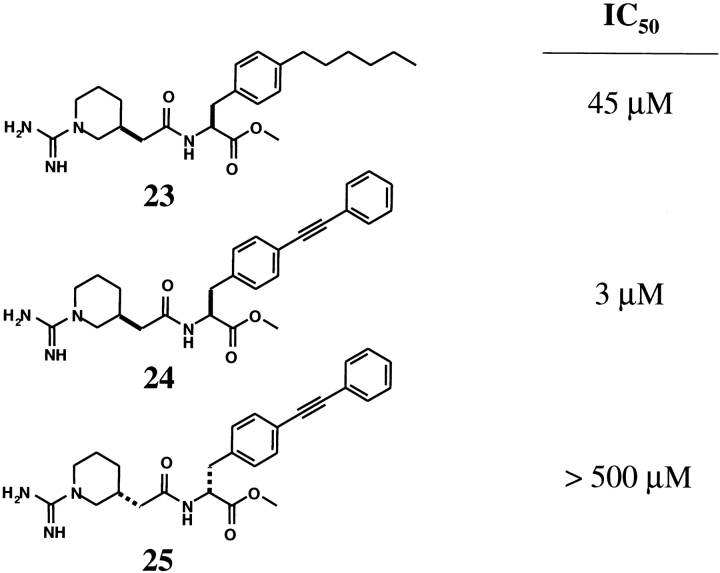
In an attempt to find a nonpeptidic inhibitor that would overcome the steric blocking problems of the apa-IL-2 design, a series of acylphenylalanine derivatives was prepared. Like the apa-IL-2 peptide, these compounds were intended to mimic the R38 to F42 residues of IL-2. One member of this initial series (23 in Fig. 8 ▶) was found to be an inhibitor of the IL-2/IL-2Rα interaction in the scintillation proximity assay, with an IC50 of approximately 45 μM. Synthesis of derivatives of the lead compound ultimately resulted in the inhibitor designated 24 (Fig. 8 ▶), which exhibited an IC50 value of 3 μM (Tilley et al. 1997). Although these acylphenylalanine derivatives were designed to bind to IL-2Rα, NMR studies revealed that compound 24 bound to IL-2. Using 0.59 mM 15N-IL-2, a series of 1H-15N HSQC experiments were collected at concentrations of 24 providing 0, 1.0, 1.5, and 2 equivalent ratios of 24 to 15N-IL-2. Figure 9A ▶ shows an overlay of the 15N-IL-2 HSQC spectra in the absence (black) and presence (red) of 24. A plot of the absolute values of the 1H and 15N chemical shift changes as a function of primary sequence number is illustrated in Figure 9B ▶. The horizontal line at 115 Hz defines the threshold used to differentiate between NH positions where the chemical shifts are “highly perturbed,” from those where the shift changes are small. Figure 10A ▶ highlights the N atoms corresponding to highly perturbed signals in Figure 9B ▶. The pattern indicates that inhibitor 24 binds to the IL-2 surface, at a site which overlaps the IL-2Rα binding epitope previously identified by site directed mutagenesis, HSQC perturbation mapping, and H/D exchange protection. The specificity of 24 is illustrated by the fact that its enantiomer (25 in Fig. 8 ▶) has no effect on the IL-2 HSQC spectrum when added to a level of 2 equivalents. Accordingly, scintillation proximity assays showed that 25 was not able to inhibit the IL-2/IL-2Rα interaction (IC50 > 500 μM).
Figure 8.
Chemical structures of compounds described in the text, and their inhibitory activity as measured in the IL-2/IL2Rα SPA assay.
Figure 9.
(A) Overlay of HSQC spectra of 15N-IL-2: black = before addition; red = after the addition of 2.0 equivalents of 24. (B) Sum of the absolute value of Δδ1H and Δδ15N chemical shift differences for 15N-IL-2 resonances in the presence and absence of 2.0 equivalents of 24. 1H shift differences are in black and 15N shift differences are in white. Shift differences are calculated as Δδ = |δ2eq − δ0eq|, where δ2eq represents the chemical shift (in Hz) for the 15N-IL-2 signal in the presence of 2.0 equivalents of 24, and δ0eq represents the chemical shift for 15N-IL-2 alone. Chemical shifts were measured in 2D 1H-15N HSQC spectra (Bodenhausen and Ruben 1980) at 40°C, and pH 4.6; in a buffer containing 10 mM sodium acetate, 0.2% sodium azide, 10% D2O, and 90% H2O; using a Varian Unityplus 600 spectrometer. The largest combined shift difference (Δδ1H+Δδ15N) was measured to be 303 Hz for F42. The horizontal line at 115 Hz is the threshold above which 1H-15N resonances are defined as “highly perturbed” by the addition of 2 equivalents of 24 to 15N-IL-2.
Figure 10.
(A) Blue spheres indicate N atom positions whose NH resonances are highly perturbed upon addition of 24. These resonances include: I28, N30, Y31, K32, N33, K35, L36, T37, M39, L40, T41, F42, K43, F44, E62, K64, E68, L72, Q74, S75, and A112 backbone NHs; and the side-chain NδH2 of N33. (B) Model of 24 bound to IL-2, derived from the NMR data. Glu62 is highlighted in red.
Figure 10B ▶ illustrates a model of the IL-2/24 complex, where the inhibitor is positioned in a cleft between the B-helix and the AB-loop on IL-2. This model is guided by four features: first, the shape of the chemical shift perturbation pattern is complementary with the long axis of the compound; second, the polar/hydrophobic complementarity of the inhibitor and the IL-2 groove serve to establish the relative head-to-tail orientation shown; third, the structure–activity results for related analogs (data not shown) indicate that the carbonyl group in the middle is probably oriented toward solvent rather than interacting with the protein; fourth, the hydrogen bonding between the guanidinium group of the inhibitor and the E62 side chain carboxylic acid group provides a favorable interaction, and displaces an ordered water molecule seen in the X-ray crystal structure. In forming the model, no alteration was allowed in the crystal structure of IL-2. The model shown exhibits steric crowding between the inhibitor and the IL-2 protein, which could be relieved by a relative separation of the B-helix and AB-loop by about one Angstrom. To our knowledge, the IL-2/24 complex represents the first example of a small molecule, nonpeptide inhibitor of a cytokine/receptor interaction in which the inhibitor binds to the cytokine.
Discussion
The surface of IL-2 which interacts with IL-2R α
Peptide antibody work originally focused attention on the AB-loop of IL-2 as being the primary epitope involved in the interaction with IL-2Rα. As described herein, NMR experiments involving H/D exchange footprinting and HSQC perturbation mapping present a mutually consistent, and slightly more extensive, picture of the surface of IL-2 that interacts with IL-2Rα. This surface is in good agreement with that defined by previous human and murine IL-2 mutagenesis results.
The H/D exchange data implicate the B and B′ helices as being involved in the IL-2/IL-2Rα interaction. In agreement with this, human IL-2 mutagenesis studies show that mutation of E62 of the B-helix strongly reduces the affinity of the IL-2/IL-2Rα complex (G. Ju, unpubl.). The characterization of this IL-2 surface by H/D exchange, although designed for maximal reliability, is somewhat incomplete, due to the single, long incubation time used. A more complete characterization of the protected surface might be possible by such H/D exchange methods (Brandt and Woodward 1987; Paterson et al. 1990), but would be consumptive of protein, requiring multiple samples of complexes for incubation at varying times. Because HSQC perturbation provided an alternative method for mapping the IL-2/IL-2Rα interaction surface, the more extensive H/D exchange experiments were not pursued. One surprise in the H/D exchange data was the presence of an antiprotected NH at F117. This may be due a conformational change resulting in increased solvent access to the side of IL-2 opposite from the IL-2Rα interaction site. Mutation of F117 to A117 reduced bioactivity only moderately, to 35% of wild-type human IL-2 (G. Ju, unpubl.), suggesting that this residue does not play a major role in binding to the IL-2Rα.
The HSQC perturbation mapping strategy proved to be a very efficient and economical technique for identification of the binding region on IL-2, using a single NMR sample of the IL-2/IL-2Rα complex. The IL-2/IL-2Rα interaction surface that was defined by HSQC perturbation mapping agrees well with that defined by mutagenesis of IL-2. These data reinforce the importance of residues at the crossing of the AB-loop and the CD-loop in the IL-2/IL-2Rα interaction.
Although no high-resolution crystal structure of the IL-2/IL-2Rα complex has been reported, the structure of the IL-4/IL-4Rα complex has been solved (Hage et al. 1999). Despite the fact that IL-2R and IL-4R are members of the same cytokine receptor family, the surfaces of IL-2 and IL-4 that interact with their respective receptor α components, are different. In the case of IL-2, the binding surface determined by both NMR and mutagenesis appears to be centered mainly at the AB-loop, with some interactions on both sides, including the B/B′ helices and the CD-loop. In the case of IL-4, the crystal structure and mutagenesis data suggest that the interaction surface is centered at the C-helix, with some additional interactions from the A-helix and the B/B′-helices. Therefore, it may be difficult to draw conclusions about the details of the IL-2/IL-2Rα interaction by analogy to the IL-4/IL-4Rα crystal structure.
Small molecule inhibitors of cytokine/receptor interactions
Preventing an extracellular protein messenger from productively interacting with its membrane-bound receptor represents a powerful strategy for inhibition of receptor-mediated signal transduction. The primary advantage of this therapeutic strategy is that blocking a single extracellular interaction can prevent an entire cascade of undesired intracellular responses. In contemplating the development of a small molecule inhibitor of this type, one must confront two key questions. First, can a molecule in the size range of 0.5 to 1 kD disrupt a sufficient number of interactions between two large proteins to be able to compete effectively, particularly when the natural ligand possesses a relatively high binding affinity? This appears to be theoretically possible for the Human Growth Hormone/Growth Hormone Receptor system (HGH/GHR), which has been studied in structural (de Vos et al. 1992) and thermodynamic (Clackson and Wells 1995) detail. The hormone/receptor interface between HGH and GHR involves approximately 30 amino acid side chains from each protein, yet it has been shown that localized interactions involving primarily two amino acids (W104, W169) of the GHR can account for most of the total binding energy of the HGH/GHR complex (Clackson and Wells 1995). Actual examples of effective small-molecule interactions in hormone/receptor systems include peptide and small molecule agonists capable of mimicking the interactions made by Erythropoietin (Livnah et al. 1996; Qureshi et al. 1999; Goldberg et al. 2002) and Thrombopoietin (Cwirla et al. 1997); a 270-Dalton compound that has been shown to restore high affinity between HGH (T175-to-G175 mutant) and the extracellular domain of GHR (W104-to-G104 mutant) (Guo et al. 2000); and small molecule mimics of insulin (Zhang et al. 1999) and granulocyte-colony-stimulating factor (Tian et al. 1998).
The second question concerns the choice of targeting the hormone or the receptor for small-molecule binding. At the systemic level, consideration of details such as location, accessibility, and relative populations, can complicate the issue of target selection. It may be that the best choice of target can only be confirmed by in vivo testing. However, at the molecular level, the question can be evaluated by studying the binding surface of each protein. Initially, there may be a conceptual bias that the receptor is the more attractive target. The receptor is generally larger than its protein ligand, and might therefore be expected to possess a “pocket” into which protruding regions of the ligand insert (Fig. 11 ▶, top left). This notion was reinforced in our earlier studies of IL-2 in which an antibody raised against a protruding segment of IL-2 was able to inhibit the interaction of the ligand with its counterpart IL-2Rα. An inhibitor was then envisioned as a molecule that could adequately mimic this protrusion of IL-2 (Fig. 11 ▶, top right). A peptidic inhibitor (1), designed to mimic the binding epitope of IL-2, was successfully developed. However, its binding affinity for IL-2Rα was low (IC50 of 230 μM) compared to human IL-2 (IC50 ~ 10 nM). Determination of the solution structure of 1 by NMR revealed that, although the desired overall conformation was attained, only three of the four key amino acid side chains showed an inherent propensity to exactly mimic their native positions in IL-2. Furthermore, the apamin-like peptide backbone appeared to cause partial steric interference between these key side chains and the receptor.
Figure 11.
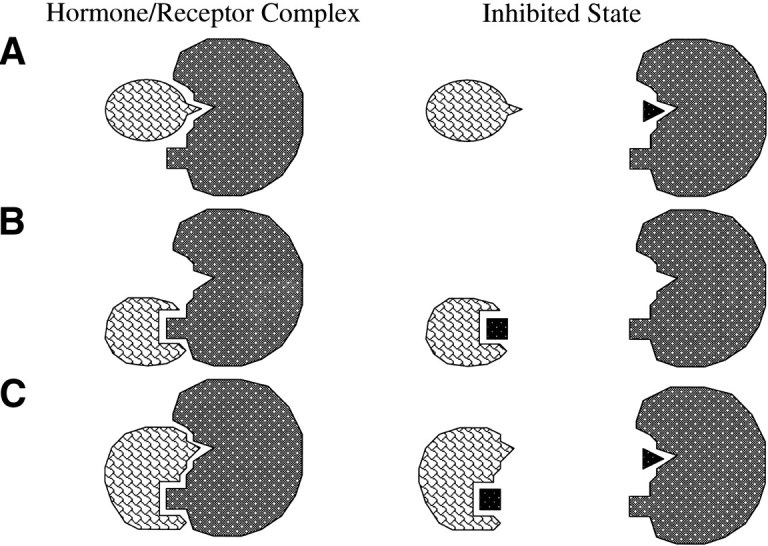
Three possible scenarios for inhibition of a hormone–receptor complex by a small molecule: (A) receptor as target; (B) hormone as target; (C) either as target.
In general, there is no reason to rule out a binding strategy in which the receptor presents protrusions, and the ligand provides complementary indentations (Fig. 11 ▶, middle left). In this case, an inhibitor might be envisioned that resembles the binding epitope of the receptor (Fig. 11 ▶, middle right). For the case of IL-2/IL-2Rα, such an inhibitor was discovered (24). Although initially developed to be a nonpeptidic version of the IL-2 epitope, 24 was shown by NMR to bind to IL-2. Chemical shift perturbations of 15N-IL-2 during titration with this inhibitor were used to map the binding site, which was shown to be the same as the receptor-binding site, thus revealing a competitive mode of inhibition. The binding model for the 24/IL-2 interaction suggests that a minor separation (approximately 1 Angstrom) is induced between the B′-helix and the AB-loop region of IL-2. The primary forces driving complex formation appear to involve hydrophobic interactions at one end, and polar interactions at the other. The ring moieties of the biphenyl acetylene of 24 can form favorable interactions with the side chains of F42 and R38. The guanidinium group of the inhibitor most likely forms a hydrogen-bond with the side chain of E62. The formation of this H-bond is supported by the observation that the affinity of 24 for IL-2 is pH dependent (Tilley et al. 1997). An approximate Kd of 2–6 mM for 24 binding to 15N-IL-2 at pH = 4.6 can be estimated from the data in Figure 9 ▶. Accurate determination of Kd at higher pH (pH = 7.5) by NMR is difficult because of limited solubility of IL-2 at this pH; however, approximate Kd measurement by NMR at pH = 7.5 clearly illustrates that a stronger complex is formed. The IC50 measured in the scintillation proximity assay at pH = 7.5 is 3 μM for 24 binding to IL-2. The dependence of Kd on pH is likely due to titration of the acid group of E62.
The most likely situation is that the binding interface is more complex than the common conception of assigning one partner the role of “lock” and the other of “key,” but instead involves a multifaceted surface (Fig. 11 ▶, bottom left), allowing for more than one type of inhibitor (Fig. 11 ▶, bottom right). As described here, the IL-2/IL-2Rα complex is the first hormone/receptor system for which both of two possible types of small molecule inhibitors have been obtained—one that binds to the receptor, and the other that binds to the cytokine ligand. It is expected that similar opportunities exist for developing inhibitors for other systems involving protein/protein interactions.
Materials and methods
Protein expression and purification
15N-labeled IL-2 was produced by metabolic labeling of IL-2 produced in the yeast Pichia pastoris. Pichia cells were transformed with an expression vector that directed the synthesis and secretion of human IL-2 under the control of a methanol-inducible promoter. The cells were grown in media containing 15N-ammonium sulfate (Cambridge Isotopes) for 2 days at 30°C. The cells were then concentrated and IL-2 expression was induced by addition of methanol to the culture. The culture medium containing secreted IL-2 was harvested. IL-2 was purified by affinity chromatography over an IL-2Rα column as described previously (Bailon et al. 1987).
NMR experiments
HSQC perturbation mapping of the IL-2/IL-2R α complex
A sample of 1 mM 15N-IL-2 in 10 mM deuterated sodium acetate, 0.02% sodium azide, 90% H2O, and 10% D2O (acetate buffer) was used to collect 15N-1H HSQC spectra (Bodenhausen and Ruben 1980) at pH 4.6, 5.5, and 6.1 on a Varian Unity 500-MHz NMR spectrometer. This set of spectra was used to transfer NH resonance assignments from pH 4.6 to pH 6.1. The pH 6.1 spectrum from this titration series is shown in black in Figure 3 ▶. The red spectrum in Figure 3 ▶ represents the HSQC spectrum of the 15N-IL-2/IL-2Rα complex in acetate buffer. The 15N-IL-2/IL-2Rα complex was formed by mixing 375 μL of 0.9 mM 15N-IL-2 in pH 6.1 acetate buffer with 255 μL of 1.5 mM IL-2Rα to obtain a final 630 μL solution that was 0.56 mM 15N-IL-2 and 0.6 mM IL-2Rα. The HSQC spectra were recorded at 40°C. 1H chemical shifts were referenced relative to H2O defined as 4.62 ppm relative to DSS. 15N chemical shifts were referenced relative to a separate sample of 15NH4Cl in 1 M HCl where the 15N resonance is defined as 24.93 ppm relative to ammonia.
H/D exchange experiments
The 15N-IL-2/IL-2Rα complex described above was lyophylized from acetate buffer. The lyophylate was then dissolved in D2O and incubated at 23.5°C for 70 h. After the incubation period, the sample was titrated to pH 4.6 and transferred to a 5 mm NMR tube. An HSQC spectrum was collected 0.5 h after dropping the pH from 6.1 to 4.6 and 70.5 h after dissolution of the lyophylate into D2O/acetate buffer. Control experiments were run in which a sample of 1 mM 15N-IL-2 in acetate buffer was lyophylized and redissolved in D2O. The D2O solution was incubated for 70 h at pH 6.1 and then titrated to pH 4.6. An HSQC spectrum was collected at 70.5 h after dissolution into D2O, which corresponds to 0.5 h after pH adjustment. HSQC spectra were also collected at later time points to follow the time course of the disappearance of NH resonance intensities in the samples with and without receptor.
HSQC perturbation mapping of the 15N-IL-2/ 24 complex
Chemical shifts were measured from 1H-15N HSQC spectra (Bodenhausen and Ruben 1980) at 40°C, and pH 4.6; in acetate buffer, using a Varian Unityplus 600 spectrometer.
Binding assays
For the evaluation of compounds as IL-2/IL-2Rα binding inhibitors, we employed a scintillation proximity assay (SPA). This assay system employs biotinylated soluble IL-2Rα conjugated to streptavidin-coated scintillant-impregnated beads. Equilibrium binding of 125I-labeled IL-2 to these beads is measured in the presence and absence of inhibitor titrations. IC50 determinations were made from Hill plots of the inhibition curves. In this assay format, 24 exhibited an IC50 of 3 μM, whereas 25 gave only a 14% inhibition of binding at a highest concentration of 500 μM and IL-2 had an IC50 of 12.7 nM.
Further experiments were performed to determine whether the observed activity was competitive by measuring the specific binding of varying concentrations of 125I-IL-2 to IL-2Rα-SPA beads, in the presence of varying concentrations of 24 or IL-2. Scatchard analyses of the 125I-IL-2 binding at each inhibitor concentration were performed and the resultant Kd values plotted versus inhibitor concentration in a Schild plot. The resultant linear plot indicates that the observed antagonistic effect is consistent with a competitive reversible inhibition. Schild regressions of the effects of unlabeled IL-2 and compound 24 on 125I-IL-2 binding to soluble IL-2Rα. Plots are derived from Scatchard analyses of 125I-IL-2 binding to the receptor in the presence of the plotted concentrations of unlabelled IL-2 or compound. DR represents the dose ratio and is defined as the Kd of 125I-IL-2 binding in the presence of inhibitor divided by the Kd in the absence of an inhibitor. For apamin-like peptides, a solid-phase assay was used to measure competition with IL-2 biotin for binding to IL-2Rα.
Acknowledgments
We thank Peter Kim for the suggestion of using apamin as an α-helical scaffold in the design of peptides which mimic the AB-loop of IL-2.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0232803.
References
- Bailon, P., Danho, W., Kenney, R.F., Fredricks, J.E., Smith, C., Familletti, P.C., and Smart, J.E. 1987. Receptor-affinity chromatography: A one-step purification for recombinant interleukin-2. Bio/Technology 5 1195–1198. [Google Scholar]
- Bodenhausen, G. and Ruben, D.J. 1980. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 69 185–189. [Google Scholar]
- Brandhuber, B.J., Boone, T., Kenney, W.C., and McKay, D.B. 1987. Three-dimensional structure of interleukin-2. Science 238 1707–1709. [DOI] [PubMed] [Google Scholar]
- Brandt, P. and Woodward, C. 1987. Hydrogen exchange kinetics of bovine pancreatic trypsin inhibitor β-sheet protons in trypsin-bovine pancreateic trypsin inhibitor, trypsinogen-bovine pancreatic trypsin inhibitor, and trypsinogen-isoleucylvaline-bovine pancreatic trypsin inhibitor. Biochemistry 26 3156–3162. [DOI] [PubMed] [Google Scholar]
- Clackson, T. and Wells, J.A. 1995. A hot spot of binding energy in a hormone–receptor interface. Science 267 383–386. [DOI] [PubMed] [Google Scholar]
- Collins, L., Tsien, W.-H., Seals, C., Hakimi, J., Weber, D., Bailon, P., Hoskings, J., Greene, W.C., Toome, V., and Ju, G. 1988. Identification of specific residues of human interleukin 2 that affect binding to the 70-kDa subunit (p70) of the interleukin 2 receptor. Proc. Natl. Acad. Sci. 85 7709–7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cwirla, S.E., Balasubramanian, P., Duffin, D.J., Wagstrom, C.R., Gates, C.M., Singer, S.C., Davis, A.M., Tansik, R.L., Mattheakis, L.C., Boytos, C.M., et al. 1997. Peptide agonist of the thrombopoietin receptor as potent as the natural cytokine. Science 276 1696–1699. [DOI] [PubMed] [Google Scholar]
- Danho, W., Markofske, R., Swistok, J., Michalewsky, J., Gabriel, T., Jenson, J., Tsien, W.-H., and Gately, M. 1987. Neutralizing rabbit antibodies to synthetic human interleukin-2 (IL-2) peptides. In Proceedings of the Tenth American Peptide Symposium, pp. 537–539. ESCOM, Leiden, The Netherlands.
- Danho, W., Markofske, R., Swistok, J., Hakimi, J., Kondas, J., Powers, G., Biondi, D., Varnell, T., Fry, D.C., Greeley, D.N., et al. 1996. Design, synthesis, and conformational analysis of an IL-2/apamin hybrid: A peptide with IL-2 receptor antagonist activity. Drug Design Discov. 13 162. [Google Scholar]
- de Vos, A.M., Ultsch, M., and Kossiakoff, A.A. 1992. Human growth hormone and extracellular domain of its receptor: Crystal structure of the complex. Science 255 306–312. [DOI] [PubMed] [Google Scholar]
- Emerson, S.D., Madison, V.S., Palermo, R.E., Waugh, D.S., Scheffler, J.E., Tsao, K.-L., Kiefer, S.E., Liu, S.P., and Fry, D.C. 1995. Solution structure of the ras-binding domain of c-raf-1 and identification of its ras interaction surface. Biochemistry 34 6911–6918. [DOI] [PubMed] [Google Scholar]
- Fry, D.C., Madison, V.S., Greeley, D.N., Felix, A.M., Heimer, E.P., Frohman, L., Campbell, R.M., Mowles, T.F., Toome, V., and Wegrzynski, B.B. 1992. Solution structures of cyclic and dicyclic analogues of growth hormone releasing factor as determined by two-dimensional NMR and CD spectroscopies and constrained molecular dynamics. Biopolymers 32 649–666. [DOI] [PubMed] [Google Scholar]
- Greeley, D., Danho, W., Madison, V., and Fry, D. 1992. NMR studies of peptide fragments and analogs of interleukin-2. Biophys. J. 61 469A. [Google Scholar]
- Goldberg, J., Jin, Q., Ambroise, Y., Satoh, S., Desharnais, J., Capps, K., and Boger, D.L. 2002. Erythropoetin mimetics derived from solution phase combinatorial libraries. J. Am. Chem. Soc. 124 544–555. [DOI] [PubMed] [Google Scholar]
- Guo, Z., Zhou, D., and Schultz, P. 2000. Designing small-molecule switches for protein–protein interactions. Science 288 2042–2045. [DOI] [PubMed] [Google Scholar]
- Hage, T., Sebold, W., and Reinemer, P. 1999. Crystal structure of the interleukin-4/receptor a chain complex reveals a mosaic binding interface. Cell 97 271–281. [DOI] [PubMed] [Google Scholar]
- Hakimi, J., Mould, D., Waldmann, T.A., Queen, C., Anasetti, C., and Light, S. 1997. Development of Zenapax: A humanized anti-Tac antibody. In Antibody Therapeutics (eds. W.J. Harris and J. Adair), pp. 277–300. CRC Press, Boca Raton, FL.
- Kaempfer, R. 1994. Regulation of the human interleukin-2/interleukin-2 receptor system: A role for immunosuppression. Proc. Soc. Exp. Biol. Med. 206 176–180. [DOI] [PubMed] [Google Scholar]
- Kraulis, P. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Livnah, O., Stura, E., Johnson, D.L., Middleton, S.A., Mulcahy, L.S., Wrighton, N.C., Dower, W.J., Jolliffe, L.K., and Wilson, I.A. 1996. Functional mimicry of a protein hormone by a peptide agonist: The EPO receptor complex at 2.8 Angstroms. Science 273 464–471. [DOI] [PubMed] [Google Scholar]
- Marion, D., Driscoll, P.C., Kay, L.E., Wingfield, P.T., Bax, A., Gronenborn, A.M., and Clore, G.M. 1989. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: Application to interleukin 1β. Biochemistry 28 6150–6156. [DOI] [PubMed] [Google Scholar]
- Matsuo, H., Walters, K., Teruya, K., Tanaka, T., Gassner, G.T., Lippard, S.J., Kyogoku, Y., and Wagner, G. 1999. Identification by NMR spectroscopy of residues at contact surfaces in large, slowly exchanging macromolecular complexes. J. Am. Chem. Soc. 121 9903–9904. [Google Scholar]
- McKay, D. 1992. Response to: Unraveling the structure of IL-2. Science 257 412–413.17832837 [Google Scholar]
- Mott, H.R., Driscoll, P.C., Boyd, J., Cooke, R.M., Weir, M.P., and Campbell, I.D. 1992. Secondary structure of human interleukin-2 from 3D heteronuclear NMR experiments. Biochemistry 31 7741–7744. [DOI] [PubMed] [Google Scholar]
- Mott, H.R., Baines, B., Hall, R.M., Cooke, R.M., Driscoll, P.C., Weir, M.P., and Campbell, I.D. 1995. The solution structure of the F42A mutant of human interleukin 2. J. Mol. Biol. 247 979–994. [DOI] [PubMed] [Google Scholar]
- Paterson, Y., Englander, S.W., and Roder, H. 1990. An antibody binding site on cytochrome c defined by hydrogen exchange and two-dimensional NMR. Science 249 755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease, J.H.B. and Wemmer, D.E. 1988. Solution structure of apamin determined by nuclear magnetic resonance and distance geometry. Biochemistry 27 8491–8498. [DOI] [PubMed] [Google Scholar]
- Qureshi, S.A., Kim, R.M., Kontestis, Z., Biazzo, D.E., Motamedi, H., Rodrigues, R., Boice, J.A., Calaycay, J.R., Bednarek, M.A., Griffin, P., et al. 1999. Mimicry of a erythropoietin by a nonpeptide molecule. Proc. Natl. Acad. Sci. 96 12156–12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve, K., Nachman, M., Spence, C., Bailon, P., Campbell, E., Tsien, W.-H., Kondas, J.A., Hakimi, J., and Ju, G. 1991. Localization in human interleukin 2 of the binding site to the a chain (p55) of the interleukin 2 receptor. Proc. Natl. Acad. Sci. 88 4636–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprang, S.R. and Bazan, J.F. 1993. Cytokine structural taxonomy and mechanisms of receptor engagement. Curr. Opin. Struct. Biol. 3 815–827. [Google Scholar]
- Tian, S.-S., Lamb, P., King, A.G., Miller, S.G., Kessler, L., Luengo, J.I., Averill, L., Johnson, R.K., Gleason, J.G., Pelus, L.M., et al. 1998. A small nonpeptidyl mimic of granulocyte-colony-stimulating factor. Science 281 257–259. [DOI] [PubMed] [Google Scholar]
- Tilley, J.W., Chen, L., Fry, D.C., Emerson, S.D., Powers, G.D., Biondi, D., Varnell, T., Trilles, R., Guthrie, R., Mennona, F., et al. 1997. Identification of a small molecule inhibitor of the IL-2/IL-2Rα receptor interaction which binds to IL-2. J. Am. Chem. Soc. 119 7589–7590. [Google Scholar]
- van Nuland, N.A.J., Kroon, G., Dijkstra, K., Wolters, G.K., Scheek, R.M., and Robillard, G.T. 1993. The NMR determination of the IIAmtl binding site in HPr of the Escherichia coli phosphoenol pyruvate-dependent phosphotransferase system. FEBS Lett. 315 11–15. [DOI] [PubMed] [Google Scholar]
- Waldmann, T. 1993. The IL-2/IL-2 receptor system: A target for rational immune intervention. Immunol. Today 14 264–270. [DOI] [PubMed] [Google Scholar]
- Wemmer, D. and Kallenbach, N.R. 1983. Structure of apamin in solution: A two-dimensional nuclear magnetic resonance study. Biochemistry 22 1901–1906. [DOI] [PubMed] [Google Scholar]
- Yu, H., Rosen, M.K., Shin, T.B., Seidel-Dugan, C., Brugge, J.S., and Schreiber, S.L. 1992. Structure of the SH3 domain of src and identification of its ligand binding site. Science 258 1665–1668. [DOI] [PubMed] [Google Scholar]
- Zhang, B., Salituro, G., Szalkowski, D., Li, Z., Zhang, Y., Royo, I., Vilella, D., Diez, M.T., Pelaez, F., and Ruby, C. 1999. Discovery of a small molecule insulin mimetic with antidiabetic activity in mice. Science 284 974–977. [DOI] [PubMed] [Google Scholar]
- Zurawski, S.M., Vega, F., Doyle, E.L., Huyghe, B., Flaherty, K., McKay, D.B., and Zurawski, G. 1993. Definition and spatial location of mouse interleukin-2 residues that interact with its heterotrimeric receptor. EMBO J. 12 5513–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]