Abstract
Nondenaturing electrospray mass spectrometry (ESI-MS) has been used to reveal the presence of potential ligands in the ligand-binding domain (LBD) of orphan nuclear receptors. This new approach, based on supramolecular mass spectrometry, allowed the detection and identification of fortuitous ligands for the retinoic acid-related orphan receptor β (RORβ) and the ultraspiracle protein (USP). These fortuitous ligands were specifically captured from the host cell with the proper stoichiometry. After organic extraction, these molecules have been characterized by classic analytical methods and identified as stearic acid for RORβ and a phosphatidylethanolamine (PE) for USP, as confirmed by crystallography. These molecules act as “fillers” and may not be the physiological ligands, but they prove to be essential to stabilize the active conformation of the LBD, enabling its crystallization. The resulting crystal structures provide a detailed picture of the ligand-binding pocket, allowing the design of highly specific synthetic ligands that can be used to characterize the function of orphan nuclear receptors. An additional advantage of this new method is that it is not based on a functional test and that it can detect low-affinity ligands.
Keywords: Nuclear receptor; orphan receptor; ROR; USP; fortuitous ligand; electrospray mass spectrometry, noncovalent interactions
Nuclear receptors (NRs) are ligand-inducible transcription regulators involved in many important physiological processes such as development, reproduction, cell growth and differentiation, apoptosis, and key metabolic pathways. Classic NRs, which comprise a DNA-binding domain (DBD) and a ligand-binding domain (LBD), activate the transcription of target genes in response to the binding of small lipophilic molecules such as steroid and thyroid hormones, vitamin D, and retinoids to their LBD. Crystallographic studies of NR DBDs and LBDs have strongly helped to understand their mechanism of action (Renaud and Moras 2000). In addition, the past 10 years have witnessed a burst in the cloning of new members of this superfamily using various strategies. However, because the natural ligands and the physiological function of these putative receptors were unknown at the start, these new members of the superfamily were called orphan NRs (Giguère 1999). In some cases, physiological ligands have been found (Forman et al. 1997; Kliewer et al. 1997, 1999; Krey et al. 1997). In addition, high-affinity synthetic agonists and antagonists can be rationally designed on the basis of the three-dimensional structure of the receptor LBD, even if the physiological ligands are unknown.
Up to now the only method that leads to the structure determination of a nuclear receptor LBD is crystallography, which requires getting crystals, usually in the presence of a ligand to stabilize the active conformation. Recent studies have demonstrated that it was possible in some cases to crystallize a NR LBD taking advantage of the presence of a fortuitous ligand captured by the heterologously produced LBD from the expression host (Bourguet et al. 2000; Billas et al. 2001; Stehlin et al. 2001). This is particularly convenient when no ligand is known for a given receptor. Indeed, most orphan receptors are known only through their amino acid sequence and finding their physiological ligands and functions is very challenging.
With the introduction of electrospray ionization mass spectrometry (ESI-MS: Kebarle 2000), a new technique has emerged to study the biomolecular noncovalent interactions (Ganem et al. 1991; Katta and Chait 1991) allying sensitivity with high resolution detection. Since the initial reports, ESI-MS has had a great impact on the field of molecular biology and its ability to characterize increasingly heavy or fragile noncovalent complexes has been widely illustrated (Przybylski and Glocker 1996; Loo 1997; Jorgensen et al. 1998; Pramanik et al. 1998; Last and Robinson 1999; Tito et al. 2001). Indeed, even if the complex is transferred to a solvent-free environment, electrospray ionization allows the transfer of intact weakly associated noncovalent complexes in the gas phase, suggesting that at least some elements of the conformation in solution survive the electrospray ionization process, allowing the characterization of many noncovalent complexes in terms of existence, stoichiometry, and specificity (McLafferty et al. 1998; Loo 2000). Recent studies also report that ESI-MS can be used to determine the relative binding affinities in solution (Loo et al. 1997; Ayed et al. 1998) or to probe cooperativity in the binding of a ligand to an enzyme (Rogniaux et al. 2001). In the field of nuclear receptors, supramolecular mass spectrometry is still not a widely spread technique. For instance, it was successfully used to characterize the oligomeric state of the estrogen receptor LBD (Witkowska et al. 1996) or the influence of several ligands or metal ions on the stability of protein–protein or protein–DNA complexes, as in the case of the human vitamin D receptor (Veenstra et al., 1998), the retinoid X receptor (Craig et al. 1999, 2001), and the glucocorticoid receptor (Low et al. 2002).
In this paper, we report an ESI-MS study of the LBD of two orphan receptors, ultraspiracle protein (USP) and retinoic acid-related orphan receptor β (RORβ), for which the crystal structure was published recently (Billas et al. 2001; Stehlin et al. 2001). RORβ is a mammalian orphan receptor expressed exclusively in areas of the central nervous system, which seems to be involved in the processing of sensory information, but its physiological function and its cognate ligand still have to be determined. On the other hand, the USP is the insect ortholog of the vertebrate retinoid X receptor (RXR). It plays a crucial role as the heterodimerization partner of the ecdysone receptor (EcR) by stimulating the binding of ecdysteroids to EcR. It is not known how USP mediates its action on its partner receptor, whether it is ligand induced, and which are the physiologically relevant ligands of USP. For both receptors, ESI-MS has been used to control the homogeneity of the protein before crystallization in terms of sequence integrity and interactions with small molecules present in the expression system/purification environment. In both cases, nondenaturing ESI-MS revealed that unexpected small organic molecules were copurified with the proteins, whereas other analytical techniques [Fast Atom Bombardment (FAB) and thin layer chromatography (TLC) in the case of USP, gas chromatography-mass spectrometry (GC-MS) in the case of RORβ] have then been used to identify these molecules. The presence of an unexpected ligand in the binding pocket was then confirmed in both cases by the examination of the electron density map obtained from the x-ray diffraction data (Billas et al. 2001; Stehlin et al. 2001). Because crystallization of macromolecules requires, in particular, the chemical homogeneity of the sample, the characterization of fortuitous ligands and the estimation of their relative abundance by mass spectrometry will be an important tool for the study of orphan receptors in general.
Results
Detection of fortuitous ligands bound to the orphan receptors RORβ and USP by ESI-MS
Because ESI-MS is a method of choice for the study of noncovalent protein–ligand complexes, it seemed appropriate to look for ligands in the Escherichia coli-produced LBDs of the orphan receptors RORβ and USP. Such fortuitous ligands may have been captured by the protein either during its heterologous production or during its purification.
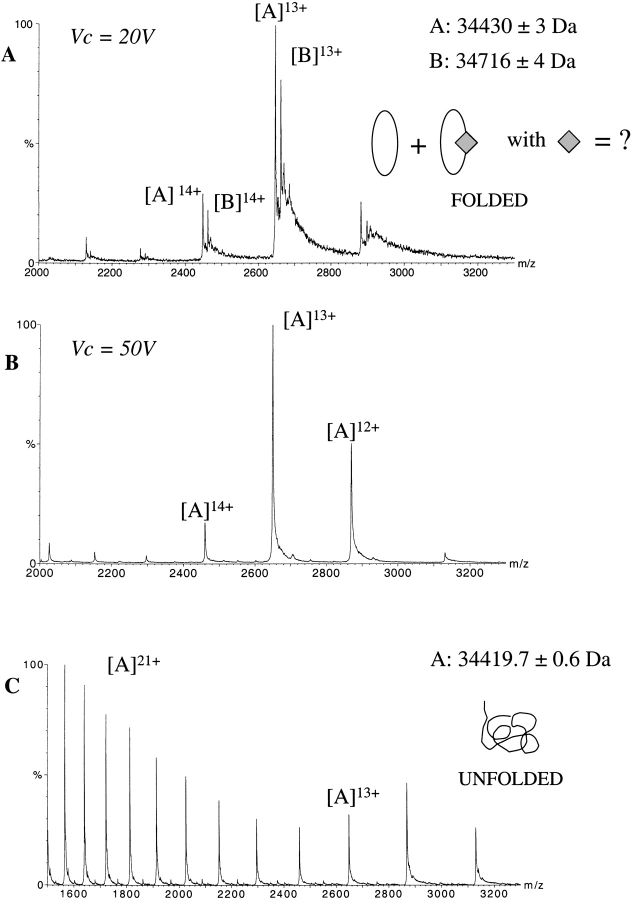
Figure 1A ▶ displays the electro-spray ionization (ESI) mass spectrum obtained for RORβ under nondenaturing conditions (20 mM AcONH4 at pH 6.5). Surprisingly, two series (A and B) of mass/charge (m/z) peaks corresponding to multiply charged ions (12 to 14 protonations) are observed. From series A, a molecular mass of 34,430 ± 3 Da is measured in good agreement with the expected mass of the RORβ monomer (34,425.3 Da). The molecular mass calculated from series B (34,716 ± 4 Da) is 286 Daltons larger than the mass measured from peak series A. The measurement performed under denaturing conditions (1% formic acid in 1:1 water/acetonitrile) reveals that this mass increment is due to noncovalent binding, as it is not present anymore when the protein is not properly folded. A single species is then observed corresponding to the RORβ monomer alone (Fig. 1C ▶). Unlike classic protein characterization techniques (gel electrophoresis, chromatography), nondenaturing ESI-MS shows that ~40% of the RORβ protein was noncovalently bound to one or several ligands of 286 Da.
Figure 1.
Detection of a fortuitous ligand in RORβ by non denaturing ESI-MS. ESI mass spectrum of RORβ in 25 mM NH4OAc at pH 6.5 at Vc = 20 V (A) and Vc = 50 V (B). Two series A and B of multiply-charged ions (12 to 14 protonations) are detected. The mass difference (ΔM) between the mass measurements shows the presence of an unexpected bound molecule of ~286 Da. Increasing the accelerating cone voltage from 20 V to 50 V resulted in more energetic collisions with residual gaseous molecules in the interface of the mass spectrometer, which led to the dissociation of the complex. Analysis of this preparation under denaturing conditions (50% CH3CN, 1% HCOOH) shows that RORβ is pure and homogenous and thus that the extra mass of 286 Da is due to a noncovalent attachment of a small organic molecule to the protein (C). apoRORβ (oval); fortuitous ligand (diamond).
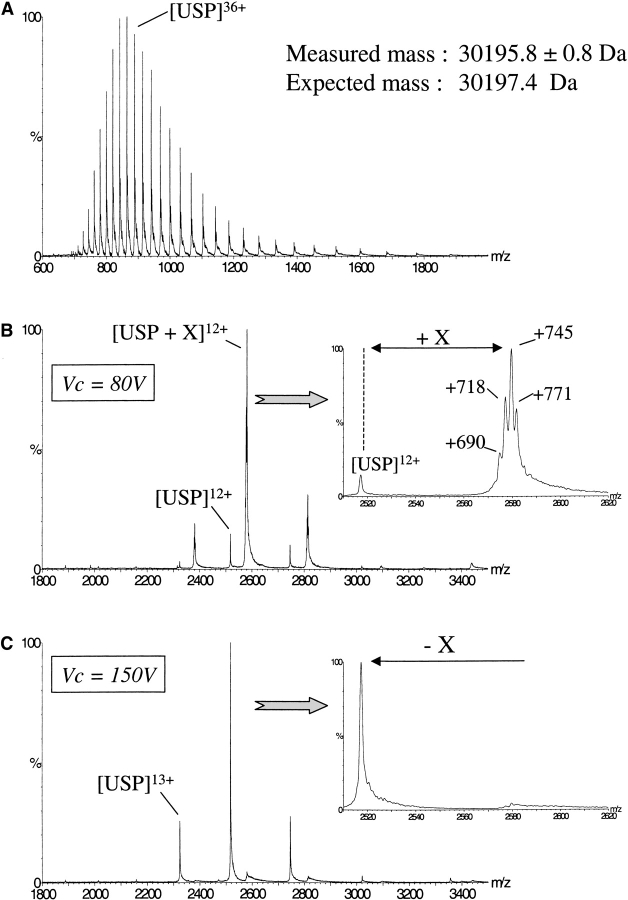
In the case of USP, ESI-MS analysis performed under denaturing conditions (Fig. 2A ▶) shows that the protein is pure, intact, and homogenous (measured mass = 30,195.8 ± 0.8 Da; expected mass = 30,197.4 Da). By using nondenaturing conditions (i.e., in 50 mM AcONH4 at pH 7), two series of m/z peaks are again observed (Fig. 2B ▶). The minor one (~10%) correspond to the expected USP monomer (MM = 30,196 ± 1 Da). The major one (90%) revealed the presence of a protein displaying a molecular mass larger by ~745 Da than the mass of the USP apoprotein. In a way similar to the observation made for RORβ, this mass increment is not detected when the protein is not properly structured and thus reflects a noncovalent binding of one or several molecules to the protein. An enlarged view of one single charged state (e.g., the most abundant one, 12+) reveals a heterogeneous mass distribution strongly suggesting that several ligands displaying mass differences of 26–28 Da bind to USP.
Figure 2.
Detection of a fortuitous ligand in USP by nondenaturing ESI-MS. Comparison of the masses obtained by ESI-MS under denaturing conditions (50% CH3CN, 1% HCOOH; A) and nondenaturing conditions (50 mM AcONH4 at pH 6.5; B) reveals noncovalent binding of one or several molecules with an average molecular weight of X = 745 Da. A zoom at the 12+ charge state shows that this noncovalent binding is heterogeneous and that several molecules are actually bound to USP with molecular masses of ~690, 718, 745, and 771 Da. Increasing the kinetic energy of the ions from Vc = 80V to Vc = 150V (C) led to the dissociation of the molecules initially bound to USP, resulting in a single ion series corresponding to USP apoprotein.
Under nondenaturing conditions, a narrow distribution of low charge states is observed while denaturing conditions yield a broad distribution. A low amount of charging as well as a small number of charge states are often explained by the conservation to some extent of the protein structure during the transfer of the ions into the gas phase. The restriction of charging may be due to fewer exposed basic sites or more severe coulombic restraints in the protein suggesting a still compact protein structure in the gas phase. In fact, several studies have demonstrated that ESI-MS can be used to follow protein conformational changes by looking at the charge state distribution (Witkowska et al. 1996; Dobo and Kaltashov 2001). In the case of USP where several ligands are bound to the receptor, the observation of one main charged state distribution suggests that the various ligands have not induced any significant structural heterogeneity.
Detection of noncovalent protein–ligand complexes requires working under conditions where the internal energy of the ions does not lead to the dissociation of the complex (Rostom and Robinson 1999; Tahallah et al. 2001). Experimentally, this is achieved by reducing the declustering voltage (Vc), which controls the kinetic energy of the generated ions through gas phase collisions in the atmospheric pressure/vacuum interface. It is a critical parameter for the observation of noncovalent complexes and has to be adjusted in each case such as to find the best compromise for removing solvent molecules without disrupting the macromolecular complex. A declustering voltage of 20 V was necessary to preserve the noncovalent RORβ–ligand complex in the gas phase. The resulting mass precision of ±4 Da is then not high enough to assign an exact molecular mass for the RORβ ligand. It is, however, sufficient to assign a binding stoichiometry of 1:1 ratio between RORβ and the ligand, assuming that a ligand displaying a molecular mass smaller than 150 Da is not likely. Below Vc = 20 V, the ratio between the ligand-bound and ligand-free protein stays unchanged (data not shown). This strongly suggests that the energy communicated to the ions in the atmospheric pressure/vacuum interface is minimized enough to preserve the noncovalent complex and that the observed free apoprotein was present in solution and not resulting from gas phase dissociation. On the other hand, increasing the declustering voltage to Vc = 50 Volts leads to the complete dissociation of the ligand from the protein and to the detection of a single species corresponding to the ligand-free RORβ (Fig. 1B ▶). The observation that the gas phase generated loss of 286 Da occurred in one step, suggests again that a single molecule of 286 Da was bound to the receptor.
In contrast to RORβ, the USP ligands appear to be quite resistant to gas phase collision. Indeed, until Vc = 100 V, the complex is still the major species. A voltage of 150 V is necessary to completely dissociate the USP–ligand complexes, as shown in Figure 2C ▶. In this case, the gas phase-generated peaks are symmetrical and narrow as compared to the peaks of the charged state distribution recorded at lower voltage, confirming that the origin of the asymmetry and broadness of the peaks is caused by the ligands bound to the protein. Differences between the declustering voltages responsible for gas phase dissociation of the two protein–ligand complexes cannot be directly related to solution affinities (Li et al. 1994; Wu et al. 1997; Potier et al. 1998). In fact, a relationship between the Vc values and the contribution of electrostatic interactions involved in noncovalent complexes seems to emerge: The lower the electrostatic contribution to complex formation, the lower the Vc value has to be settled to keep the noncovalent complex intact until the detector (Robinson et al. 1996; Loo 1997; Rogniaux et al. 1999; see also discussion).
Characterization of the molecules noncovalently bound to RORβ and USP
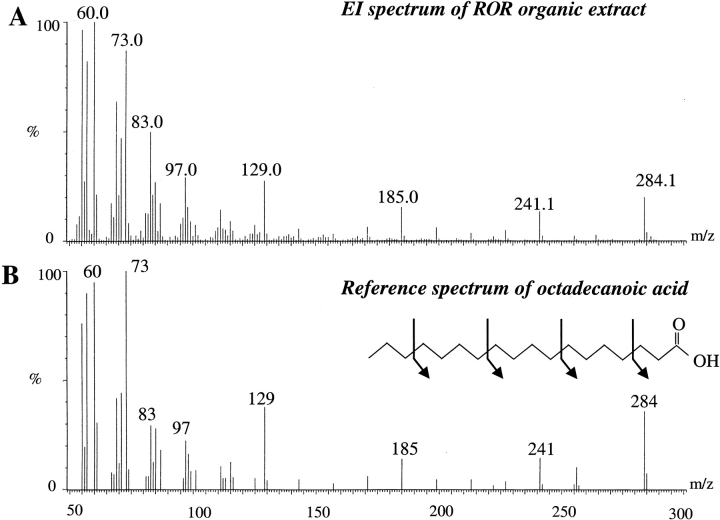
The characterization of the ligand from the knowledge of its molecular mass can only be achieved if it is measured precisely. As stated above in the case of RORβ, the exact molecular mass of the ligand obtained from measurements under native conditions cannot be determined as accurately as required. Attempts to determine the measurements from denaturing ESI-MS technique by looking at the low portion of the spectra (m/z = 100–800) were equally unsuccessful, both in the positive and negative ionization modes. Therefore, extraction of the RORβ LBD ligand was carried out with dichloromethane under acidic conditions. The organic fraction was then analyzed by GC-MS and the obtained spectrum is displayed in Figure 3 ▶. By giving as input the masses measured from this spectrum into an internal GC-MS library (Micromass), the ligand was identified as stearic acid. It displays a molecular ion M+ at m/z = 284 and characteristic fragment ions at m/z = 241, 185, 129, 73, and 60 as shown in Figure 3 ▶.
Figure 3.
Identification of the ligand of RORβ by GC-MS after organic extraction. Electron ionization (EI) spectra of the organic fraction of the RORβ preparation (A) and EI reference spectrum of the octadecanoic acid (B). The arrows show the fragmentation along the hydrocarbon backbone as occurring in the mass spectrometer source.
As suggested by these ESI-MS results, an obvious way to get a fully liganded protein sample is to add stearic acid throughout the purification process. In this case, nondenaturing ESI-MS indicates that RORβ has quantitatively bound one stearic acid molecule (data not shown). This result shows that ESI-MS technique can be fully integrated in a strategy aiming at obtaining a homogeneous fully liganded protein as a prerequisite for crystallization, even if the identified fortuitous ligand is not the physiological one.
In the case of USP, ESI-MS analysis was also carried out under denaturing conditions both in positive and negative ionization modes. Mass peaks corresponding to the free ligands were only observed in the negative mode at m/z corresponding to 690, 716, and 745, suggesting an acidic character for the ligands. Exact mass measurement of the ligands has been obtained using fast atom bombardment (FAB) mass spectrometry under high resolution conditions (Table 1). Ions of PEG 2000 at m/z = 696.4382 and m/z = 740.4644 were used as internal calibrant. Assuming that the ligands are composed of the most common biochemical elements (C, N, O, S, P, H), a crude formula can be proposed (C37H71N1O8P1 ± C2H4). Taken together, the results obtained from FAB and native ESI-MS measurements strongly suggest phosphatidyl lipid molecules to be the ligands bound to USP. This is in agreement with the crystallographic data showing residual electron density in the ligand-binding pocket of the receptor.
Table 1.
Experimental molecular masses of USP ligands measured by FAB-MS
| Experimental molecular masses | Crude formula |
| 688.4918 | C37H71N1O8P1 |
| 690.5071 | C37H73N1O8P1 |
| 716.5237 | C39H75N1O8P1 |
| 718.5391 | C39H77N1O8P1 |
| 744.5556 | C41H79N1O8P1 |
To better characterize the ligands, the organic fraction of the purified USP preparation was extracted using 2:1 chloroform/methanol and deposited on a thin layer chromatography (TLC) plate. A unique spot displaying a retention time corresponding to phosphatidylethanolamine (PE) was observed. Methanolyse of this PE molecule and analysis by gas chromatography show a fatty acid composition of C16:0 (26%), C16:1:9 (21%), C18:1:11 (46%), and traces of C14:0 and C18:3 that can explain the mass differences of 26–28 Da observed by mass spectrometry. The presence of PE in USP is consistent with the available phospholipids found in E. coli, of which 70% are PE, 18% phosphatidylglycerol, and 12% cardiolipin molecules (Voet and Voet 1995). The analysis of the phosphatidyl lipid content of an E. coli culture performed using TLC plates shows the presence of PE and cardiolipin in the soluble fraction. However, USP seems to bind selective PE molecules.
Discussion
An important question concerning the function of orphan nuclear receptors is whether or not their activity is mediated through ligand binding. Some orphan receptors have been reported to constitutively activate transcription, such as constitutive androstonol receptors (CARs), estrogen-related receptors (ERRs), and RORs, but this constitutive activity may in fact be only apparent. On one hand, the crystal structure of the RORβ LBD (Stehlin et al. 2001) revealed that a fortuitous fatty acid ligand from the E. coli expression host, identified as stearic acid in the present study, stabilizes the active LBD conformation in vitro, suggesting that the in vivo constitutive transcriptional activity of the receptor is only apparent and results from the binding of an endogenous ligand, as confirmed by the loss of activity in site-directed mutants that fill up the ligand-binding pocket. On the other hand, the crystal structure of the ERR3 LBD along with functional data on mutants demonstrated that the transcriptional activity of this receptor is really ligand independent, the active conformation being observed in the absence of any ligand (Greschik et al. 2002).
In our experience, we usually observe that orphan nuclear receptor LBDs are difficult to crystallize in the absence of an added high-affinity ligand, because the ligation state and the conformational state of the heterologously produced LBD cannot be controlled. If a fortuitous ligand is present but the binding is not quantitative, the resulting chemical and conformational heterogeneity will hamper crystallization of the LBD. The work described here shows how the complete characterization of the E. coli-produced LBDs by mass spectrometry is an important helpful step to solve this problem. In particular, we demonstrated for two orphan receptors (RORβ and USP) that noncovalent binding of small organic molecules occurs during expression or purification. In the case of RORβ and USP, nondenaturing ESI-MS allowed detection of the binding of these molecules and to characterize them as being a stearic acid and a PE molecules, respectively. Bourguet et al. (2000) already showed that unexpected binding of oleic acid occurred in a mutant RXR LBD. Mass spectrometry was also used to characterize docosahexaenoic acid, a potential ligand that has been shown to activate RXR in cell-based assays (de Urquiza et al. 2000) or to characterize the homogeneity of cellular retinol-binding protein (CRBP) apo-preparations expressed in different media in view of the possible binding of fatty acids (Elviri et al. 2001). Supramolecular mass spectrometry was also essential to demonstrate the presence of a new endogenous ligand in the AI-2 sensor protein LuxP, raising the potential biological role for boron in bacterial quorum sensing (Chen et al. 2002). The identification of such fortuitous ligands may not give direct clues on the nature of the true physiological ligands, as for instance in the case of RORβ where stearic acid does not activate the receptor. However, its presence stabilizes the LBD, which can then be crystallized (Stehlin et al. 2001). In turn, the knowledge of the three-dimensional structure of the ligand-binding pocket is a good starting point for the design of high-affinity ligands. These ligands can then be used in the functional characterization of the orphan receptor.
Detection of small organic fortuitous ligands that do not display any spectroscopic property might be very challenging. ESI-MS is a method of choice for the study of noncovalent protein–ligand complexes in terms of existence and stoichiometry. However, this technique is not yet routine and seems to be dependent on the nature of the interactions involved in the stability of the complex (Rogniaux et al. 1999). As pointed out by several investigators, the relative abundance of some complexes may be dramatically affected by the desorption process, as electrostatic forces are strengthened in a solvent-free environment, whereas complexes whose formation in solution is mainly driven by the hydrophobic effect (entropic effect arising from the exclusion of water molecules upon complex formation) appear to be weakened in the gas phase (Li et al., 1993, 1994). In the case of NRs, the complexes are sufficiently stable to be detected by mass spectrometry, which can be explained by the location of the ligand-binding site deep inside the LBD.
In conclusion, based on the examples of RORβ and USP, we have shown that mass spectrometry will be an essential tool for the full characterization of the heterologously produced orphan nuclear receptor LBDs. In both cases, ESI-MS was shown to be the only technique able to detect the presence of a fortuitous ligand bound to the LBD. As the number and diversity of both clone products and host expression systems increase each day, it becomes more and more important to have in hand a technique that enables the rapid and detailed characterization of the material derived from gene expression before performing biochemical or structural studies.
Materials and methods
Protein expression and purification
The rat RORβ LBD (residues 201–459) was overproduced as an amino-terminal His6-tagged fusion protein in E. coli and grossly purified by affinity chromatography on a Co2+ chelating column. The Heliothis virescens USP LBD (residues 205–466) was overproduced as an amino-terminal His6-tagged fusion protein in E. coli BL21(DE3) strain. The purification procedure included an affinity chromatography step on a Co2+ chelating column followed by a gel filtration on a Superdex 200 16/60 column.
RORβ ligand extraction and analysis
The RORβ LBD purified fraction (3 mL at 4 mg/mL) was acidified by addition of 6 drops of 1 M HCl and the ligand was extracted with 4 mL of dichloromethane. The organic fraction was concentrated by SpeedVac and injected into a gas chromatography column for GC-MS analysis (see below).
USP ligand extraction and analysis
USP ligand extraction was performed by addition of 20 volumes of 2:1 chloroform/methanol to 300 μL of 11 mg/mL USP LBD purified fraction. After 20 min, the organic fraction was extracted, dried under nitrogen, and deposited on a TLC plate precoated with Silicagel (LK5, 150 Å, 20 by 20 cm, Whatman). Elution was carried out with chloroform/ethanol/water/triethylamine (30/35/7/28). After drying, the plates were stained with primulin for visualization of phosphatidyl lipids.
Fatty acid composition of the PE
Transesterification of the PE isolated on TLC plate was performed with 1 mL of trifluoroborane in methanol at 100°C during 15 min. After addition of 1 mL of water and 2 mL of pentane, the organic phase was extracted and dried under nitrogen. The free methylated fatty acids were then solubilized in 50 μL of hexane and injected into a gas chromatography column (Carbowax, 30 m by 0.25 mm, Altech).
Electrospray mass spectrometry
All studies were performed using an electrospray time-of-flight mass spectrometry (ESI-TOF) mass spectrometer (LCT, Micromass, Manchester). RORβ and USP samples were first desalted by five dilution–concentration steps using centricon-10 concentrators (Amicon) against 20 mM and 100 mM NH4OAc at pH 6.5, respectively, diluted at a final concentration of 10−5 M and continuously infused into the ion source at a flow rate of 4 μL/min using a Harvard Model 11 syringe pump (Harvard Apparatus). To preserve the noncovalent complexes, relatively mild interface conditions were used, especially the declustering voltage (Vc), which controls the kinetic energy of the ions in the interface, was set to 20 V. Data were acquired in the positive mode and calibration was performed using the multiply charged ions produced by a separate injection of myoglobin dissolved in 1:1 water/acetonitrile with 1% formic acid. Average molecular masses were calculated using MassLynx v3.4 (Micromass).
Gas chromatography-MS
GC-MS analyses were carried out on a Fisons MD800 quadrupole mass spectrometer coupled to a Fisons 8065 gas chromatograph equipped with an on-column injector. A fused silica capillary column (30 m by 250 μm) coated with methylsilicone (HP-5MS; 0.1 μm film thickness) was used with helium as carrier gas. One microliter of sample was injected at 80°C and the oven temperature subsequently programmed to 300°C at 10°C/min. Mass spectra (electron ionization) were recorded at 70 eV over a mass range of 50 to 550 m/z with a cycle time of 1 scan/s.
Fast Atom Bombardment
FAB-MS analyses were performed on an AutoSpec mass spectrometer from Micromass using a mixture of m-nitrobenzylalcohol and glycerol as matrices. High resolution (10,000) mass measurement was achieved using PEG 2000 as internal reference.
Acknowledgments
We thank Erich Greiner (Freiburg University) and Eva-Maria Franken (Bayer AG) for the pRSET-B/rRORβ(201–459) and pET-15b/hvUSP(205–466) expression vectors, respectively. We are grateful to Claude Leray from the Etablissement Français du Sang (EFS) for the TLC experiments.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0232503.
References
- Ayed, A., Krutchinsky, A.N., Ens, W., Standing, K.G., and Duckworth, H.W. 1998. Quantitative evaluation of protein–protein and ligand–protein equilibria of a large allosteric enzyme by electrospray ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 12 339–344. [DOI] [PubMed] [Google Scholar]
- Billas, I.M.L., Moulinier, L., Rochel, N., and Moras, D. 2001. Crystal structure of the ligand-binding domain of the ultraspiracle protein USP, the ortholog of retinoid X receptors in insects. J. Biol. Chem. 276 7465–7474. [DOI] [PubMed] [Google Scholar]
- Bourguet, W., Andry, V., Iltis, C., Klaholz, B., Potier, N., van Dorsselaer, A., Chambon, P., Gronemeyer, H., and Moras, D. 2000. Heterodimeric complex of RAR and RXR nuclear receptors ligand-binding domains: Purification, crystallization and preliminary X-ray diffraction analysis. Protein Expr. Purif. 19 284–288. [DOI] [PubMed] [Google Scholar]
- Chen, X., Schauder, S., Potier, N., van Dorsselaer, A., Pelczer, I., Bassler, B.L., and Hughson, F.M. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415 545–549. [DOI] [PubMed] [Google Scholar]
- Craig, T.A., Benson, L.M., Tomlinson, A.J., Veenstra, T.D., Naylor, S., and Kumar, R. 1999. Analysis of transcription complexes and effects of ligands by microelectrospray ionization mass spectrometry. Nat. Biotechnol. 17 1214–1218. [DOI] [PubMed] [Google Scholar]
- Craig, T.A., Benson, L.M., Naylor, S., and Kumar, R. 2001. Modulation effects of zinc on the formation of vitamin D receptor and retinoid X receptor alpha–DNA transcription complexes: Analysis by microelectrospray mass spectrometry. Rapid Commun. Mass Spectrom. 15 1011–1016. [DOI] [PubMed] [Google Scholar]
- de Urquiza, A.M., Liu, S., Sjöberg, M., Zetterström, R.H., Griffiths, W., Sjövall, J., and Perlmann, T. 2000. Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science 290 2140–2144. [DOI] [PubMed] [Google Scholar]
- Dobo, A. and Kaltashov, I.A. 2001. Detection of multiple protein conformational ensembles in solution via deconvolution of charge-state distributions in ESIMS. Anal. Chem. 73 4763–4773. [DOI] [PubMed] [Google Scholar]
- Elviri, L., Zagnoni, I., Careri, M., Cavazzini, D., and Rossi, G.L. 2001. Non-covalent binding of endogenous ligands to recombinant cellular retinol-binding proteins studied by mass spectrometric techniques. Rapid Commun. Mass Spectrom. 15 2186–2192. [DOI] [PubMed] [Google Scholar]
- Forman, B.M., Chen, J., and Evans, R.M. 1997. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc. Natl. Acad. Sci. 94 4312–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, B., Li, Y.T., and Henion, J.D. 1991. Detection of noncovalent receptor–ligand complexes by electrospray mass spectrometry. J. Am. Chem. Soc. 113 6294–6296. [Google Scholar]
- Giguère, V. 1999. Orphan nuclear receptors: From gene to function. Endocrine Rev. 20 689–725. [DOI] [PubMed] [Google Scholar]
- Greschik, H., Wurtz, J.M., Sanglier, S., Bourguet, W., van Dorsselaer, A., Moras, D., and Renaud, J.P. 2002. Structural and functional evidence for ligand-independent transcriptional activation by the estrogen-related receptor 3. Mol. Cell 9 303–313. [DOI] [PubMed] [Google Scholar]
- Jorgensen, T.J.D., Roepstorff, P., and Heck, A.J.R. 1998. Direct determination of solution binding constants for noncovalent complexes between bacterial cell wall peptide analogues and vancomycin group antibiotics by electrospray ionization mass spectrometry. Anal. Chem. 70 4427–4432. [Google Scholar]
- Katta, V. and Chait, B.T. 1991. Observation of the heme-globin complex in native myoglobin by electrospray ionization mass spectrometry. J. Am. Chem. Soc. 113 8534–8535. [Google Scholar]
- Kebarle, P. 2000. A brief overview of the present status of the mechanisms involved in electrospray mass spectrometry. J. Mass Spectrom. 35 804–817. [DOI] [PubMed] [Google Scholar]
- Kliewer, S.A., Sundseth, S.S., Jones, S.A., Brown, P.J., Wisely, G.B., Koble, C.S., Devchand, P., Wahli, W., Willson, T.M., Lenhard, J.M., et al. 1997. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ. Proc. Natl. Acad. Sci. 94 4318–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer, S.A., Lehmann, J.M., and Willson, T.M. 1999. Orphan nuclear receptors: Shifting endocrinology into reverse. Science 284 757–760. [DOI] [PubMed] [Google Scholar]
- Krey, G., Braissant, O., L’Horset, F., Kalkhoven, E., Perroud, M., Parker, M.G., and Wahli, W. 1997. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand essay. Mol. Endocrinol. 11 779–791. [DOI] [PubMed] [Google Scholar]
- Last, A.M. and Robinson, C.V. 1999. Protein folding and interactions revealed by mass spectrometry. Curr. Opin. Chem. Biol. 3 564–570. [DOI] [PubMed] [Google Scholar]
- Li, Y.T., Hsieh, Y.L., Henion, J.D., Senko, M.W., McLafferty, F.W., and Ganem, B. 1993. Mass spectrometric studies on noncovalent dimers of leucine peptides. J. Am. Chem. Soc. 115 8409–8413. [Google Scholar]
- Li, Y.T., Hsieh, Y.L., Henion, J.D., Ocain, T.D., Schiehser, G.A., and Ganem, B. 1994. Analysis of the energetics of gas phase immunophilin–ligand complexes by ion spray mass spectrometry. J. Am. Chem. Soc. 116 7487–7493. [Google Scholar]
- Loo, J.A. 1997. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom. Rev. 16 1–23. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Electrospray ionization mass spectrometry: A technology for studying non-covalent macromolecular complexes. Int. J. Mass Spectrom. 200 175–186. [Google Scholar]
- Loo, J.A., Hu, P., McConnell, P., Mueller, W.T., Sawyer, T.K., and Thanabal, V., 1997. A study of Src SH2 domain protein–phosphopeptide binding interactions by electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 8 234–243. [Google Scholar]
- Low, L.Y., Hernandez, H., Robinson, C.V., O’Brien, R., Grossmann, J.G., Ladbury, J.E., and Luisi, B. 2002. Metal-dependent folding and stability of nuclear hormone receptors DNA-binding domains. J. Mol. Biol. 319 87–106. [DOI] [PubMed] [Google Scholar]
- McLafferty, F., Guan, Z., Haupts, U., Wood, T., and Kelleher, N. 1998. Gaseous conformational structures of cytochrome c. J. Am. Chem. Soc. 120 4732–4740. [Google Scholar]
- Potier, N., Donald, L.J., Chernushevich, I., Ayed, A., Ens, W., Arrowsmith, C.H., Standing, K.G., and Duckworth, H.W. 1998. Study of a noncovalent trp repressor: DNA operator complex by electrospray ionization time-of-flight mass spectrometry. Protein Sci. 7 1388–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik, B.N., Bartner, P.L., Mirza, U.A., Liu, Y.H., and Ganguly, A.K. 1998. Electrospray ionization mass spectrometry for the study of non-covalent complexes: an emerging technology. J. Mass Spectrom. 33 911–920. [DOI] [PubMed] [Google Scholar]
- Przybylski, M. and Glocker, M.O. 1996. Electrospray mass spectrometry of biomolecular complexes with noncovalent interactions: New analytical perspectives for supramolecular chemistry and molecular recognition processes. Angew. Chem. Int. Ed. Engl. 35 807–826. [Google Scholar]
- Renaud, J.P. and Moras, D. 2000. Structural studies on nuclear receptors. Cell. Mol. Life Sci. 57 1748–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, C.V., Chung, E.W., Kragelund, B.B., Knudsen, J., Aplin, R.T., Poulsen, F.M., and Dobson, C.M. 1996. Probing the nature of noncovalent interactions by mass spectrometry. A study of protein–CoA ligand binding and assembly. J. Am. Chem. Soc 118 8646–8653. [Google Scholar]
- Rogniaux, H., Barbanton, J., Barth, P., Biellmann, J.F., van Zandt, M., Chevrier, B., Howard, E., Mitschler, A., Potier, N., Urzhumtseva, L., et al. 1999. Binding of aldose reductase inhibitors: Correlation of crystallographic and mass spectrometry studies. J. Am. Soc. Mass Spectrom. 10 635–647. [DOI] [PubMed] [Google Scholar]
- Rogniaux, H., Sanglier, S., Strupat, K., Azza, S., Roitel, O., Ball, V., Tritsch, D., Branlant, G., and van Dorsselaer, A. 2001. Mass spectrometry as a novel approach to probe cooperativity in multimeric enzymatic systems. Anal. Biochem. 291 48–61. [DOI] [PubMed] [Google Scholar]
- Rostom, A.A. and Robinson, C.V. 1999. Disassembly of intact multiprotein complexes in the gas phase. Curr. Opin. Struc. Biol. 9 135–141. [DOI] [PubMed] [Google Scholar]
- Stehlin, C., Wurtz, J.M., Steinmetz, A., Greiner, E., Schüle, R., Moras, D., and Renaud, J.P. 2001. X-ray structure of the orphan nuclear receptor RORβ ligand-binding domain in the active conformation. EMBO J. 20 5822–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahallah, N., Pinkse, M., Maier, C.S., and Heck, A.J. 2001. The effect of the source pressure on the abundance of ions on noncovalent protein assemblies in an electrospray ionization orthogonal time-of-flight instrument. Rapid Commun. Mass Spectrom. 15 596–601. [DOI] [PubMed] [Google Scholar]
- Tito, M.A., Miller, J., Walker, N., Griffin, K.F., Williamson, E.D., Despeyroux-Hill, D., Titball, R.W., and Robinson, C.V. 2001. Probing molecular interactions in intact antibody:antigen complexes, a electrospray time-of-flight mass spectrometry approach. Biophys. J. 81 3503–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra, T.D., Johnson, K.L., Tomlinson, A.J., Craig, T.A., Kumar, R., and Naylor, S. 1998. Zinc-induced conformational changes in the DNA-binding domain of the vitamin D receptor determined by electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 9 8–14. [DOI] [PubMed] [Google Scholar]
- Voet, D. and Voet, J.G. 1995. Biochemistry, 2nd ed. John Wiley & Sons, New York.
- Witkowska, H.E., Green, B.N., Carlquist, M., and Shackleton, C.H. 1996. Intact noncovalent dimer of estrogen receptor ligand-binding domain can be detected by electrospray ionization mass spectrometry. Steroids 61 433–438. [DOI] [PubMed] [Google Scholar]
- Wu, Q., Gao, J., Joseph-McCarthy, D., Sigal, G.B., Bruce, J.E., Whitesides, G.M., and Smith, R.D. 1997. Carbonic anhydrase-inhibitor binding: From solution to the gas phase. J. Am. Chem. Soc. 119 1157–1158. [Google Scholar]