Abstract
Two mutant proliferating cell nuclear antigens from the hyperthermophilic archaeon Pyrococcus furiosus, PfuPCNA(D143A) and PfuPCNA(D143A/D147A), were prepared by site-specific mutagenesis. The results from gel filtration showed that mutations at D143 and D147 drastically affect the stability of the trimeric structure of PfuPCNA. The PfuPCNA(D143A) still retained the activity to stimulate the DNA polymerase reaction, but PfuPCNA(D143A/D147A) lost the activity. Crystal structures of the mutant PfuPCNAs were determined. Although the wild-type PCNA forms a toroidal trimer with intermolecular hydrogen bonds between the N- and C-terminal domains, the mutant PfuPCNAs exist as V-shaped dimers through intermolecular hydrogen bonds between the two C-terminal domains in the crystal. Because the mutated residues are involved in the intermolecular ion pairs through their side chains in the wild-type PfuPCNA, these ion pairs seem to play a key role in maintaining the toroidal structure of the PfuPCNA trimer. The comparison of the crystal structures revealed intriguing conformational flexibility of each domain in the PfuPCNA subunit. This structural versatility of PCNA may be involved in the mechanisms for ring opening and closing.
Keywords: DNA replication, sliding clamp, X-ray diffraction, Archaea, hyperthermophile
DNA replication and repair processes play central roles for the sustenance of life. Extensive studies on the molecular mechanism of DNA replication revealed the existence of a molecule that retains DNA polymerase on the template DNA strand for processive DNA synthesis (reviewed in Waga and Stillman 1998; Hingorani and O’Donnell 2000; Bruck and O’Donnell 2001; Trakselis and Benkovic 2001). This processivity factor of the DNA polymerases forms a dimer or trimer depending on the organism’s domain of life. The bacterial DNA polymerase III β subunit exists as dimer, whereas the proliferating cell nuclear antigens (PCNA) from eukarya and archaea form homotrimers. The gp43 protein from bacteriophages also forms a homotrimeric structure. These proteins, often called sliding clamps, capture dsDNA within the central hole of the ring shaped dimer or trimer and directly interact with the replicative DNA polymerases. The eukaryotic PCNA also anchors various proteins involved in DNA repair, cell cycle control, and apoptotic processes (reviewed in Jonsson and Hubscher 1997; Kelman 1997; Kelman and Hurwitz 1998; Tsurimoto 1998, 1999; Warbrick 1998, Warbrick 2000; Paunesku et al. 2001). From these studies, it is now well recognized that the molecular analyses of PCNA are very important to elucidate the structure–function relationships of this multifunctional molecule.
The proteins related to the genetic information system in Archaea, the third domain of life, are structurally more similar to eukaryotic proteins than to those from Bacteria (reviewed for DNA replication in Edgell and Doolittle 1997; Ishino and Cann 1998; Cann and Ishino 1999; Leipe et al. 1999). We cloned a gene encoding a sequence homologous to the eukaryotic PCNA from the hyperthermophilic euryarchaeote, Pyrococcus furiosus, expressed it in Escherichia coli, and characterized the purified gene product (Cann et al. 1999). The protein interacted with both DNA polymerases I (Pol BI) and II (Pol D) in this organism and enhanced their DNA synthesizing activities in vitro, and therefore, we designated it as PfuPCNA. We have also determined the crystal structure of PfuPCNA and compared it with those of the human and yeast PCNAs (Matsumiya et al. 2001).
The three dimensional structures of the DNA sliding clamps have been determined for eukaryotes (Krishna et al. 1994; Gulbis et al. 1996), bacteriophages (Shamoo and Steitz 1999; Moarefi et al. 2000), and a bacterium (Kong et al. 1992) to date, in addition to the archaeal PCNA. All of these structures share the toroidal morphology with a pseudo-sixfold symmetry, and the archaeal and eukaryotic PCNAs are especially similar among them. The structural similarity of the PCNAs in Archaea and Eukarya is supported by the experiments showing that PfuPCNA functionally interacts with calf thymus DNA polymerase δ and human replication factor C (RFC) in vitro (Ishino et al. 2001). The Thermococcus PCNA also stimulates the calf thymus DNA polymerase activity (Henneke et al. 2000).
Although the overall structure of PfuPCNA resembles those of the human and yeast PCNAs, the mode of the intermolecular interactions between neighboring molecules differs between the archaeal and eukaryotic PCNAs. In the PfuPCNA trimer, the number of intermolecular main chain hydrogen bonds is lower, and the side chains of acidic and basic amino acids form an intermolecular ion pair network. We previously proposed that the formation of the ion pair network at the boundary between the subunits compensates for the weakening of the hydrogen bonds, to maintain the trimeric structure of PfuPCNA (Matsumiya et al. 2001). In this article, we report the biochemical properties of two mutant PfuPCNAs, in which one and two aspartates at the intermolecular interface are replaced with alanines, and examine the effects of the mutations on the molecular structures.
Results
Mutations of Asp143 to Ala and Asp147 to Ala in PfuPCNA
On the basis of the crystal structure of PfuPCNA (Matsumiya et al. 2001), the residues participating in the formation of the intermolecular ion pair network are distributed on each side of the intermolecular interfaces. Positively charged residues in the N-terminal domain (Arg82, Lys84, and Arg109) and negatively charged residues in the C-terminal domain (Glu139, Asp143, and Asp147) are participated in the ion pair network of the intermolecular interfaces of the PCNA ring structure. We substituted Asp143 and Asp147 with Ala to investigate the roles of the ion pair network in the interaction between the subunits. The plasmid pTPCNAL73 (Matsumiya et al., 2001) was used as a template and a PCR-mediated site-specific mutagenesis was performed. Two rounds of PCR were performed to introduce the two mutations into the gene, corresponding to Asp143Ala and Asp147Ala. The overexpression and purification procedures for the wild-type PfuPCNA were applied to obtain the desired mutant PCNAs, PfuPCNA (D143A) and PfuPCNA(D143A/D147A). These mutant PCNA proteins displayed the same profiles as those of the wild-type PfuPCNA in the ion-exchange chromatography, and were purified to homogeneity (Fig. 1 ▶).
Figure 1.
Purification of mutant PfuPCNA proteins. Recombinant PfuPCNA proteins purified as described in the text were loaded onto a 12.5% polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. Molecular masses indicated on the left side were from the marker proteins (New England Biolabs).
Oligomerization states of the mutant PfuPCNAs in solution
To examine the oligomerization state of the PfuPCNA (D143A) and PfuPCNA(D143A/D147A) mutants, the elution profiles of the gel filtration chromatography were compared for the wild-type and two mutant PfuPCNAs (Fig. 2 ▶). The chromatogram of the wild-type PfuPCNA shows a large main peak, which corresponds to the trimer from the estimated molecular weight. In contrast, PfuPCNA(D143A/D147A) exhibited a single peak at the position for the monomer. In the case of PfuPCNA(D143A), the peak for the monomeric form was accompanied by a small peak, which may correspond to the trimer. These results suggest that the intermolecular interaction becomes weaker by neutralization of the negative-charged residues that participate in forming the intermolecular ion pairs, as we predicted from the crystal structure of PfuPCNA.
Figure 2.
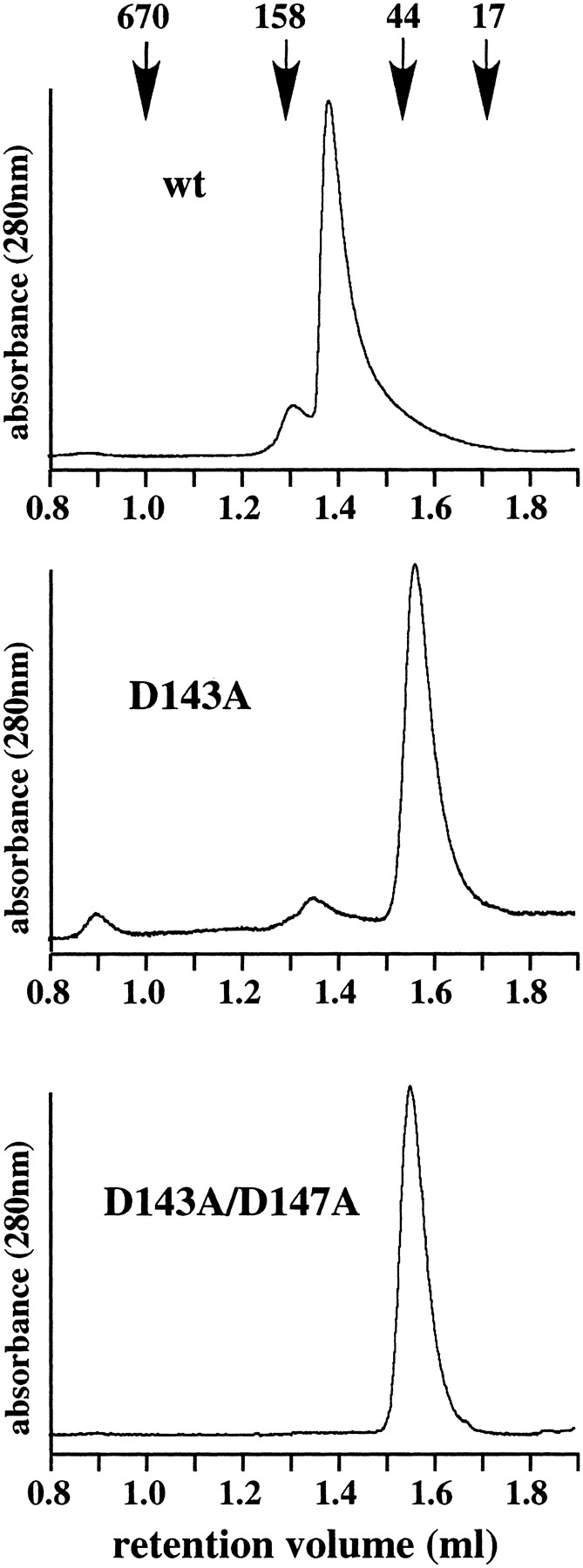
Investigation of trimer formation by gel filtration analysis. The wild-type and mutant PfuPCNA proteins were subjected to a gel filtration column. Elution of the proteins was monitored by absorbance at 280 nm. The molecular masses on the top were from a gel filtration standard (BioRad), containing thyroglobulin (670 kD), γ-globulin (158 kD), ovalbumin (44 kD), myoglobulin (17 kD).
Comparison of biological activities
PCNA generally requires the Replication Factor C (RFC) complex for ring opening and loading onto double stranded DNA in an ATP-dependent manner (reviewed in Ellison and Stillman 2001; O’Donnell et al. 2001; Trakselis et al. 2001; Jeruzalmi et al. 2002). It is well known that the ATPase activity of RFC is stimulated by PCNA and DNA in vitro, including the case of the P. furiosus RFC and PCNA (Cann et al. 2001). In this study, the ATPase activity of PfuRFC was measured in the presence of PfuPCNA, PfuPCNA(D143A), or PfuPCNA(D143A/D147A) with DNA. The two mutant PfuPCNAs stimulated the ATPase activity of PfuRFC to the same extent as the wild-type PfuPCNA, even though the toroidal structures of the mutant PCNAs are distinctly unstable, as described above (data not shown). This result indicates that the appropriate conformation is maintained in the mutant PfuPCNA monomers for the functional interaction with PfuRFC.
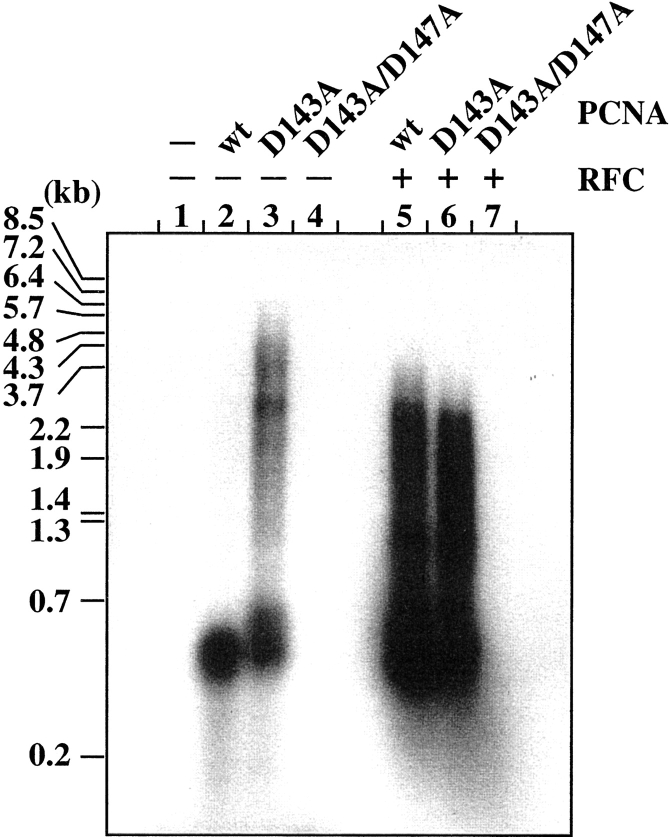
For the eukaryotic PCNA and the bacterial Pol III β subunit, trimer or dimer formation is essential for their activities as processivity factors for DNA polymerases. We examined the stimulation activity of mutant PfuPCNAs on the DNA synthesis reaction by P. furiosus Pol I in the presence or absence of the PfuRFC complex to determine the effect of the mutation on the function as the sliding clamp under both RFC-assisted and self-loading conditions. As shown in Figure 3 ▶, PfuPCNA(D143A/D147A) did not show any stimulation of DNA synthesis by Pol I, in either the presence or absence of PfuRFC (lanes 4 and 7). In contrast, the wild-type and PfuPCNA(D143A) proteins enhanced the processivity of Pol I in both the presence and absence of PfuRFC (lanes 2, 3, 5, and 6). It is noteworthy that PfuPCNA(D143A) stimulated the Pol I reaction more extensively than the wild-type PfuPCNA, especially under the RFC-free condition. These results indicate that PfuPCNA (D143A) can form the active ring structure in solution, although the monomeric form is dominant in the gel filtration. The stronger activity of PfuPCNA (D143A) may be caused by the weaker intermolecular ion pair interaction, which made the trimeric ring much easier to open and close for loading onto and unloading from the DNA strand.
Figure 3.
Effects of the mutations in PfuPCNA on the DNA synthesis reaction by P. furiosus Pol I. The reaction mixtures with indicated PfuPCNA proteins in each lane, both in the presence and absence of PfuRFC, were analyzed by 1% alkali agarose gel electrophoresis, and the products were visualized by autoradiography. The sizes indicated on the left were from BstI-digested λ phage DNA labeled by 32P at each 5′ end.
Subunit interactions in the crystal structures of the mutant PfuPCNAs
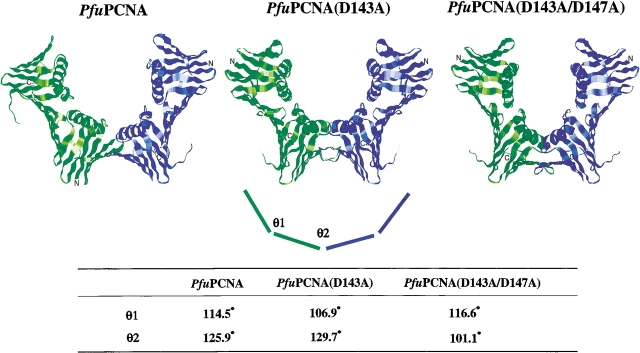
The crystals of the mutant PfuPCNAs were obtained by the vapor diffusion method using ammonium sulfate as the precipitant, as in the case of the wild-type PfuPCNA (Matsumiya et al. 2001). Interestingly, the crystalline shape along with the molecular packing of each mutant was different. Whereas the wild-type PfuPCNA crystals were hexagonal rods with the space group P63 (Matsumiya et al. 2001), the crystals of PfuPCNA (D143A) and PfuPCNA (D143A/D147A) in this study were bipyramidal blocks with the space group P43212 and parallelepiped plates with the space group P212121, respectively. The structural motifs of the mutant PfuPCNA molecules were identical with that of the wild-type PfuPCNA. However, the intermolecular interaction modes in the mutant crystals differed from that in the wild-type crystal (Fig. 4 ▶). The wild-type PfuPCNA formed a toroidal trimer as observed in solution with a head-to-tail alignment, in which the C-terminal domain (tail) is connected to the N-terminal domain (head) of the adjacent molecule via antiparallel β-sheet main-chain hydrogen bonds. This head-to-tail alignment has been observed in all of the crystals of circular sliding clamps (Kong et al. 1992; Krishna et al. 1994; Gulbis et al. 1996; Shamoo and Steitz 1999; Moarefi et al. 2000). In the case of the mutant crystals, the PfuPCNA molecules form V-shaped dimers with a novel tail-to-tail alignment (Fig. 4 ▶). The intermolecular interaction of the wild-type PfuPCNA is composed of main-chain β-sheet hydrogen bonds and side-chain ion pairs (Matsumiya et al. 2001, 2002). The ion pair network is formed between positively charged residues at the N-terminal domain (Arg82, Lys84, and Arg109) and negatively charged residues at the C-terminal domain (Glu139, Asp143, and Asp147). The mutant PCNAs prepared in this study lack the carboxyl groups involved in the ion pair network at the intermolecular interface, and therefore, the neutralized interface may connect with either N- or C-terminal domains. From this observation, it can be predicted that the asymmetrical distribution of the electrical charges at the two edges of the PfuPCNA molecule restricts the contact of the subunits selectively, in a head-to-tail manner. Given the fact that there are six intermolecular β-sheet hydrogen bonds (N•••O ≤ 3.3 Å) in the mutant crystals, while the wild-type has only four hydrogen bonds per intermolecular interface (Fig. 5 ▶), the tail-to-tail connection should be more stable than the head-to-tail form in the case of the mutant PfuPCNA.
Figure 4.
Subunit interactions in the three PfuPCNA crystals and comparison of interdomain angles. Wild-type PfuPCNA forms a troidal trimer with C-terminal domain to N-terminal domain alignment (two molecules in the trimer are shown here). Both two mutant PfuPCNAs form V-shaped dimers with C-terminal to C-terminal alignment. Interdomain angles at the intramolecular interface and the intermolecular interface were represented by θ1 and θ2, respectively.
Figure 5.
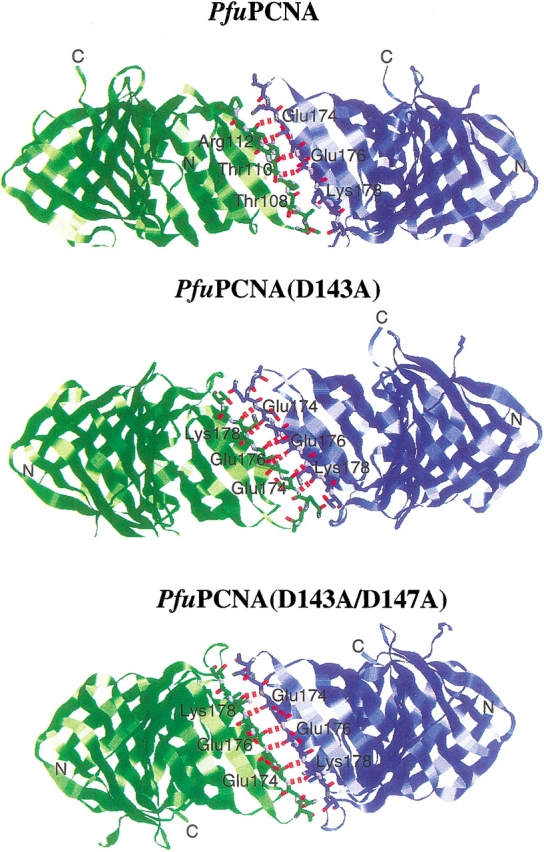
Hydrogen bonds at the intermolecular interfaces in the three PfuPCNA crystals. Hydrogen bonds (N...O ≤ 3.3 Å) observed in the trimer (wild-type) or dimer (mutants) of PfuPCNAs between the subunits are shown as red dashed lines.
Domain movement in the mutant PCNA structures
The sliding clamp molecules are composed of two (PCNA and gp43) or three (Pol III β subunit) structurally similar domains in tandem. These domains are rather rigid, and the conformational change of the clamp can be attributed to the motion around the interdomain β-sheet hydrogen bonds. This domain movement may change the clamp structure from closed ring to open ring. The motion is divided into two components: hinged motion and swing motion, which may open the ring by in-plane and out-of-plane modes, respectively. The former motion is evaluated from the angle between the lines defined by the midpoints of the βD and βI strands of each domain, and the latter is estimated from the dihedral angle between the farthest β-strands in two neighboring domains (e.g., the intramolecular torsion angle is defined by βI1 and βD2, and the intermolecular torsion angle is defined by βI2 and βD1 in the next molecule for the toroidal PCNA). The interdomain angles of the eukaryotic and archaeal PCNA cyclic trimers fall between 114.5°– 119.4° and 120.0°–125.9° at the intramolecular and intermolecular interfaces, respectively (Matsumiya et al. 2001). Small differences are observed in the crystal structure of PfuPCNA complexed with an RFCL PIP-box peptide (Matsumiya et al. 2002). In the V-shaped (open ring) structure of the PfuPCNA (D143A) crystal, the intramolecular angle shrinks to 106.9° and the intermolecular angle widens to 129.7°. In contrast, in the PfuPCNA (D143A/D147A) crystal, the intermolecular angle shrinks to 101.1° while the intramolecular angles of 116.5°–116.6° are similar to those of the wild-type PfuPCNA crystals (Fig. 4 ▶).
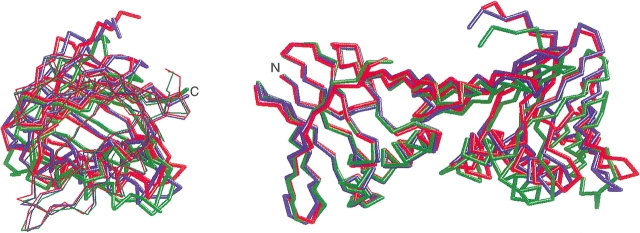
The C-terminal domain of the wild-type PfuPCNA swings counterclockwise in the complex with the RFCL PIP-box peptide, compared with that in the uncomplexed form. Along with the swing motion, the interdomain torsion angle decreases from 59.7° in the uncomplexed form to 52.7° in the complexed form (Matsumiya et al. 2002). A similar motion is also predicted in the human PCNA–p21WAF1/CIP1 C-terminal complex (Gulbis et al. 1996), by comparing the crystal structure with that of the uncomplexed form of yeast PCNA (Krishna et al. 1994) from 59.1° in yeast PCNA to 53.7°–54.4° in the human PCNA-p21 complex. In the crystals of the mutant PfuPCNAs (dimer form), in which the N-terminal domain is not fixed, the interdomain torsion angles are lower than that of the uncomplexed wild-type PfuPCNA (trimer form; Fig. 6 ▶): they are 55.1° for PfuPCNA (D143A) and 51.8°–52.1° for PfuPCNA(D143A/D147A). In the case of the Pol III β subunit of E. coli, the difference in the interdomain dihedral angles in the closed ring dimer and the monomeric mutant is not remarkable. The only difference is at the interface between the N-terminal domain and the center domain of chain A of the β subunit–full-length δ subunit complex (62.7°–64.6° in wild type to 57.1° in the complex), but this decrease in the dihedral angle is probably compensated by the increase in the center domain–C-terminal domain interface by δ subunit binding. Because in the β–δ (1–140) complex, the center domain–C-terminal domain torsion angle (54.9°) is smaller than that of the wild type, the increase in the β–δ (full-length) complex may reflect the flexibility at the interdomain interfaces.
Figure 6.
Domain movement in the mutant PfuPCNA crystals. The monomer structures of the two mutant PfuPCNA crystals were superimposed on the wild-type structure. The wild-type, D143A, and D143A/D147A are indicated in blue, green, and red, respectively. A side view (left) and a top view (right) are shown. In the side view, the N-terminal and the C-terminal domains are shown with a thin and a thick line, respectively.
Discussion
In this study, we have shown that the intermolecular ion pair network is the key factor for PfuPCNA to function as the processivity factor, by securing the proper trimeric structure. The results from gel filtration and DNA elongation reactions revealed that the proper trimerization is partially and completely inhibited in the mutant PCNAs, PfuPCNA (D143A) and PfuPCNA (D143A/D147A), in which the negatively charged side chains of the C-terminal domain were eliminated. On the other hand, the mutant PCNAs formed the crystals of the open ring dimer with a tail-to-tail alignment, probably due to the disruption of the specific ion pair network at the intermolecular interface. The alignment of the D143A subunits in the active trimer form should be in the head-to-tail manner, because a molecule with a tail-to-tail alignment at one interface, as seen in the crystal, must contain an electrostatically unfavorable head-to-head interface (where the positively charged residues meet together) at one of the two other interfaces. If the positively charged residues at the N-terminal domain were neutralized, then the head-to-head alignment could be realized. When both the head-to-head and tail-to-tail alignments were allowed in trimer formation, two of the three subunits would expose the interaction sites for the PCNA binding proteins on the front face, and the binding site in the other one subunit would be exposed toward the rear. This trimer could hold the PCNA binding proteins on both faces simultaneously (e.g., Pol on the front and clamp loader on the rear). This assembly mode may be forbidden by the electrostatic repulsion between the PfuPCNA subunits. The charged residues at the intermolecular interfaces in PfuPCNA have dual functions: to maintain the trimeric structure, and to determine the head-to-tail alignment in the trimer form. The residues involved in this intermolecular ion pair (corresponding to R82, K84, R109, D143, and D147) are highly conserved among PCNAs from thermophilic archaea, and therefore, the contribution of intermolecular ion pairs to the formation of stable trimeric PCNA seems to be in common in the archaeal PCNA. In contrast, the above residues are not conserved among PCNAs from eukaryotes. The yeast PCNA and the bacterial Pol III β subunit use hydrophobic contacts in conjunction with the hydrogen bonds to maintain the toroidal structure (Krishna et al. 1994; McAlear et al. 1994; Ayyagari et al. 1995; Jonsson et al. 1995; Stewart et al., 2001). As the hydrophobic interaction lacks the polarity characteristic of an electrostatic interaction, shape complementarity may be the key factor in determining the head-to-tail alignment.
The structural differences of the wild-type PfuPCNA (Matsumiya et al. 2001), its complex with the PIP-box peptide from the large subunit (RFCL) of PfuRFC (Matsumiya et al. 2002), and the mutant PfuPCNA in this study induced by crystal packing force revealed the conformational flexibility of the interdomain or intermolecular interface of the PCNA molecules. This conformational versatility of PfuPCNA may contribute to the functional change of the ring structure. A comparison of the interdomain angles between the dimeric wild-type and the monomeric mutant of the E. coli Pol III β subunit support the in-plane open ring model (Jeruzalmi et al. 2001). The proposed model for the β subunit includes an expansion at the interface between the N-terminal and center domains, although the δ subunit, the “wrench” of the clamp loader binds on the interface between the central and C-terminal domains (Jeruzalmi et al. 2001). The dynamic structural model for the replisome in bacteriophage T4, gp45 (clamp)–gp44/62 (clamp loader)– gp43 (DNA polymerase), has recently been determined using a stopped-flow fluorescence energy transfer technique (Alley et al. 2000; Trakselis et al. 2001). The proposed model for the opening and closing of the gp45 clamp includes an in-plane opening at the initial stage, an out-of-plane, partially closed conformation upon DNA capture, and an in-plane, slightly open final form complexed with gp43 (Trakselis et al. 2001).
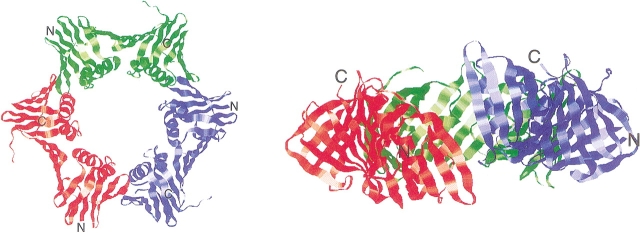
The interdomain angles of the mutant PfuPCNAs were similar to or narrower than those of the wild-type PfuPCNA, in contrast to the case of the mutant E. coli Pol III β subunit. It does not appear that this structural difference supports the in-plane mode for the ring opening of PfuPCNA. Based on the monomeric structure of mutant PfuPCNAs, a hypothetical trimer was built (Fig. 7 ▶). The ring is opened with an out-of-plane distortion in this model. The structural differences between the PfuPCNA and PfuPCNA–PIP box peptide, and also between yeast PCNA and the human PCNA–p21 peptide complex (Matsumiya et al. 2002) support the idea that the out-of-plane movement, derived from the conformational flexibility between the domains, may be in common in the archaeal and eukaryotic PCNA.
Figure 7.
An open ring model of PfuPCNA trimer based on structural comparisons between wild-type and mutant PfuPCNA crystals. A ring structure model was built by superposing the monomer structure of D143A/D147A on the intermolecular interfaces of the wild-type structure. Two orthogonal views (a top view, left, and a side view, right) are shown. Three subunits are indicated by different colors.
Molecules from hyperthermophilic archaea are very suitable materials for detailed structural analyses, not only for the primary determination of the crystal structure, but also in more dynamic studies using different crystal types of the same proteins under various conditions. Novel information will be obtained from the structural analysis of each crystal. We solved a crystal structure of the small subunit complex of the PfuRFC recently (Oyama et al. 2001). The structural analyses of the PfuPCNA–PfuRFC complex will also provide important knowledge for the clamp-opening mechanism.
Materials and methods
Preparation of mutant PfuPCNA proteins
The plasmid pTPCNAL73 (Matsumiya et al. 2001), in which the gene for PfuPCNA (Met73Leu) is inserted into the pET21a vector (Novagen), was used for the polymerase chain reaction (PCR)-mediated mutagenesis. Because no difference was observed between wild-type and M73L mutant PCNA in our biologic analyses (Matsumiya et al. 2001), PfuPCNA(M73L) is called “wild-type PfuPCNA” in this study. In the first stage, two primers, PCNA MUTF2 (5′-TTCTTGGAGAAGTCCTAAAAGCTGCTGTTAA AGATGCCTCTCTAGTGAGTGACAG-3′) and PCNAMUTR2 (5′-CTGTCACTCACTAGAGAGGCATCTTTAACAGCAGCTTTTAGGACTTCTCCAAGAA-3′), which have the codons corresponding to alanine instead of aspartate at position 143 (underlined), were used for the PCR reaction. The PCR conditions involved 30 cycles of denaturation for 30 sec at 95°C, annealing for 1 min at 55°C, and extension for 10 min at 68°C with Pyrobest DNA polymerase (Takara Shuzo). The amplified product was treated with DpnI for 1 h at 37°C and was introduced into E. coli JM109 cells. The plasmids isolated from the transformants were sequenced, and the plasmid encoding the Asp143Ala mutant was selected. The mutant plasmid was designated as pTPCNAA143. In the second stage, the same procedure was adopted with pTPCNAA143 as the template and the primers PCNAMUTF1 (5′-TTCTTGGAGAAGTCCTAAAAGCTGCTGTTAAAGCTGCCTCTCTAGTGAGTGACAG-3′) and PCNAMUTR1 (5′-CTGTCACTCACTAGAGAGGCAGCTTTAACAGCAGCTTTTAGGACTTCTCCAAGAA-3′), which contain two alanine codons instead of aspartates at positions 143 and 147 (underlined) as the primers. The resultant plasmid was designated as pTPCNAA147. The mutant PfuPCNAs, PfuPCNA(D143A) and PfuPCNA(D143A/D147A), were overproduced in E. coli BL21(DE3) cells and were purified according to the protocol described for the wild-type PfuPCNA (Cann et al. 1999). The purified mutant PfuPCNA solutions were concentrated to 20 mg/mL by ultrafiltration and were stored at 4°C.
Gel filtration analysis
Solutions (1.0 mg/mL; 36 μM) of the mutant PfuPCNAs were subjected to gel filtration on a Superdex 200 PC 3.2/30 column (Amersham Pharmacia Biotech) in a 50 mM Tris-HCl, pH 8.0, 150 mM NaCl solution. The molecular weights of the solutes were estimated from the retention volume, using those of the Gel Filtration Standards (Bio-Rad) as the standards.
Assay of primer extension stimulation
The effects of the mutant PfuPCNAs on the DNA synthesis activity were assayed by using P. furiosus Pol I (Komori and Ishino 2000) and M13 single-stranded DNA annealed with a deoxyoligonucleotide primer, as described previously, in the presence or absence of the PfuRFC complex (Cann et al. 1999, 2001).
Crystallization and data collection
Single crystals of the mutant PfuPCNAs were prepared by the hanging drop vapor diffusion method. A 1.0 μL aliquot of the protein solution and 1.0 μL of precipitation buffer were mixed and equilibrated against 500 μL of precipitation buffer at 20°C. The compositions of the precipitation buffers were as follows: 100 mM sodium citrate pH 4.5, 0.6 M ammonium sulfate and 40% (v/v) glycerol for PfuPCNA(D143A), and 100 mM sodium citrate pH 5.5, 2.6 M ammonium sulfate, 11% (v/v) 2-methyl-2,4-pentanediol and 4% (v/v) glycerol for PfuPCNA(D143A/D147A). The colorless octahedrons of PfuPCNA(D143A) or colorless plates of PfuPCNA(D143A/D147A) were obtained in a week. The crystals could be flash-cooled without cryoprotection. X-ray diffraction data were collected at 100 K on the BL24XU beamline of SPring-8 using 0.836 Å radiation and an R-AXIS V imaging plate detector (Rigaku Corporation). The data were processed by MOSFLM (Leslie 1992), scaled, and converted for crystal structure determination with SCALA and TRUNCATE in the CCP4 program suite (Collaborative Computational Project, Number 4, 1994).
Structure determination
The crystal structures of the mutant PfuPCNAs were determined by the molecular replacement method using CNS (Brünger et al. 1998), with the structure of PfuPCNA (Matsumiya et al., 2001; PDB code 1GE8) as the search model. In the case of PfuPCNA(D143A), the solution with the highest correlation factor after the rotation and translation search yielded a reasonable model for the structure refinement. In the case of PfuPCNA(D143A/D147A), the two molecules in the asymmetric unit were placed by two cycles of the translation search after the rotation search. Because the two molecules formed a dimer around an apparent twofold axis, a noncrystallographic symmetry (NCS) restraint was applied in the structure refinement stage. The molecular structures were manually built using O (Jones et al. 1991), and were refined by the simulated annealing method using CNS. The residues Met1, Met121–Leu125, and Glu248–Glu249 could not be modeled because of the poor electron densities at these regions. The results of the crystal structure analyses are summarized in Table 1. The molecular coordinates and structure factors have been deposited in the Protein Data Bank under the ID codes 1IZ4 (for D143A) and 1IZ5 (for D143A/D147A).
Table 1.
Details of crystal structure analyses
| D143A | D143A/D147A | |
| Data collection | ||
| Space group | P43212 | P212121 |
| Unit cell (Å) a = | 64.994 | 64.289 |
| Unit cell (Å) b = | 85.348 | |
| Unit cell (Å) c = | 156.395 | 118.030 |
| Number of molecules in the asymmetric unit | ||
| 1 | 2 | |
| Resolution range (Å) | 50.00–2.00 | 50.00–1.80 |
| Total reflections | 91731 | 214024 |
| Unique reflections | 22310 | 60694 |
| Completeness (%) | 95.9 | 99.6 |
| Average redundancy | 4.1 | 3.5 |
| Average I/σ (I) | 10.0 | 4.7 |
| Rmeas | 0.069 | 0.086 |
| Highest resolution bin (Å) | 2.11–2.00 | 1.90–1.80 |
| Completeness (%) | 97.4 | 98.8 |
| Average redundancy | 4.0 | 6.2 |
| Average I/σ (I) | 1.6 | 1.4 |
| Rmeas | 0.529 | 0.602 |
| Structure refinement | ||
| Resolution range (Å) | 35.00–2.00 | 35.00–1.80 |
| Reflections used for refinement | 20084 | 54547 |
| Reflections used for cross validation | 2211 | 5952 |
| R | 0.2698 | 0.2715 |
| Rfree | 0.2986 | 0.2975 |
| Rmsd of bond lengths (Å) | 0.00609 | 0.00598 |
| Rmsd of bond angles (°) | 1.247 | 1.198 |
| Rmsd of dihedral angles (°) | 25.5 | 25.4 |
| Rmsd of improper angles (°) | 0.708 | 0.715 |
| Average B factor (Å2) | 29.8 | 33.3 |
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Acknowledgments
We thank Y. Katsuya for help with the use of the SPring-8 BL24XU beamline. This work was supported in part by New Energy and Industrial Technology Development Organization (NEDO) in the Japanese Government.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0234503.
References
- Alley, S.C., Abel-Santos, E., and Benkovic, S.J. 2000. Tracking sliding clamp opening and closing during bacteriophage T4 DNA polymerase holoenzyme assembly. Biochemistry 39 3076–3090. [DOI] [PubMed] [Google Scholar]
- Ayyagari, R., Impellizzeri, K.J., Yoder, B.L., Gary, S.L., and Burgers, P.M. 1995. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol. Cell. Biol. 15 4420–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck, I. and O’Donnell, M. 2001. The ring-type polymerase sliding clamp family. Genome Biol. 2 3001.1–3001.3. [DOI] [PMC free article] [PubMed]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J-S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D54 905–921. [DOI] [PubMed] [Google Scholar]
- Cann, I.K.O. and Ishino, Y. 1999. Archaeal DNA replication: Identifying the pieces to solve a puzzle. Genetics 152 1249–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann, I.K.O., Ishino, S., Hayashi, I., Komori, K., Toh, H., Morikawa, K., and Ishino, Y. 1999. Functional interactions of a homolog of proliferating cell nuclear antigen with DNA Polymerases in Archaea. J. Bacteriol. 181 6591–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann, I., Ishino, S., Yuasa, M., Daiyasu, H., Toh, H., Morikawa, K., and Ishino, Y. 2001. Biochemical analysis of replication factor C from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183 2614–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D50 760–763. [DOI] [PubMed] [Google Scholar]
- Edgell, D.R. and Doolittle, W.F. 1997. Archaea and the origin(s) of DNA replication proteins. Cell 89 995–998. [DOI] [PubMed] [Google Scholar]
- Ellison, V. and Stillman, B. 2001. Opening of the clamp: An intimate view of an ATP-driven biological machine. Cell 106 655–660. [DOI] [PubMed] [Google Scholar]
- Gulbis, J.M., Kelman, Z., Hurwitz, J., O’Donnell, M., and Kuriyan, J. 1996. Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell 87 297–306. [DOI] [PubMed] [Google Scholar]
- Henneke, G., Raffin, J.P., Ferrari, E., Jonsson, Z.O., Dietrich, J., and Hubscher, U. 2000. The PCNA from Thermococcus fumicolans functionally interacts with DNA polymerase δ. Biochem. Biophys. Res. Commun. 276 600–606. [DOI] [PubMed] [Google Scholar]
- Hingorani, M.M. and O’Donnell, M. 2000. Sliding clamps: A tailored fit. Curr. Biol. 10 R25–R29. [DOI] [PubMed] [Google Scholar]
- Ishino, Y. and Cann, I.K.O. 1998. The euryarchaeotes, a subdomain of Archaea, survive on a single DNA polymerase: Fact or farce? Genes Genet. Syst. 73 323–336. [DOI] [PubMed] [Google Scholar]
- Ishino, Y., Tsurimoto, T., Ishino, S., and Cann, I.K.O. 2001. Functional interactions of an archaeal sliding clamp with mammalian clamp loader and DNA polymerase δ. Genes Cells 6 699–706. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi, D., Yurieva, O., Zhao, Y., Young, M., Stewart, J., Hingorani, M., O’Donnell, M., and Kuriyan, J. 2001. Mechanism of processivity clamp opening by the δ subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell 106 417–428. [PubMed] [Google Scholar]
- Jeruzalmi, D., O’Donnell, M.M., and Kuriyan, J. 2002. Clamp loaders and sliding clamps. Curr. Opin. Struct. Biol. 12 217–224. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J-Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47 110–119. [DOI] [PubMed] [Google Scholar]
- Jonsson, Z.O. and Hubscher, U. 1997. Proliferating cell nuclear antigen: More than a clamp for DNA polymerase. Bioessays 19 967–975. [DOI] [PubMed] [Google Scholar]
- Jonsson, Z.O., Podust, V.N., Podust, L.M., and Hubscher, U. 1995. Tyrosine 114 is essential for the trimeric structure and the functional activities of human proliferating cell nuclear antigen. EMBO J. 14 5745–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman, Z. 1997. PCNA: Structure, functions, and interactions. Oncogene 14 629–640. [DOI] [PubMed] [Google Scholar]
- Kelman, Z. and Hurwitz, J. 1998. Protein–PCNA interactions: A DNA-scanning mechanism? Trends Biochem. Sci. 23 236–238. [DOI] [PubMed] [Google Scholar]
- Komori, K. and Ishino, Y. 2000. Functional interdependence of DNA polymerizing and 3′ → 5′ exonucleolytic activities in Pyrococcus furiosus DNA polymerase I. Protein Eng. 13 41–47. [DOI] [PubMed] [Google Scholar]
- Kong, X.P., Onrust, R., O’Donnell, M., and Kuriyan, J. 1992. Three dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell 69 425–437. [DOI] [PubMed] [Google Scholar]
- Krishna, T.S.R., Kong, X.P., Gary, S., Burgers, P.M., and Kuriyan, J. 1994. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79 1233–1243. [DOI] [PubMed] [Google Scholar]
- Leipe, D.D., Aravind, L., and Koonin, E.V. 1999. Did DNA replication evolve twice independently? Nucleic Acids Res. 27 3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, A.G.W. 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26.
- Matsumiya, S., Ishino, Y., and Morikawa, K. 2001. Crystal structure of an archaeal DNA sliding clamp: Proliferating cell nuclear antigen from Pyrococcus furiosus. Protein Sci. 10 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumiya, S., Ishino, S., Ishino, Y., and Morikawa, K. 2002. Physical interaction between proliferating cell nuclear antigen and replication factor C from Pyrococcus furiosus. Genes Cells 7 911–922. [DOI] [PubMed] [Google Scholar]
- McAlear, M.A., Howell, E.A., Espenshade, K.K., and Holm, C. 1994. Proliferating cell nuclear antigen (pol30) mutations suppress cdc44 mutations and identify potential regions of interaction between the two encoded proteins. Mol. Cell. Biol. 14 4390–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moarefi, I., Jeruzalmi, D., Turner, J., O’Donnell, M., and Kuriyan, J. 2000. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J. Mol. Biol. 296 1215–1223. [DOI] [PubMed] [Google Scholar]
- O’Donnell, M., Jeruzalmi, D., and Kuriyan, J. 2001. Clamp loader structure predicts the architecture of DNA polymerase III holoenzyme and RFC. Curr. Biol. 11 R935–R946. [DOI] [PubMed] [Google Scholar]
- Oyama, T., Ishino, Y., Cann, I.K.O., Ishino, S. and Morikawa, K. 2001. Atomic structure of the clamp loader small subunit from Pyrococcus furiosus. Mol. Cell 8 455–463. [DOI] [PubMed] [Google Scholar]
- Paunesku, T., Mittal, S., Protic, M., Oryhon, J., Korolev, S.V., Joachimiak, A., and Woloschak, G.E. 2001. Proliferating cell nuclear antigen (PCNA): Ringmaster of the genome. Int. J. Radiat. Biol. 77 1007–1021. [DOI] [PubMed] [Google Scholar]
- Shamoo, Y. and Steitz, T.A. 1999. Building a replisome from interacting pieces: Sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell 99 155–166. [DOI] [PubMed] [Google Scholar]
- Stewart, J., Hingorani, M.M., Kelman, Z., and O’Donnell, M. 2001. Mechanism of β clamp opening by the δ subunit of Escherichia coli DNA polymerase III holoenzyme. J. Biol. Chem. 276 19182–19189. [DOI] [PubMed] [Google Scholar]
- Trakselis, M.A. and Benkovic, S. J. 2001. Intricacies in ATP-dependent clamp loading: Variations across replication systems. Structure 9 999–1004. [DOI] [PubMed] [Google Scholar]
- Trakselis, M.A., Alley, S.C., Abel-Santos, E., and Benkovic, S.C. 2001. Creating a dynamic picture of the sliding clamp during T4 DNA polymerase holoenzyme assembly by using fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. 98 8368–8375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto, T. 1998. PCNA, a multifunctional ring on DNA. Biochim. Biophys. Acta 1443 23–39. [DOI] [PubMed] [Google Scholar]
- ———. 1999. PCNA binding proteins. Front. Biosci. 4 849–858. [DOI] [PubMed] [Google Scholar]
- Waga, S. and Stillman, B. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67 721–751. [DOI] [PubMed] [Google Scholar]
- Warbrick, E. 1998. PCNA binding through a conserved motif. BioEssays 20 195–199. [DOI] [PubMed] [Google Scholar]
- ———. 2000. The puzzle of PCNA’s many partners. BioEssays 22 997–1006. [DOI] [PubMed] [Google Scholar]