Abstract
Protein disulfide isomerase (PDI, EC 5.3.4.1), an enzyme and chaperone, catalyses disulfide bond formation and rearrangements in protein folding. It is also a subunit in two proteins, the enzyme collagen prolyl 4-hydroxylase and the microsomal triglyceride transfer protein. It consists of two catalytically active domains, a and a′, and two inactive ones, b and b′, all four domains having the thioredoxin fold. Domain b′ contains the primary peptide binding site, but a′ is also critical for several of the major PDI functions. Mass spectrometry was used here to follow the folding pathway of bovine pancreatic ribonuclease A (RNase A) in the presence of three PDI mutants, F449R, Δ455–457, and abb′, and the individual domains a and a′. The first two mutants contained alterations in the last α helix of domain a′, while the third lacked the entire domain a′. All mutants produced genuine, correctly folded RNase A, but the appearance rate of 50% of the product, as compared to wild-type PDI, was reduced 2.5-fold in the case of PDI Δ455–457, 7.5-fold to eightfold in the cases of PDI F449R and PDI abb′, and over 15-fold in the cases of the individual domains a and a′. In addition, PDI F449R and PDI abb′ affected the distribution of folding intermediates. Domains a and a′ catalyzed the early steps in the folding but no disulfide rearrangements, and therefore the rate observed in the presence of these individual domains was similar to that of the spontaneous process.
Keywords: Mass spectrometry, PDI, protein folding, RNase A
Protein disulfide isomerase (PDI, EC 5.3.4.1), an abundant protein of the eukaryotic endoplasmic reticulum, catalyses disulfide bond formation and rearrangement in newly synthesized polypeptides translocated into this cell compartment (for reviews, see Freedman et al. 1994; Freedman and Klappa 1999), and also assists in protein folding by acting as a molecular chaperone (LaMantia and Lennarz 1993; Cai et al. 1994; Otsu et al. 1994; Puig and Gilbert 1994; Puig et al. 1994; Rupp et al. 1994; Hayano et al. 1995; Klappa et al. 1997; Yao et al. 1997; Winter et al. 2002). Furthermore, it functions as the β subunit in two heteromeric proteins, the enzyme collagen prolyl 4-hydroxylase (Pihlajaniemi et al. 1987; Kivirikko and Myllyharju 1998; Kivirikko and Pihlajaniemi 1998) and the microsomal triglyceride transfer protein (Wetterau et al. 1990, 1991).
PDI is a modular protein consisting of domains a, b, b′, and a′, and an acidic C-terminal extension c (Kemmink et al. 1997). Domains a and a′ are similar to thioredoxin, and they both contain the sequence CGHC, which represents two independently acting catalytic sites (Freedman et al. 1994). Domains a, b, and a′ possess the thioredoxin fold, and b′ is also presumed to have this fold, so that PDI consists of four thioredoxin-like modules, two active and two inactive (Kemmink et al. 1997, 1999; Dijkstra et al. 1999). Domain b′ contains the primary peptide binding site and is capable of binding short peptides by itself, whereas the binding of longer polypeptides requires the presence of additional domains (Darby et al. 1998a; Klappa et al. 1998). The minimum domain requirement for the assembly of a prolyl 4-hydroxylase tetramer is fulfilled by domain pair b′a′ (Pirneskoski et al. 2001). Domain a is capable of replacing a′ in the binding to “scrambled” RNase but not in the prolyl 4-hydroxylase subunit function (Pirneskoski et al. 2001). Extension c can be deleted from the polypeptide without affecting any of its major functions (Koivunen et al. 1999). Mutations introduced into the C-terminal end of domain a′ have been shown to impair several PDI functions (Koivunen et al. 1999). One such mutation, F449R, introduced into the last α helix, markedly reduced the isomerase and reductase activities of the PDI polypeptide and destroyed its ability to function as a subunit in prolyl 4-hydroxylase assembly (Koivunen et al. 1999). However, it was subsequently found that binding of the peptide Δ-somatostatin to this mutant could only be demonstrated after treatment with proteinase K, which removed the entire a′ domain, suggesting that a structural change in this domain indirectly affected peptide binding to b′ (Klappa et al. 2000). Interestingly, a three–amino-acid deletion in the same region, Δ455–457, reduced the chaperone activity to about one third when measured as the ability to fold denatured rhodanese, but resulted in only a modest decrease in the isomerase and reductase activities and left the protein subunit functions intact (Koivunen et al. 1999).
This work studied the influence of three mutations, F449R, Δ455–457, and deletion of the entire domain a′ together with extension c, PDI abb′, (Fig. 1 ▶), on the protein folding activity of the PDI polypeptide. In addition, the folding capabilities of the individual a and a′ domains (Fig. 1 ▶) were investigated. The aim was to obtain a further understanding of the mechanisms of PDI-assisted protein folding at the molecular and structural levels.
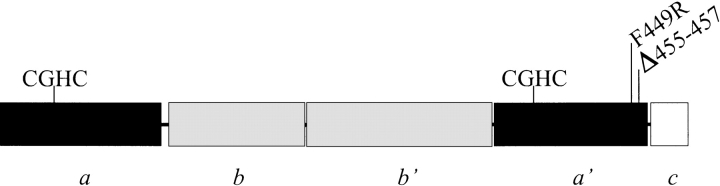
Figure 1.
Schematic representation of the human protein disulfide isomerase polypeptide. Domains a and a′ are indicated by black bars and their catalytic sites by one-letter amino-acid symbols. Domains b and b′ are indicated by gray bars and extension c by a white bar. The sites of the F449R and Δ455–457 mutations are indicated above the a′ domain.
The experiments were performed with bovine pancreatic ribonuclease A (RNase A), a widely employed model substrate. Mass spectrometry has been shown to be very powerful for studies of the oxidative folding of RNase A and of disulfide-containing proteins more generally, under quasiphysiological conditions (Ruoppolo et al. 1996, 1997; Vinci et al. 2000). PDI catalyzes the entire RNase A folding by enhancing the formation and reduction of mixed disulfides with glutathione and the formation of intramolecular disulfides (Ruoppolo et al. 1997). The distribution of isomeric species among the one-disulfide-bond intermediates that accumulate during the early stages is similar in uncatalyzed and PDI-assisted folding (Vinci et al. 2000). It is thus evident that catalysis of the isomerization of disulfide bonds has no significant effect on the folding in its early stages, whereas this catalysis becomes significant for the overall pathway at the later stages, where the important function of PDI is to catalyze disulfide rearrangements within kinetically trapped, structured intermediates. In this study, the use of mass spectrometry allowed us to (1) follow the formation of intermediates, and thus the RNase A folding pathway, in the presence of wild-type PDI and its mutants, (2) quantify the intermediates formed, and (3) compare the rates of formation of the intermediates.
Results
Circular dichroism spectrum analysis of wild-type PDI and its mutants
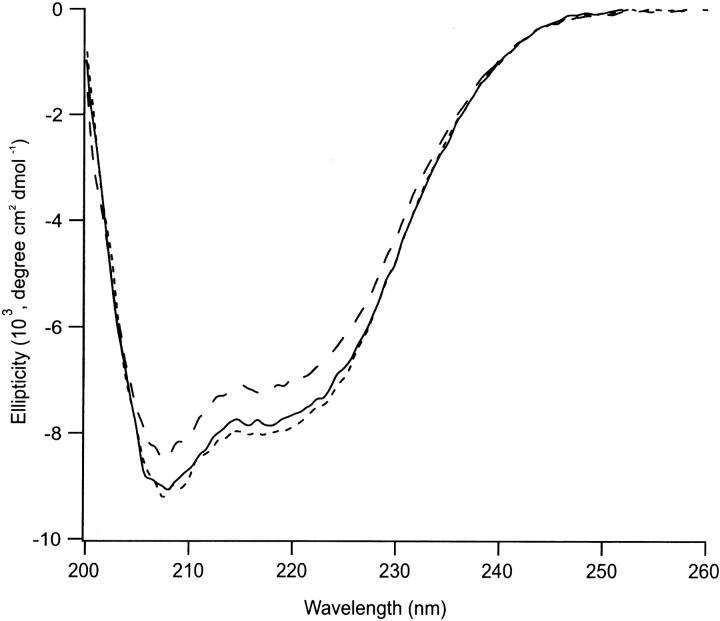
Circular dichroism (CD) spectrum analyses of wild-type PDI and its Δ455–457 and F449R mutants in the far ultraviolet (UV) region showed the latter two to be folded proteins, their spectra being very similar, if not identical, to that of the wild-type PDI (Fig. 2 ▶). It has been reported previously, however, that the F449R mutant is more sensitive to proteinase K than the wild-type, suggesting that this mutant exhibits some altered structural features (Klappa et al. 2000).
Figure 2.
CD spectra of wild-type protein disulfide isomerase (PDI) and its mutants Δ455–457 and F449R in the far ultraviolet (200–260 nm) region. Wild-type PDI is indicated by a solid line, Δ455–457 by a dashed line, and F449R by a dotted line.
Effect of wild-type PDI on RNase A folding
The reduced and denatured RNase A was incubated in the presence of 10 μM wild-type PDI. Aliquots of the folding reaction were withdrawn at different time intervals, and the intermediates present were trapped by alkylation of the free thiol groups and analyzed by electrospray ionization mass spectrometry (ESIMS) to identify the disulfide-bonded intermediates formed (Ruoppolo et al. 1997, 2000). It is worth underlining that under the present alkylation conditions the trapping of free thiols is quantitative (Gray 1993; Ruoppolo et al. 1996, 1997, 2000; Vinci et al. 2000). The carboxyamidomethylation reaction used to trap the free SH groups increased the molecular mass of the intermediates by a fixed amount of 57 Da for each free SH group, thus allowing the separation by mass of intermediates containing different numbers of disulfide bonds. In addition, the relative concentration of each intermediate could be determined by measuring the total ion current produced by each species, provided that the different components were endowed with comparable ionization capabilities (Ruoppolo et al. 1997, 2000; Vinci et al. 2000).
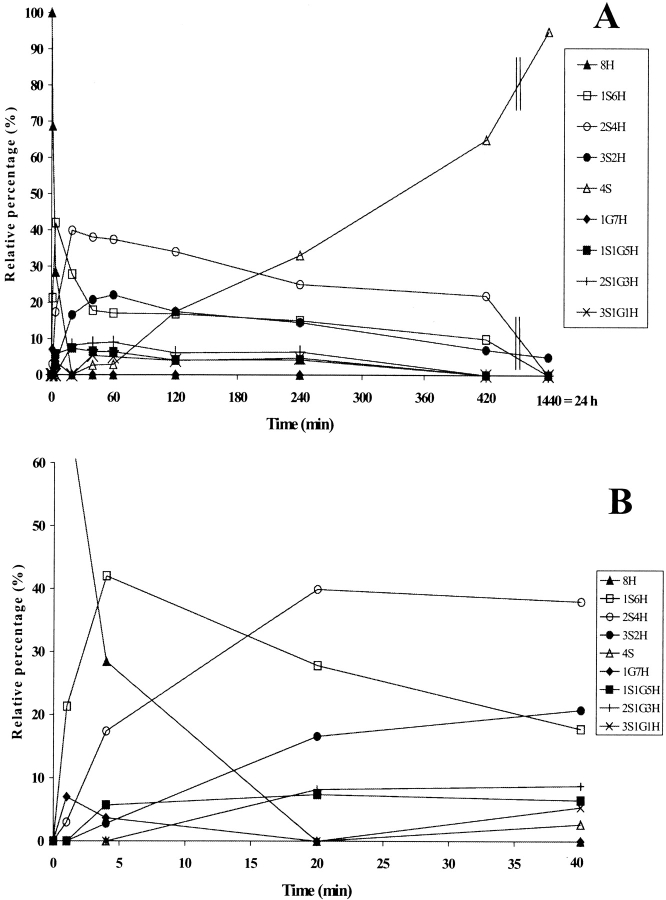
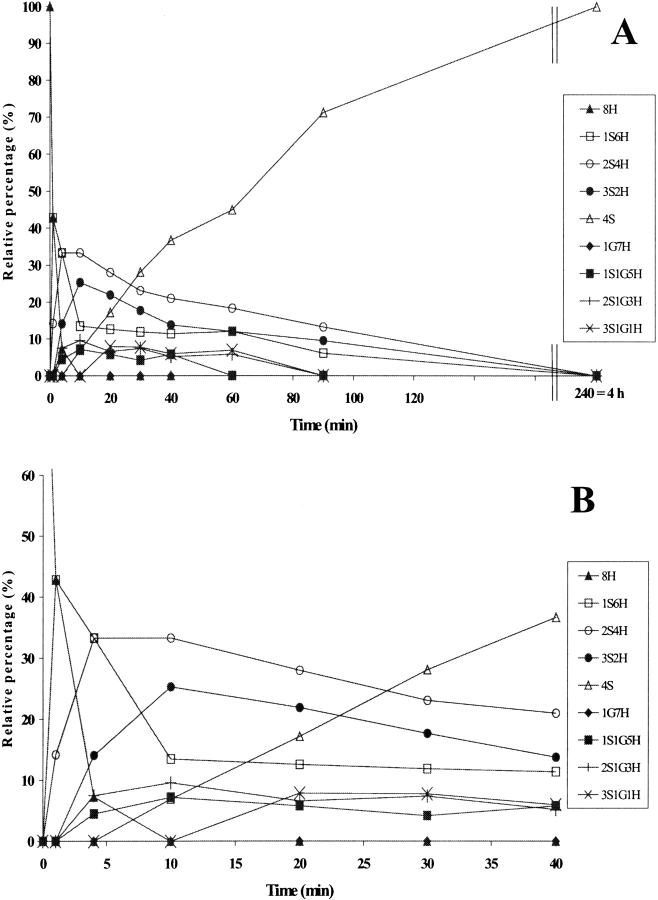
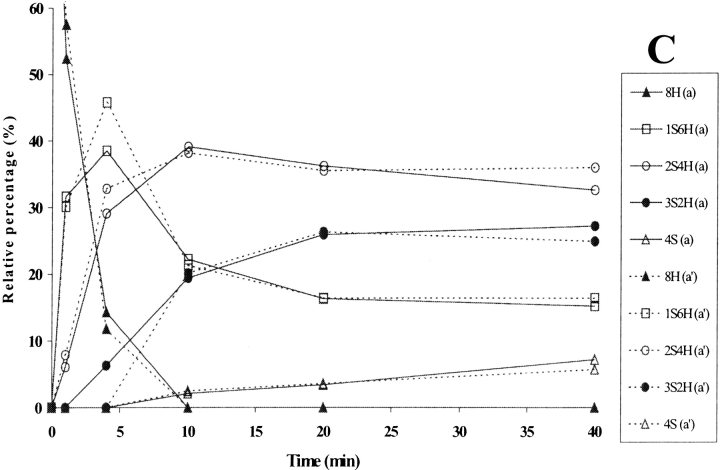
The relative intensities of the intermediates formed during PDI-assisted folding are shown in Fig. 3C ▶, where they are compared with the spontaneous RNase A folding described elsewhere (Ruoppolo et al. 1997) and reported here for the sake of clarity (Figs. 3A,B ▶). The reduced species 8H (Fig. 3C ▶) rapidly disappeared within 5 min in catalyzed folding, as compared with 20 min in the spontaneous process (Fig. 3B ▶). The 1S6H intermediates (Fig. 3C ▶) predominated only during the very first few minutes, while the 2S4H intermediates predominated from 5 min up to about 20 min, at which point the relative concentration of the 4S species increased. The fully oxidized protein accumulated to an extent of 100% at 40 min, whereas in the spontaneous process the fully oxidized species reached a value of about 90% only after 24 h (Fig. 3A ▶). The predominance of the 2S4H species over 3S2H observed in PDI-catalyzed folding was also seen in the spontaneous process under various sets of redox conditions (compare Fig. 3A ▶; Torella et al. 1994; Orrù et al. 2000). A larger number of intermediates containing three disulfides had been expected on a purely statistical basis, but the isomerization reactions seemed to occur at the level of the 2S4H species, which therefore accumulated and predominated in the course of the process (Orrù et al. 2000). Species containing mixed disulfides with glutathione did not accumulate to a concentration exceeding 15% in either the catalyzed or the spontaneous process (compare Figs. 3A,B ▶). PDI thus increased the rate of the whole folding process (Table 1) but did not alter the distribution of intermediates, our data confirming results obtained under different conditions (Ruoppolo et al. 1997).
Figure 3.
(A) Time-course analysis of the folding of RNase A in the presence of 1.5 mM GSH/0.3 mM GSSG (taken from Ruoppolo et al. 1997). (B) Early stages of the time-course shown in A. (C) Time-course analysis of folding in the presence of wild-type protein disulfide isomerase (PDI). PDI was incubated with 1.5 mM GSH/0.3 mM GSSG for 10 min at 25°C and then added to RNase A (1 mg/mL = 73 μM) at a concentration of 10 μM. Percentages of intermediates were derived by electrospray ionization mass spectrometry analysis. The differences between folding experiments performed completely independent of each other were about 5%. For the sake of clarity, error bars are not shown. nS represents intramolecular disulfide bonds, nG mixed disulfides with glutathione, and nH free thiols.
Table 1.
Folding rate of RNase A a
| Catalyst | Appearance of 50% of 4S (min) | 4Sb |
| Spontaneous folding | 400 | 1 |
| PDI | 27 | 17 |
| PDIΔ455–457 | 70 | 4.3 |
| PDI F449R | 220 | 1.5 |
| PDI abb′ | 200 | 1.9 |
| PDI domain a | >420 | 0.6 |
| PDI domain a′ | >420 | 0.6 |
a Derived from data in Figures 3 ▶–8 ▶. Reactions were carried out in the presence of 10 μM catalyst. See figure legends for detailed conditions.
b The rate of formation of 4S species relative to the spontaneous folding at a time point when 35% of 4S had been formed. This rate was found to be linear in all the reactions except that catalyzed by wild-type PDI. This nonlinearity was probably the result of PDI being limited in the other reaction steps.
Effects of the PDI mutants Δ455–457, F449R, and abb′ on RNase A folding
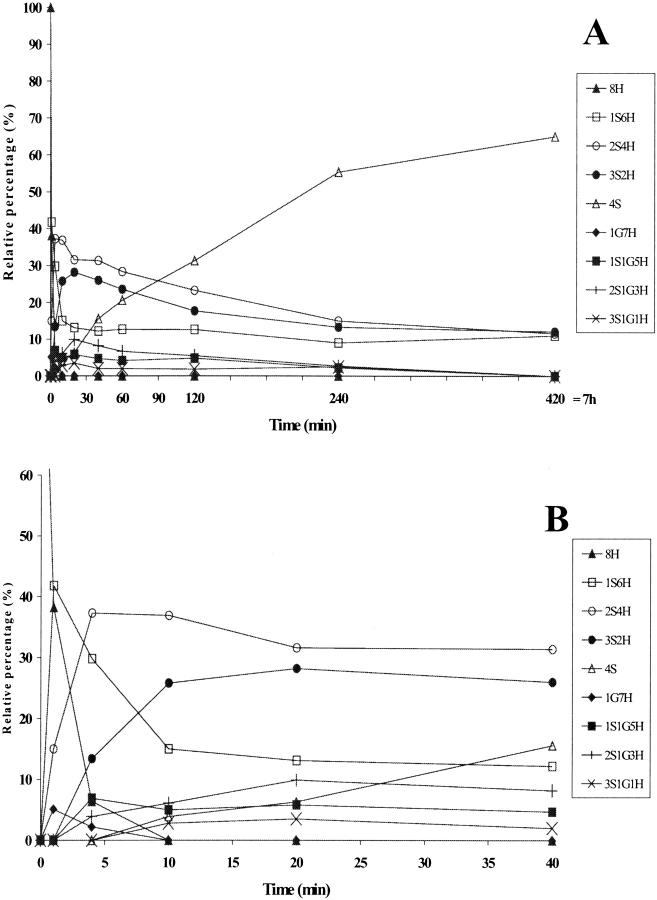
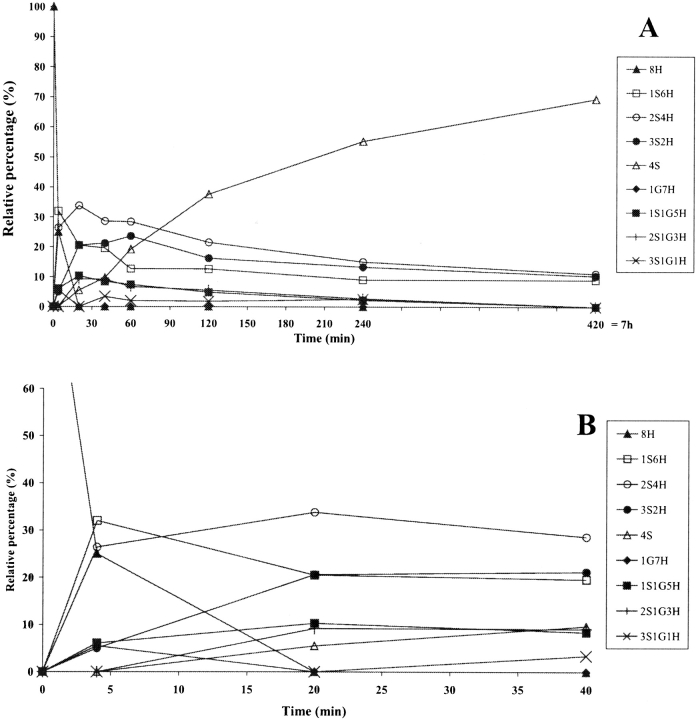
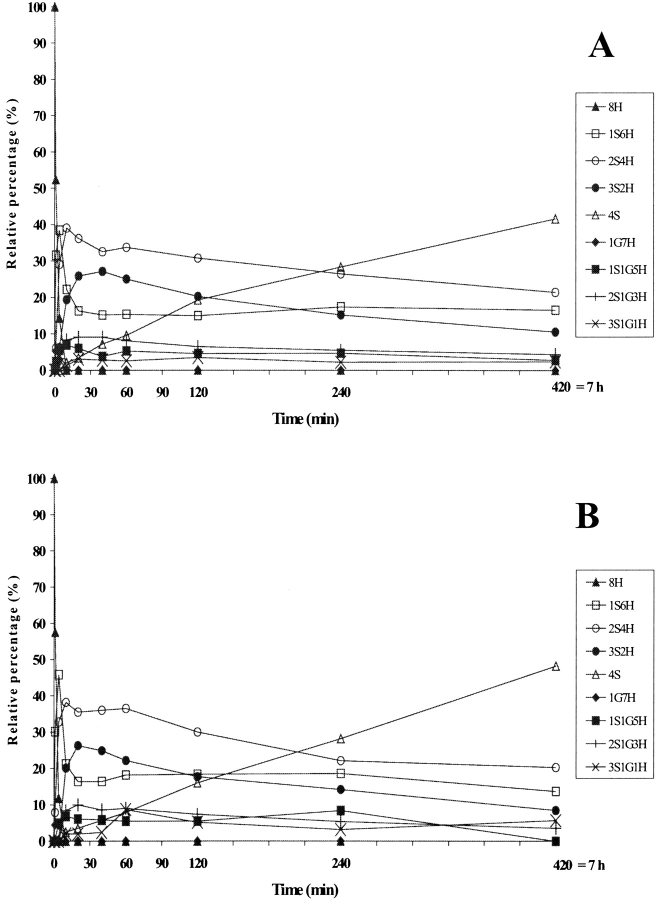
In the folding catalyzed by a 10 μM concentration of any of the three PDI mutants, the completely reduced species 8H disappeared within the first 10 min (Figs. 4A ▶, 5A ▶, 6A ▶), although it did so more slowly than in the presence of the wild-type PDI (5 min, Fig. 3C ▶). The 1S6H species appeared during the very first few minutes (Figs. 4A ▶, 5A ▶, 6A ▶) and had disappeared only by 240 min in the case of Δ455–457 (Fig. 4B ▶), while it was present even at 420 min in the cases of F449R and abb′ (Figs. 5B ▶, 6B ▶). This species was seen only during the first 10 min in the folding catalyzed by the wild-type PDI (Fig. 3C ▶). The 2S4H intermediates predominated until about 25 min in the presence of Δ455–457 (Fig. 4A ▶) and 90 min in the presence of the other two mutants (Figs. 5A ▶, 6A ▶), while the 4S species became predominant after these time points, reaching 100% by 240 min in the case of Δ455–457 (Fig. 4B ▶), but never reaching a value higher than 65–70% even after 420 min in the cases of the other two mutants (Figs. 5B ▶, 6B ▶). The appearance of 50% of the 4S species was 2.5 times slower in the presence of Δ455–457 and 7.5–8 times slower in the presence of F449R and abb′ than in the folding catalyzed by the wild-type (Table 1). Intermediates containing mixed disulfides with glutathione accumulated at a level of about 10% in the cases of all three mutants, but had longer lifetimes in the presence of F449R and abb′ than in the folding catalyzed by the wild-type PDI or the Δ455–457 mutant (Figs. 3 ▶–6 ▶).
Figure 4.
(A) Time-course analysis of folding in the presence of protein disulfide isomerase (PDI) Δ455–457, the early stages being shown in (B). PDI Δ455–457 was incubated with GSH/GSSG and the intermediates were analyzed as described for wild-type PDI in Figure 3 ▶. For the sake of clarity, error bars are not shown. nS represents intramolecular disulfide bonds, nG mixed disulfides with glutathione, and nH free thiols.
Figure 5.
(A) Time-course analysis of folding in the presence of PDI F449R, the early stages being shown in (B). PDI F449R was incubated with GSH/GSSG and the intermediates were analyzed as described in Figure 3 ▶. For the sake of clarity, error bars are not shown. nS represents intramolecular disulfide bonds, nG mixed disulfides with glutathione, and nH free thiols.
Figure 6.
(A) Time-course analysis of folding in the presence of protein disulfide isomerase (PDI) abb′, the early stages being shown in (B). PDI abb′ was incubated with GSH/GSSG and the intermediates were analyzed as described in Figure 3 ▶. For the sake of clarity, error bars are not shown. nS represents intramolecular disulfide bonds, nG mixed disulfides with glutathione, and nH free thiols.
The presence of PDI Δ455–457 involved increases in the rate of disappearance of the reduced protein and the rate of formation of species containing intramolecular disulfides by comparison with spontaneous folding (Figs. 3B ▶, 4A ▶). The final product, that is, the fully oxidized protein, reached 100% after 4 h in the presence of Δ455–457, while this level was reached only in 24 h in the spontaneous process. F449R and abb′ accelerated the early stages of folding (Figs. 3B ▶, 5B ▶, 6B ▶), conversely a steady state was observed later in the process. Altogether, the data showed that PDI Δ455–457 led to an overall 4.5 to sixfold increase in the folding rate depending on the method used to evaluate the speed and PDI F449R and PDI abb′ to a twofold increase (Table 1).
Folding experiments were also performed using a 50-μM concentration of the F449R and abb′ mutants to increase the level of isomerase activity (data not shown). These experiments showed an increase of about threefold in the rate of appearance of 50% of the 4S species as compared with spontaneous folding, while the increases in the presence of 10 μM concentrations were about twofold (above). This rate was still about 4–5 times less than that observed for wild-type PDI, however. The distribution of folding intermediates was very similar to that seen with 10 μM concentrations of the two mutants (data not shown).
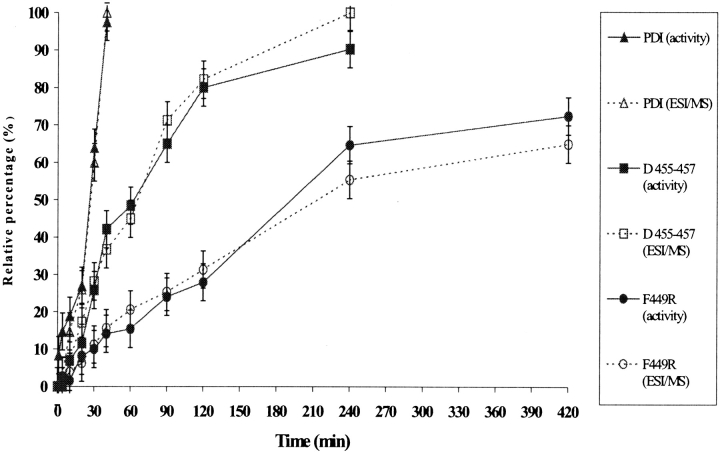
The rates of appearance of the 4S species under different folding conditions were compared with the recovery of RNase A activity as assayed on aliquots of the folding mixtures from the same experiments (Fig. 7 ▶). A perfect correlation was found between the percentage of fully oxidized protein and the recovery of enzyme activity, demonstrating that the PDI mutants generated genuine native RNase A rather than significant amounts of isomers containing nonnative conformations.
Figure 7.
Appearance of the 4S species in the RNase A folding, as detected by electrospray ionization mass spectrometry (ESIMS) analysis under different conditions, relative to the recovery of enzyme activity from aliquots of the same folding reactions. The continuous and dotted lines refer to the activity data and ESIMS data, respectively.
Effects of PDI domains a and a′ on RNase A folding
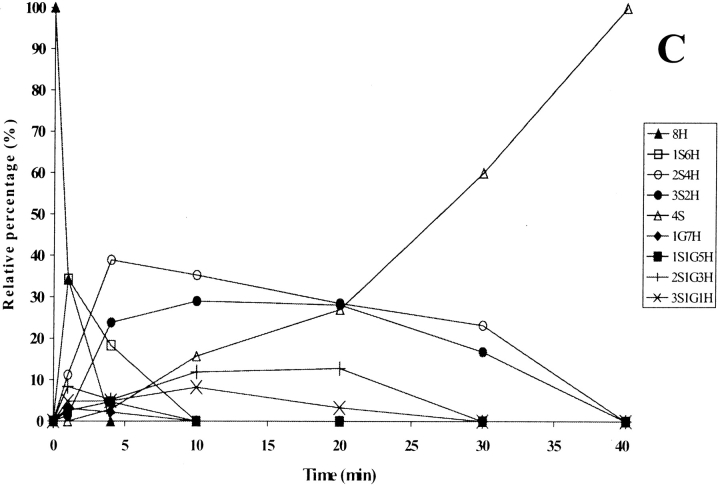
In the folding catalyzed by 10 μM concentrations of domains a and a′, the completely reduced species 8H disappeared within the first 10 min in both cases (Figs. 8A,B ▶), while it did so within 5 min in the presence of wild-type PDI (Fig. 3C ▶). The 1S6H species appeared at almost the same level within the very first few minutes in the cases of both domains, and was present even at the late stages (420 min; Figs. 8A,B ▶), as was also seen in spontaneous oxidation (Fig. 3A ▶) and the folding catalyzed by PDI F449R and PDI abb′ (Figs. 5A ▶, 6A ▶). The 2S4H intermediates predominated until about 240 min in the presence of either of the two domains (Figs. 8A,B ▶), while the 3S2H species appeared at an earlier time than in the spontaneous process (compare Fig. 8C ▶, Fig. 3B ▶), suggesting initial rapid formation of intermediates, which were then present until the late stages. The 4S species appeared at the same rate in the presence of domain a and a′, the formation rate of 50% of the 4S species being over 15 times less than that observed for catalysis by the wild-type PDI (Table 1). This species predominated from 240 min onward, but never reached a value of 50%, even after 420 min. When the folding speed of domains a and a′ was compared to that of PDI at a time point when 35% of the 4S species had been formed and the reaction rate was linear, the decrease was 28-fold (Table 1). It should be noted that the isolated domains contain only half the number of active sites to be found in wild-type PDI, and that the experiments were carried out with the same molar protein concentration.
Figure 8.
Time-course analysis of folding in the presence of protein disulfide isomerase (PDI) domain a (A) and a′ (B), the major species accumulating at the early stages being shown in (C). The continuous and dotted lines refer to intermediates present in (A) and (B), respectively. The PDI domains were incubated with GSH/GSSG and the intermediates analyzed as described in Figure 3 ▶. For the sake of clarity, error bars are not shown. nS represents intramolecular disulfide bonds, nG mixed disulfides with glutathione, and nH free thiols.
Altogether, these data show that domains a and a′ led to no increase in the folding rate relative to the spontaneous process (Table 1), but they did catalyze the disappearance of the reduced protein and the formation of species containing intramolecular disulfides at the early stages, as previously observed in the folding of bovine pancreatic trypsin inhibitor (BPTI; Darby and Creighton 1995). At the later stages, when the rearrangements of disulfide bonds in the quasinative species became significant (Vinci et al. 2000), the two domains showed negligible catalysis. Domains a and a′ produced identical effects on the distribution of intermediates accumulating in the course of folding, our data thus confirming that there is no substantial difference in catalytic activity between them (Darby and Creighton 1995).
To conclude, the rates of formation of the 4S species were in the order wild-type PDI >Δ455–457 > abb′ ~F449R > spontaneous process > domain a = a′.
Effect of PDI mutants and domains a and a′ on proportions of folding intermediates
Although the number of intermediates involved rendered a full kinetic analysis of the data unattainable, it was possible to calculate the proportions of the folding intermediates 1S, 2S, and 3S at a time point when 35% of the 4S species had been formed. At this point, the reactions were in temporary kinetic equilibrium as the intermediate levels were comparatively stable indicating a balance in the forward and reverse reactions (Table 2).
Table 2.
Proportions of the RNase A folding intermediatesa
| Catalyst | 1S/2S | 2S/3S |
| Spontaneous folding | 0.6 | 1.8 |
| PDI | 0b | 1.1 |
| PDI Δ455–457 | 0.5 | 1.5 |
| PDI F449R | 0.5 | 1.3 |
| PDI abb′ | 0.6 | 1.3 |
| PDI domain a | 0.7 | 1.9 |
| PDI domain a′ | 0.8 | 1.9 |
a Proportions of the folding intermediates were calculated in the presence of 10 μM catalyst at a time point when 35% of the 4S species had been formed. The levels of the 1S, 2S, and 3S intermediates were relatively stable at this point, indicating that the forward and reverse reactions were balanced and that the species were in temporary kinetic equilibrium.
b No 1S could be detected because of its rapid conversion to 2S.
In the reaction catalyzed by the wild-type PDI, the 1S species were converted to 2S so rapidly that no 1S could be detected at the time point selected (Table 2, Fig. 3C ▶). The 1S/2S ratio was slightly smaller for the mutants Δ455–457 and F449R than for abb′ and spontaneous folding, whereas domains a and a′ had slightly higher ratios than in spontaneous folding (Table 2). The 2S/3S ratio was lowest for the wild-type PDI, slightly higher for mutants F449R and abb′, and even higher for Δ455–457 (Table 2). All these ratios were distinctly lower than in spontaneous folding, whereas the ratios for domains a and a′ were no better than in the uncatalyzed reaction (Table 2).
Discussion
Despite the fact that PDI is a well-studied protein, molecular interpretations of its modes of action are still under debate. Such interpretations have been made more difficult by the fact that PDI is multifunctional in the cell, as mirrored by the range of catalytic functions seen in vitro. It consists of multiple domains, all of which may contribute to the differing functions to various extents. Furthermore, while the structures of some of the individual domains are known (Kemmink et al. 1997, 1999; Dijkstra et al. 1999), no high-resolution structure is available for full-length PDI. This compromises our ability to understand the synergistic interactions between the domains.
We studied here the influence on the refolding of RNase A of three PDI mutations, two of which, Δ455–457 and F449R, have previously been shown to be critical for all PDI functions (Koivunen et al. 1999), even though neither of them directly occupies the catalytic or peptide binding sites (Fig. 1 ▶). The Δ455–457 mutant had previously been show to retain the abilities to form active prolyl 4-hydroxylase tetramers, to catalyze the DTT-dependent reduction of insulin and to bind peptide substrates as per the wildtype protein, but exhibited only 67% of the activity of the native protein in a continuous spectroscopic RNase assay (Koivunen et al. 1999; Klappa et al. 2000). The F449R mutant was unable to form active prolyl 4-hydroxylase tetramers or to bind peptide substrates and exhibited only 21% of the activity of the native protein in the spectroscopic RNase assay, although retaining 43% of its reductase activity (Koivunen et al. 1999; Klappa et al. 2000). The third mutant lacked the entire domain a′ and also the extension c, which can be deleted without affecting any of the major PDI functions (Koivunen et al. 1999).
Analysis of folding in the presence of the Δ455–457 mutant showed a 2.5 to fourfold decrease in the rate of the overall reaction relative to the wild type depending on the method used to evaluate the speed (Table 1). Calculation of the amounts of the folding intermediates at a time point when only 35% of the 4S species had been formed (Table 2) showed that PDI Δ455–457 differed from the wild type in that its 1S/2S ratio was higher, thus indicating that there were still earlier folding intermediates present. Also, this mutant was slower than PDI in catalyzing the 2S to 3S step. These data agree with the previous findings (Koivunen et al. 1999), indicating that this mutant retains full reductase and oxidase activities but has lost a significant proportion of its isomerase activity, resulting in less efficient catalysis of the steps involving disulfide rearrangements which occur at a late stage. Because these late isomerizations are the rate-limiting steps in the overall process, the loss of this activity explains the decrease in the rate of generation of native RNase.
The F449R and abb′ mutants showed substantial differences in the distribution of the intermediates relative to the wild type. They accelerated the formation of intermediates at the very early stages, but the intermediates were still present at longer incubation times, when a steady state was observed. Both mutants were clearly very ineffective in catalyzing the last steps of disulfide isomerization, but they functioned reasonably well in catalyzing the formation of species containing one, two, and three intramolecular disulfides. These mutants appeared to have greatly reduced isomerase activities, making them unable to interconvert the partially oxidized intermediates to the fully oxidized protein at the stages in the reaction at which the isomerization of disulfide bonds in partially structured intermediates becomes significant. The PDI mutants studied here clearly differed from each other in the catalysis of the final step, as the rate of formation of the 4S species was 2–3 times slower for F449R and abb′ than for Δ455–457, whereas the 1S/2S values were equal (Tables 1 and 2). Interestingly, the 2S/3S values of F449R and abb′ were even lower than that of Δ455–457, indicating faster formation of the 3S species. It thus seems that F449R and abb′ were specifically disabled with regard to catalysis of the final step of folding. These two mutants were still distinctly better than the isolated domains a and a′, however (Tables 1 and 2), thus indicating that there is still some interplay left between their domains.
Modeling of the Δ455–457 and F449R mutations based on the known structure of domain a (Kemmink et al. 1997) and the preliminary structure of a′ (Dijkstra et al. 1999) suggests that the deletion of residues 455–457 removes one turn from the final α helix of domain a′, while mutation F449R introduces a charge onto the interior buried face of the same helix. Comparison of the CD spectra of the mutants with the wild type argues against any major changes in the overall folding of the polypeptides, as the spectra were very similar, if not identical. However, the F449R mutant has previously been shown to be more sensitive to digestion by proteinase K than the wild type, suggesting the presence of some altered structural features, increased structural mobility, or a decrease in conformational stability (Klappa et al. 2000). It has been suggested that these features may prevent the binding of peptides to domain b′ (Klappa et al. 2000), but our data are not consistent with this suggestion, as F449R was no less efficient than abb′ (Table 1), which effectively binds scrambled RNase (Klappa et al. 1998). Our data on the catalysis of RNase A folding by the three mutants thus suggest that alterations in the last α helix of domain a′ affect the folding capability of the PDI polypeptide, most probably by causing conformational changes that reduce the ability of domain a′ to participate in the catalysis of the isomerase reaction in the case of Δ455–457 and destroy this ability entirely in the case of F449R. All three mutants retained some oxidase activity, however. The net effect of this was a substantial decrease in the rate of formation of native RNase, but not of many of the folding intermediates. Our data agree with those reported for the rearrangement of disulfides in the folding of BPTI (Darby and Creighton 1995). The activity of PDI abb′ in BPTI folding was only 6–15% of that of the wild type, while PDI in which the two cysteines at the second catalytic site had been mutated to a SGHA sequence still had more than half the activity of the wild type, indicating that a′ has regions in addition to the active site, which are important for disulfide bond isomerization activity (Darby and Creighton 1998a).
The isolated domains a and a′ accelerated the early stages of folding relative to the spontaneous process, but had a negligible effect on disulfide bond rearrangements (Tables 1 and 2). It is possible that the oxidase activity of these domains may generate a higher proportion of folding intermediates, which are kinetically challenged upon reaching the native state, or that isomerization of a quasinative intermediate in a glutathione buffer in the absence of PDI proceeds either via reduction and then oxidation or via the formation of a mixed disulfide with glutathione, and that in either case the isolated a or a′ domains can efficiently catalyze the reverse of the first step, preventing the second step from occurring. Our data are consistent with the finding that all PDI domains are required for disulfide bond rearrangements in the folding of BPTI (Darby et al. 1998a). Another explanation for the poor isomerase activity of the isolated a and a′ domains could be that PDI is efficient in catalyzing reactions involving two disulfide bonds in the substrate simultaneously. This suggestion is supported by the finding that PDI and dimeric DsbC (Darby et al. 1998b) are able to catalyze disulfide rearrangements, whereas monomeric DsbA is not (Zapun et al. 1995).
PDI displays a unique capacity to isomerize disulfide bonds that have been reported only for similar multidomain proteins, such as ERp57 (Oliver et al. 1997), ERp72 (Mazzarella et al. 1990), and P5 (Lundström-Ljung et al. 1995). The present data suggest that this property resides in the global structure of PDI and possibly also in an ability to differentiate between structured, partially structured, and misfolded intermediates, which facilitates disulfide isomerization linked to conformational changes. PDI and ERp57 have been found in mixed disulfide complexes with their substrates in vivo (Molinari and Helenius 1999). Under the present reaction conditions, it might have been possible to observe PDI-RNase A mixed complexes, but they were not seen, most probably because of the measurement calibrations, which favored the detection of lower molecular weight substances. Recent findings in yeast suggest the existence of a direct pathway of oxidizing equivalents from Ero1 via PDI to the folding substrate (Tu et al. 2000). These yeast experiments cast doubt on the importance of PDI for the reshuffling of disulfide bonds, suggesting that its critical function in vivo may be to act as an oxidase. However, our data with RNase A and the previous observations with BPTI (Darby et al. 1998a) strongly indicate that the rate-limiting step for native disulfide bond formation, at least in vitro, is the isomerization of disulfide bonds, often in quasinative structures, and that a fully functional PDI is crucial to this process. Furthermore, it is evident that the multidomain structure of PDI is needed for this function, with many of the features required for the catalysis of disulfide rearrangements remaining as yet undefined. Progressive reengineering of the PDI molecule should prove an important approach to gaining an understanding of this process, but careful interpretation of the results is required because of the multifunctionality of this protein and its interdomain interactions.
Materials and methods
Reagents
Reduced DTT, EDTA, reduced glutathione (GSH), oxidized glutathione (GSSG), and RNase A were from Sigma Chemical Co. Tris and iodoacetamide (IAM) were from Fluka, prepacked Sephadex G-25M PD10 from Amersham Pharmacia Biotech, and guanidinium chloride from Pierce. All other reagents were of the highest grade commercially available.
The concentrations of RNase A solutions were determined by reference to the absorption of 0.695 at 278 nm recorded for a 1-mg/mL solution (Schaffer et al. 1975), while concentrations of the wild-type PDI and its mutants were determined by the method of Bradford (1976) with bovine serum albumin as a standard. The concentrations of the a and a′ domains were determined from their absorbance at 280 nm, using molar absorbance coefficients of 19,060 and 15,220, respectively (Darby and Creighton 1995).
Expression vectors
Expression vectors for the PDI mutants Δ455–457 and F449R were generated as described (Koivunen et al. 1999). Expression vectors for PDI abb′ and the a domain were synthesized by PCR using primers corresponding to amino acids 1–350 and 4–123 in the mature polypeptide, respectively. The purified PCR product of PDI abb′ was cloned into the BamHI-HindIII site of pQE30 (Qiagen) and that of the domain a into the BamHI-SalI site of pQE31 (Qiagen). An expression vector for the a′ domain coding for residues 348–462 cloned into pET12a (Novagen; Darby and Creighton 1995) was obtained from Dr. Johan Kemmink.
Protein production and purification
PDI mutants Δ455–457, F449R, abb′ and the a domain were produced in Escherichia coli SG13009 (pREP4; Qiagen) and purified based on the histidine tags at their N termini. The cells were harvested 3 h after induction and sonicated in a buffer of 50 mM Tris-HCl, 0.1% Triton X-100, pH 8, supplemented with a cocktail tablet of complete EDTA-free proteinase inhibitor (Boehringer Mannheim). After centrifugation at 60000 g at 4°C, the supernatants were applied to Ni-NTA columns, which were washed with a buffer containing 60 mM imidazole and eluted with a buffer of 0.5 M imidazole. The fractions containing the proteins were pooled. PDIΔ455–457, F449R, and domain a fractions were dialyzed against 50 mM Tris-HCl, pH 8, and applied to a Q Sepharose column (Amersham Pharmacia Biotech) equilibrated with the same buffer. The column was washed with this buffer until O.D. 280 was <0.01. The protein was eluted with a linear gradient of 0 to 0.4 M NaCl, and the fractions containing the protein were pooled, dialyzed against 0.1 M Tris-HCl, 1 mM EDTA, pH 7.5, and concentrated. PDI abb′ fractions from the Ni-NTA column were dialyzed against 20 mM Tris-HCl, 0.3 M NaCl, pH 8, and applied to a Sephacryl S-200 high resolution gel filtration column (Amersham Pharmacia Biotech) equilibrated and eluted with the same buffer. Fractions containing the protein were pooled, dialyzed against 0.1 M Tris-HCl, 1 mM EDTA, pH 7.5, and concentrated. a′ domain was expressed in E. coli BL21 (DE3) pLysS (Novagen) and purified as described earlier (Darby and Creighton 1995).
CD spectrum analyses
CD spectra of wild-type PDI and its mutants Δ455–457 and F449R were measured in a Jasco J715 spectropolarimeter using a 1-mm path length cell at 20°C. The spectra were averages of five scans. The proteins were dissolved in 10 mM sodium phosphate, pH 7.5.
Folding reactions
RNase A was reduced and denatured as described (Torella et al. 1994). The protein solution was then separated from the excess of DTT and denaturant by HPLC desalting. The protein fraction was recovered, lyophilized, and used within 2 d. The purity of the reduced and denatured protein was checked by ESIMS. Lyophilized reduced and denatured RNase A was dissolved to a concentration of ~3 mg/mL in 1% acetic acid and then diluted in the folding buffer of 0.1 M Tris-HCl, 1 mM EDTA, pH 7.5 to a final concentration of 1 mg/mL. The desired amounts of GSH and GSSG stock solutions (25 mM made fresh daily) were added to initiate folding, typical final concentrations being 1.5 mM GSH and 0.3 mM GSSG. The pH was adjusted to 7.5 with Tris base and the reaction was carried out at 25°C in a nitrogen atmosphere. When refolding was carried out in the presence of wild-type or mutant PDI or domain a or a′, the enzyme was dissolved in 0.1 M Tris-HCl, 1 mM EDTA, pH 7.5, and preincubated in the presence of 1.5 mM GSH/0.3 mM GSSG redox buffer for 10 min at 25°C. The concentration of wild-type or mutant PDI or domain a or a′ was fixed at 10 μM. The redox conditions used were shown to be the most effective in promoting RNase A spontaneous folding (Torella et al. 1994), and the same conditions were selected for the catalyzed folding in order to give the best comparison, as different redox conditions could have enhanced the disulfide isomerase activity of PDI relative to the uncatalyzed process.
Alkylation of folding aliquots
Folding was monitored on a time-course basis by sampling aliquots of the folding mixture at appropriate intervals. The protein samples were alkylated as described (Gray 1993; Torella et al. 1994). IAM was freshly dissolved in 0.1 M Tris-HCl, 1 mM EDTA, pH 7.5 at 65°C and cooled to room temperature before use. The solution was protected from light during preparation of the reagents to minimize the photolytic production of iodine, a very potent oxidizing agent for thiols. The folding aliquots (100 μL) were added to an equal volume of a 2.2 M IAM solution. Alkylation was performed for 30 s in the dark at room temperature under a nitrogen atmosphere. Acetic acid was then added, the sample was desalted by HPLC, and the protein fraction was recovered and lyophilized. These alkylation conditions are effective in providing rapid quenching of thiol groups (Gray 1993; Torella et al. 1994).
Electrospray mass analysis
The ESIMS analyses were carried out using a Bio-Q triple quadrupole mass spectrometer equipped with an electrospray ion source (Micromass). The protein samples were dissolved in water containing 2% acetic acid and diluted 1/1 with acetonitrile. A 10 μL aliquot of the protein solution (10 pmole/μL) was injected directly into the ion source via loop injection, and data were acquired for 10 s/scan and elaborated using the Mass Lynx software provided by the manufacturer. Mass-scale calibration was performed by means of multiply charged ions from a separate injection of horse heart myoglobin (average molecular mass 16951.5 Da). Each set of folding data was obtained as means of three independent folding experiments. The differences between folding experiments performed completely independent of each other were about 5%.
Activity data
Desalted, alkylated aliquots from the refolding reaction were concentrated by lyophilization and dissolved in 300 μL of distilled water. Concentrations of proteins, including RNase A, were determined from the absorbance at 280 nm after compensating for the presence of PDI, as estimated by integrating the corresponding HPLC peaks in the desalting chromatogram. RNase A activity was determined in triplicate by mixing 100 μL of sample diluted in distilled water with 450 μL of an assay mixture containing 50 mM Tris-HCl, pH 7.5, 25 mM KCl, 5 mM MgCl2, and 200 mg/mL 2′3′cCMP. The specific activity of RNase A was determined from the rate of increase in the absorbance at 288 nm and related to a standard curve for native RNase A activity. Control experiments in which native RNase A was subjected to alkylation with IAM, desalted, and lyophilized had shown that this procedure did not affect the enzyme activity (Ruoppolo et al. 1997).
Acknowledgments
We are grateful to Dr. Lloyd Ruddock for his valuable comments on the manuscript. We thank Eeva Lehtimäki and Merja Nissilä for their expert technical assistance. This work was supported by grants from the EC (BIO4-CT96–0436) to K.I.K. and G.M., Progetto Finalizzato Biotecnologie, CNR, Progetto di Ricerca di Interesse Nazionale, PRIN, Murst, Roma, the Health Science Council of the Academy of Finland, the Finnish Centre of Excellence Programme 2000–2005 (44843), and FibroGen Inc. (South San Francisco, CA).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
PDI, protein disulfide isomerase
F449R, PDI containing a point mutation F449R
Δ455–457, PDI lacking amino acids 455–457
PDI abb′, PDI lacking domain a′ and the C-terminal extension c
RNase A, bovine pancreatic ribonuclease A
CD, circular dichroism
ESIMS, electrospray ionisation mass spectrometry
BPTI, bovine pancreatic trypsin inhibitor
GSH, reduced glutathione
GSSG, oxidized glutathione
IAM, iodoacetamide
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0242803.
References
- Bradford, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Cai, H., Wang, C.-C., and Tsou, C.-L. 1994. Chaperone-like activity of protein disulfide isomerase in the refolding of a protein with no disulfide bonds. J. Biol. Chem. 269 24550–24552. [PubMed] [Google Scholar]
- Darby, N.J. and Creighton, T.E. 1995. Functional properties of the individual thioredoxin-like domains of protein disulfide isomerase. Biochemistry 34 11725–11735. [DOI] [PubMed] [Google Scholar]
- Darby, N.J., Penka, E., and Vincentelli, R. 1998a. The multi-domain structure of protein disulfide isomerase is essential for high catalytic efficiency. J. Mol. Biol. 276 239–247. [DOI] [PubMed] [Google Scholar]
- Darby, N.J., Raina, S., and Creighton, T.E. 1998b. Contributions of substrate binding to the catalytic activity of DsbC. Biochemistry 37 783–791. [DOI] [PubMed] [Google Scholar]
- Dijkstra, K., Karvonen, P., Pirneskoski, A., Koivunen, P., Kivirikko, K.I., Darby, N.J., van Straaten, M., Scheek, R.M., and Kemmink, J. 1999. Assignment of 1H, 13C and 15N resonances of the a′ domain of protein disulfide isomerase. J. Biomol. NMR 14 195–196. [DOI] [PubMed] [Google Scholar]
- Freedman, R.B. and Klappa, P. 1999. Protein disulfide isomerase: A catalyst of thiol:disulfide interchange and associated protein folding. In Molecular chaperones and protein folding (ed. B. Bukau), pp. 437–459. Harwood Academic Press, London.
- Freedman, R.B., Hirst, T.R., and Tuite, M.F. 1994. Protein disulfide isomerase: Building bridges in protein folding. Trends Biochem. Sci. 19 331–336. [DOI] [PubMed] [Google Scholar]
- Gray, W.R. 1993. Disulfide structures of highly bridged peptides: A new strategy for analysis. Protein Sci. 2 1732–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayano, T., Hirose, M., and Kikuchi, M. 1995. Protein disulfide isomerase mutant lacking its isomerase activity accelerates protein folding in the cell. FEBS Lett. 377 505–511. [DOI] [PubMed] [Google Scholar]
- Kemmink, J., Darby, N.J., Dijkstra, K., Nilges, M., and Creighton, T.E. 1997. The folding catalyst protein disulfide isomerase is constructed of active and inactive thioredoxin modules. Curr. Biol. 7 239–245. [DOI] [PubMed] [Google Scholar]
- Kemmink, J., Dijkstra, K., Mariani, M., Scheek, R.M., Penka, E., Nilges, M., and Darby, N.J. 1999. The structure in solution of the b domain of protein disulfide isomerase. J. Biomol. NMR 13 357–368. [DOI] [PubMed] [Google Scholar]
- Kivirikko, K.I. and Myllyharju, J. 1998. Prolyl 4-hydroxylases and their protein disulfide isomerase subunit. Matrix Biol. 16 357–368. [DOI] [PubMed] [Google Scholar]
- Kivirikko, K.I. and Pihlajaniemi, T. 1998. Collagen hydroxylases and the protein disulfide isomerase subunit of prolyl 4-hydroxylases. Adv. Enzymol. Relat. Areas Mol. Biol. 72 325–398. [DOI] [PubMed] [Google Scholar]
- Klappa, P., Hawkins, H.C., and Freedman, R.B. 1997. Interactions between protein disulfide isomerase and peptides. Eur. J. Biochem. 248 37–42. [DOI] [PubMed] [Google Scholar]
- Klappa, P., Ruddock, L.W., Darby, N.J., and Freedman, R.B. 1998. The b′ domain provides the principal peptide-binding site of protein disulfide isomerase but all domains contribute to binding of misfolded proteins. EMBO J. 17 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klappa, P., Koivunen, P., Pirneskoski, A., Karvonen, P., Ruddock, L.W., Kivirikko, K.I., and Freedman, R.B. 2000. Mutations that destabilize the a′ domain of human protein-disulfide isomerase indirectly affect peptide binding. J. Biol. Chem. 275 13231–13218. [DOI] [PubMed] [Google Scholar]
- Koivunen, P., Pirneskoski, A., Karvonen, P., Ljung, J., Helaakoski, T., Notbohm, H., and Kivirikko, K.I. 1999. The acidic C-terminal domain of protein disulfide isomerase is not critical for the enzyme subunit function or for the chaperone or disulfide isomerase activities of the polypeptide. EMBO J. 18 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMantia, M. and Lennarz, W.J. 1993. The essential function of yeast protein disulfide isomerase does not reside in its isomerase activity. Cell 74 899–908. [DOI] [PubMed] [Google Scholar]
- Lundström-Ljung, J., Birnbach, U., Rupp, K., Soling, H.D., and Holmgren, A. 1995. Two resident ER-proteins, CaBP1 and CaBP2, with thioredoxin domains, are substrates for thioredoxin reductase: Comparison with protein disulfide isomerase. FEBS Lett. 357 305–308. [DOI] [PubMed] [Google Scholar]
- Mazzarella, R.A., Srinivasan, M., Haugejorden, S.M., and Green, M. 1990. ERp72, an abundant luminal endoplasmic reticulum protein, contains three copies of the active site sequences of protein disulfide isomerase. J. Biol. Chem. 265 1094–1101. [PubMed] [Google Scholar]
- Molinari, M. and Helenius, A. 1999. Glycoproteins form mixed disulfides with oxidoreductases during folding in living cells. Nature 402 90–93. [DOI] [PubMed] [Google Scholar]
- Oliver, J.D., van der Wal, F.J., Bulleid, N.J., and High, S. 1997. Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science 275 86–88. [DOI] [PubMed] [Google Scholar]
- Orrù, S., Vitagliano, L., Esposito, L., Mazzarella, L., Marino, G., and Ruoppolo, M. 2000. Effect of deamidation on folding of ribonuclease A. Protein Sci. 9 2577–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu, M., Omura, F., Yoshimori, T., and Kikuchi, M. 1994. Protein disulfide isomerase associates with misfolded human lysozyme in vivo. J. Biol. Chem. 269 6874–6877. [PubMed] [Google Scholar]
- Pihlajaniemi, T., Helaakoski, T., Tasanen, K., Myllylä, R., Huhtala, M.-L., Koivu, J., and Kivirikko, K.I. 1987. Molecular cloning of the β-subunit of human prolyl 4-hydroxylase. This subunit and protein disulfide isomerase are products of the same gene. EMBO J. 6 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirneskoski, A., Ruddock, L.W., Klappa, P., Freedman, R.B., Kivirikko, K.I., and Koivunen, P. 2001. Domains b′ and a′ of protein disulfide isomerase fulfill the minimum requirement for function as a subunit of prolyl 4-hydroxylase. The N-terminal domains a and b enhances this function and can be substituted in part by those of ERp57. J. Biol. Chem. 276 11287–11293. [DOI] [PubMed] [Google Scholar]
- Puig, A. and Gilbert, H.F. 1994. Protein disulfide isomerase exhibits chaperone and anti-chaperone activity in the oxidative refolding of lysozyme. J. Biol. Chem. 269 7764–7771. [PubMed] [Google Scholar]
- Puig, A., Lyles, M.M., Noiva, R., and Gilbert, H.F. 1994. The role of the thiol/disulfide centers and peptide binding site in the chaperone and anti-chaperone activities of protein disulfide isomerase. J. Biol. Chem. 269 19128–19135. [PubMed] [Google Scholar]
- Ruoppolo, M., Freedman, R.B., Pucci, P., and Marino, G. 1996. The glutathione dependent pathways of refolding of RNase T1 by oxidation and disulfide isomerization. Catalysis by protein disulfide isomerase. Biochemistry 35 13636–13646. [DOI] [PubMed] [Google Scholar]
- Ruoppolo, M., Lundstrom-Ljung, J., Talamo, F., Pucci, P., and Marino, G. 1997. Effect of glutaredoxin and protein disulfide isomerase on the glutathione-dependent folding of ribonuclease A. Biochemistry 36 12259–12267. [DOI] [PubMed] [Google Scholar]
- Ruoppolo, M., Vinci, F., Klink, T.A., Raines, R.T., and Marino, G. 2000. Contribution of individual disulfide bonds to the oxidative folding of ribonuclease A. Biochemistry 39 12033–12042. [DOI] [PubMed] [Google Scholar]
- Rupp, K., Birnbach, U., Lundström, J., Nguyen Van, P., and Söling, H.-D. 1994. Effects of CaBP2, the rat analog of ERp72, and of CaBP1 on the refolding of denatured reduced proteins. Comparison with protein disulfide isomerase. J. Biol. Chem. 269 2501–2507. [PubMed] [Google Scholar]
- Schaffer, S.W., Ahmed, A.K., and Wetlaufer, D.B. 1975. Salt effects in the glutathione-facilitated reactivation of reduced bovine pancreatic ribonuclease. J. Biol. Chem. 250 8483–8486. [PubMed] [Google Scholar]
- Torella, C., Ruoppolo, M., Marino, G., and Pucci, P. 1994. Analysis of RNase A refolding intermediates by electrospray/mass spectrometry. FEBS Lett. 352 301–306. [DOI] [PubMed] [Google Scholar]
- Tu, B.P., Ho-Schleyer, S.C., Travers, K.J., and Weissman, J.S. 2000. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science 290 1571–1574. [DOI] [PubMed] [Google Scholar]
- Vinci, F., Ruoppolo, M., Pucci, P., Freedman, R.B., and Marino, G. 2000. Early intermediates in the PDI-assisted folding of ribonuclease A. Protein Sci. 9 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterau, J.R., Combs, K.A., Spinner, S.N., and Aggerbeck, L.P. 1990. Protein disulfide isomerase is a component of the microsomal triglyceride transfer protein complex. J. Biol. Chem. 265 9801–9807. [PubMed] [Google Scholar]
- Wetterau, J.R., Combs, K.A., McLean, L.R., Spinner, S.N., and Aggerbeck, L.P. 1991. Protein disulfide isomerase appears necessary to maintain the catalytically active structure of the microsomal triglyceride transfer protein. Biochemistry 30 9728–9735. [DOI] [PubMed] [Google Scholar]
- Winter, J., Klappa, P., Freedman, R.B., Lilie, H., and Rudolph, R. 2002. Catalytic activity and chaperone function of human protein-disulfide isomerase are required for the efficient refolding of proinsulin. J. Biol. Chem. 277 310–317. [DOI] [PubMed] [Google Scholar]
- Yao, Y., Zhou, Y.-C., and Wang, C.-C. 1997. Both the isomerase and chaperone activities of protein disulfide isomerase are required for the reactivation of reduced and denatured acidic phospholipase A2. EMBO J. 16 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun, A., Missiakas, D., Raina, S., and Creighton, T.E. 1995. Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry 34 5075–5089. [DOI] [PubMed] [Google Scholar]