Abstract
We address the problem of clustering the whole protein content of genomes into three different categories—globular, all-α, and all-β membrane proteins—with the aim of fishing new membrane proteins in the pool of nonannotated proteins (twilight zone). The focus is then mainly on outer membrane proteins. This is performed by using an integrated suite of programs (Hunter) specifically developed for predicting the occurrence of signal peptides in proteins of Gram-negative bacteria and the topography of all-α and all-β membrane proteins. Hunter is tested on the well and partially annotated proteins (2160 and 760, respectively) of Escherichia coli K 12 scoring as high as 95.6% in the correct assignment of each chain to the category. Of the remaining 1253 nonannotated sequences, 1099 are predicted globular, 136 are all-α, and 18 are all-β membrane proteins. In Escherichia coli 0157:H7 we filtered 1901 nonannotated proteins. Our analysis classifies 1564 globular chains, 327 inner membrane proteins, and 10 outer membrane proteins. With Hunter, new membrane proteins are added to the list of putative membrane proteins of Gram-negative bacteria. The content of outer membrane proteins per genome (nine are analyzed) ranges from 1.5% to 2.4%, and it is one order of magnitude lower than that of inner membrane proteins. The finding is particularly relevant when it is considered that this is the first large-scale analysis based on validated tools that can predict the content of outer membrane proteins in a genome and can allow cross-comparison of the same protein type between different species.
Keywords: All-β membrane proteins, all-α membrane proteins, structural genomics, neural networks, hidden Markov models, topography prediction of membrane proteins
For an increasing number of organisms, particularly prokaryotes, we now know the genes and the encoded proteins (Benson et al. 2002). In the genomic era methods for large-scale analysis are necessary not only for a correct protein annotation, but also for focusing on specific protein categories or predicting their interaction (Iliopoulos et al. 2001; von Mering et al. 2002).
To understand the genetic blueprint of different organisms, protein sequences are automatically analyzed for function assignment and annotation by means of extensive homology search with PSI-BLAST or hidden Markov models (Eddy 1996; Altschul et al. 1997).
However, there is still a substantial number of uncharacterized proteins, including hypothetical proteins (with homologs of unknown function) or unique proteins (without known homologs) that deserve further characterization (Fischer and Eisenberg 1999; Iliopoulos et al. 2001).
To this aim we integrated a set of independent predictors that have been developed in our laboratory and tested their discriminating capability on the reannotated genome sequence data base of Escherichia coli K12, including 4173 protein coding genes (EcoGene; Rudd 2000). Of these, 52% and 18% are fully and partially annotated, respectively. The remaining 30% is nonannotated (without a Swiss-Prot entry or corresponding to proteins that are not functionally annotated).
In this article we test the efficiency of our programs in correctly discriminating globular from membrane proteins, and all-α from all-β membrane proteins using as a test set the 70% annotated portion of the E. coli genome.
It is presently known that proteins found in the inner membrane of bacteria are interacting with typical bundles of α helices with the lipid bilayer (and are termed all-α membrane proteins; von Heijne 1999). Conversely, in the outer membrane of Gram-negative bacteria, proteins spanning the membrane bilayer with β-strands (named all-β membrane proteins, Schultz 2000) are organized in barrel-like structures.
Prompted by the high performance of our method, we label new globular and membrane proteins on the remaining nonannotated portion of the E. coli genome. This procedure characterizes new sequences of globular, outer, and inner membrane proteins. For the sake of comparison, the same procedure is applied to the genome of the pathogenic strain of E. coli 0157:H7 and highlights new sequences of membrane proteins. New outer membrane proteins without a counterpart in K12, and possibly related to pathogenicity, are predicted. Furthermore a genome-wide analysis of other pathogens and one thermophile is also presented. From this it emerges that in the Gram-negatives taken into consideration, outer membrane proteins are generally a small fraction of the protein content, being at least one order of magnitude lower than that of inner membrane proteins.
Hunter at work
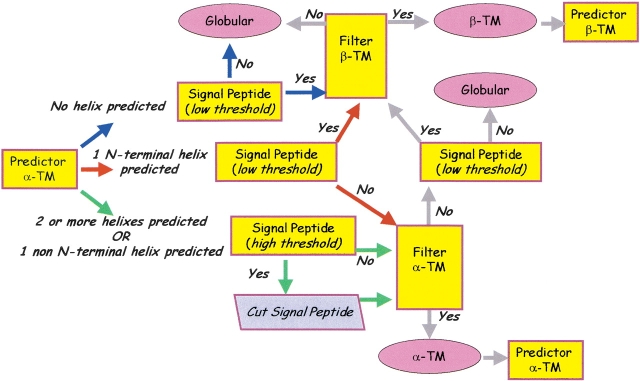
The flow chart of Hunter is shown in Figure 1 ▶. The implementation essentially reflects the general rules that can be derived from a statistical analysis of the protein chains from E. coli K12, which are well annotated in Swiss-Prot (Gasteiger et al. 2001).
Globular proteins can be endowed or not with signal peptides (189 chains out of 1704, 11%)
Similarly, all-α membrane proteins can include or not signal peptides (23 chains out of 422, 5.5%)
All-β outer membrane proteins contain signal peptides (34 chains)
Figure 1.
Hunter: The suite of predictors. The flow chart indicates the possible alternatives after the first prediction done with a neural network-based method. Chain flow limiting steps are: a signal peptide predictor (acting with two different threshold values), trained and tested on signal peptides of Gram-negatives; a hidden Markov model-based filter for outer membrane proteins; a neural network-based filter for all α transmembrane proteins. All the predictors are described in the Materials and Methods section. See text for details.
It is evident that the presence of a signal peptide in the sequences is a characteristic tag of outer membrane proteins (Nielsen et al. 1997). However, signal peptide predictors are affected by rates of false positives even when top scoring (Nielsen et al. 1999). Furthermore, not all the proteins with a signal peptide are outer membrane proteins.
On the other hand, it has been argued that enzyme function is less conserved than anticipated, and that functional annotation may be biased when performed only on a sequence homology basis (Rost 2002). An alternative approach for classification is based on structure prediction (Frishman and Mewes 1999; Jones 2000; Kelley et al. 2000; Thornton et al. 2000; Frishman et al. 2001; Turcotte et al. 2001). In this article, we choose to address the problem on a structural basis, relying on the classification obtained with methods specifically suited for predicting membrane protein topography.
We implemented a signal peptide predictor that compares well with the top-scoring SignalIP (Nielsen et al. 1999). Furthermore, we developed two well-performing predictors of the topography of inner all-α and outer all-β membrane proteins, endowed with filters that minimize the rate of false positives (proteins falsely predicted in the category). The predictor for all-β membrane proteins is similar to that already described (Jacoboni et al. 2001; Martelli et al. 2002). That for all-α inner membrane proteins is based on neural networks as other predictors of this type (Rost et al. 1995). However, it is the first predictor trained and tested only on the inner membrane proteins known with atomic resolution. The predictors and their performance are described in the Materials and Methods section.
One possibility to address the task at hand is to combine all three predictors in an efficient manner. Now the set of empirical rules that are to be taken into consideration for solving our discriminative problem are:
We need to maximize the number of protein chains endowed with signal peptides to let the outer membrane protein filter receive the maximum number of chains (only the proteins retained by this filter will be classified as an outer membrane)
We have to take into consideration that all predictors are affected by a rate of false positives and negatives, and that this is particularly so for the signal peptide predictor (so we need at least two different threshold values to filter the genome)
At the same time, we have to cope with the fact that the predictor of all-α membrane proteins wrongly predicts signal peptides as transmembrane segments in the N-terminal portion of the chain, and that it can be affected as well by false positives.
The scheme we propose to integrate our predictors (Fig. 1 ▶) is particularly suited to mitigate the number of false positives and negatives and to send the maximal number of chains endowed with a predicted signal peptide towards the outer membrane protein classifier. The discriminative power of the suite of programs resides mainly on two filters: one based on a hidden Markov model specifically developed for the outer membrane proteins of Gram-negative bacteria (Martelli et al. 2002); the other is a neural network-based filter minimizing the rate of false positives of a neural network predicting the topography of all-α membrane proteins (this work). All predictors use as input the sequence profile derived from multiple alignment of the target chain towards the nonredundant database.
The protein content of the genome is first filtered with the neural network-based predictor trained and tested on 36 membrane proteins known with atomic resolution. Depending on the number of helices predicted (zero, one, and two or more) the chain is then filtered with the signal peptide predictor. This is done both with low (more false positives) and stringent threshold (less false positives).
A chain with no transmembrane helices predicted (blue path in Fig. 1 ▶) is then filtered with the low-threshold signal peptide predictor. If the protein is without a signal peptide it is classified as globular. Otherwise, if the protein is retained by the HMM filter, the chain is classified as transmembrane all-β and eventually predicted with the neural network for computing the topography (Jacoboni et al. 2001).
When the protein is predicted to have a transmembrane helix in the N-terminal (red path in Fig. 1 ▶) it is also filtered with the low-threshold signal peptide predictor with two alternatives: if the signal peptide is present, then the protein is sent to the beta-strand filter and the end steps are those described above; if not the protein is presented to the filter for all-α membrane proteins, and if retained, it is accordingly classified; if not, it is classified as globular.
When one helix is predicted in the chain (but not in the N-terminal region) or two or more helices are predicted (green path in Fig. 1 ▶), the stringent signal peptide predictor is activated; then the protein can be classified as either globular or membrane all-α, depending on the output of the all-α transmembrane filter. When the protein is classified as all-α, the signal peptide is excised and the topography prediction is performed without the segment.
Testing the performance of Hunter
Although each of the predictors has been statistically validated during the implementation (see Materials and Methods), a more general validation of the integrated tool is necessary before proceeding into the analysis aiming at fishing new proteins in the twilight zone of the genome.
The test is performed using the subsets of proteins from E. coli K12 that are well annotated (2160) or partially annotated (760) in Swiss Prot. Test experiments were performed at three different values of sequence identity between the proteins of the testing and training tests of the predictors: ≤20%; ≤25%, ≤30%. With the exception of the number of sequences included in the training and testing sets, the performance of Hunter was rather similar at each level of sequence identity. We show the results obtained with the largest number of sequences, corresponding to sequence identity ≤30%.
The statistical validation of the predictor is shown in Tables 1 and 2, respectively. In both cases the score per protein (Q3p) is higher than 90% (95.6% and 92.6% in the case of well and partially annotated sets, respectively). Also, both the rate of proteins correctly predicted in the class (Qclass) and the probability of correct prediction in the class (Pclass) are good.
Table 1.
Predicting well and partially annotated proteins of Escherichia coli K12a with Hunter
| Prediction | ||||
| α-TM | β-TM | Globular | Total | |
| Well annotated proteins | ||||
| Annotation | ||||
| α -TM | 389 | 0 | 33 | 422 |
| β -TM | 0 | 28 | 6 | 34 |
| Globular | 50 | 3 | 1651 | 1704 |
| Total | 439 | 31 | 1690 | 2160 |
| Partially annotated proteins | ||||
| Annotation | ||||
| α -TM | 317 | 0 | 35 | 352 |
| β -TM | 0 | 14 | 4 | 18 |
| Globular | 15 | 2 | 373 | 390 |
| Total | 332 | 16 | 412 | 760 |
a Annotation of Escherichia coli K12 is according to EcoGene (Rudd 2000).
Table 2.
Scoring the performance of Hunter
| Set (E. coli K12) | Q3p (%) | Qα-TM (%) | Qβ-TM (%) | QGlobular (%) | Pα-TM (%) | Pβ-TM (%) | PGlobular (%) |
| Well-annotated proteins | 95.6 | 92.2 | 82.4 | 96.9 | 88.6 | 90.3 | 97.7 |
| Partially annotated proteins | 92.6 | 90.1 | 77.8 | 95.6 | 95.5 | 87.5 | 90.5 |
Q3p = accuracy per protein; Qclass = accuracy per protein class (transmembrane all-α, transmembrane all-β, globular); Pclass = probability of correct prediction per protein class. Statistical indexes are as defined before (Jacoboni et al. 2001). The correlation coefficients are: 0.88, 0.86, and 0.87 for transmembrane all-α, transmembrane all-β, and globular proteins, respectively.
The rate of false negatives (1− Qclass) and of false positives (1 − Pclass) for the all-α transmembrane proteins ranges from 8.8% to 9.9% and from 11.4% to 4.5% when the estimate is evaluated on the two sets, respectively; similarly, it is 17.6%–22.2% and 9.7%–12.5% for all-β outer membrane, and 3.1%–4.4%, 2.3%–9.5% for globular proteins. Evidently the rate of false negatives and positives is affected by the smaller number of membrane proteins, particularly all-β, compared to that of globular proteins.
From these figures we may roughly estimate the rate of false negatives and positives to be associated to putative numbers of proteins predicted in each of the three categories as a value averaged over the two sets. We may conclude that the outer membrane-classification may be affected by underprediction (about 20%) more than overprediction (about 10%). Under- and overprediction for the other two categories are comparable: by averaging, about 9% for inner membrane proteins and 5% for globular ones.
In conclusion, from the test it is evident that Hunter quite accurately classifies the proteins of the E. coli genome, although it misses and overpredicts some chains. This was expected, considering also the statistical validation of each predictor (see Materials and Methods).
Hunter is then used to filter the remaining portion of the genome of E. coli K12. The results are shown in Table 3. Out of 1253 proteins, 136 are classified transmembrane all-α; 18 transmembrane all-β, and 1099 globular. We also detail for the outer membrane proteins the name of the file, the Swiss-Prot ID if existing, the length of the chain, the number of predicted transmembrane beta-strands, the number and the annotation of the homologs (E-value <10−7). For the sake of clarity we include the annotation of the first homolog as detected by BLAST and the level of local and global identity (%) of the target to the homolog.
Table 3.
Fishing new globular, inner, and outer membrane proteins in the E. coli KI2 genome with Hunter
| New globular proteins | 1099 |
| New inner membrane proteins | 136 |
| New outer membrane proteins | 18 |
| EcoGene code | Swiss-Prot code | Length | No. of predicted TM strands | No. of homologs in Swiss-Prot | Annotation of homologs (first homolog, % identity of local and global alignments) |
| a Outer membrane proteins in K-islands (Perna et al. 2001). | |||||
| EG13412 | CSGF_ECOLI | 138 | 2 | 1 | Biogenesis of curli organelles (CSGF_SALTY: 90%; 90%) |
| EG12668 | UIDC_ECOLI | 416 | 18 | 0 | |
| EG11307 | YDBA_ECOLI | 2003 | 38 | 2 | Hypothetical protein (YHFJ_ECOLI: 38%; 38%) |
| EG12269 | YHJY_ECOLI | 232 | 10 | 1 | Lipase 1 (LIP1_PHOLU: 27%; 11%) |
| EG12513 | YTFM_ECOLI | 577 | 12 | 1 | Hypothetical protein (YTFM_HAEIN: 43%; 42%) |
| EG12562 | YJHT_ECOLI | 368 | 14 | 5 | Hypothetical protein (YJHT_HAEIN: 43%; 42%) |
| EG12850 | YFAL_ECOLI | 1250 | 16 | 2 | Pertactin precursor (PERT_BORBR: 25%; 15%) |
| EG13297a | YAIO_ECOLI | 257 | 14 | 0 | |
| EG13480 | YLII_ECOLI | 371 | 2 | 3 | Glucose dehydrogenase (DHGB_ACICA: 32%; 22%) |
| EG13563 | YAGX_ECOLI | 841 | 14 | 1 | Fimbrial subunit C (CFAC_ECOLI: 25%; 20%) |
| EG13605 | YAIT_ECOLI | 486 | 28 | 5 | Pertactin precursor (PERT_BORBR: 33%; 14%) |
| EG13783 | — | 96 | 4 | 24 | Outer membrane porin C (OMPC_KLEPN: 60%; 15%) |
| EG13889 | — | 882 | 12 | 2 | Adhesin aidA-I precursor (AIDA_ECOLI: 29%; 23%) |
| EG13984 | YDIY_ECOLI | 252 | 12 | 0 | |
| EG14088 | YFAZ_ECOLI | 180 | 8 | 0 | |
| EG11743 | YDDB_ECOLI | 790 | 24 | 1 | Hypothetical protein (YDDB_HAEIN: 24%; 22%) |
| EG13160a | YFEN_ECOLI | 254 | 12 | 0 | |
| EG13565 | YAGZ_ECOLI | 195 | 2 | 0 | |
Out of the 18 outer membrane proteins, six chains have no homologs in Swiss-Prot, four are annotated as hypothetical proteins, and the remaining have homologs that interestingly include an outer membrane protein C, a fimbrial subunit C, a precursor of Pertactin (a virulence factor in Bordetella pertussis; Emsley et al. 1996), and an Adhesin AIDA-I precursor.
Filtering of E. coli 0157:H7
E. coli 0157:H7 is a major food-borne infectious pathogen that causes diarrhea, hemorrhagic colitis, and hemolytic uremic syndrome. Most of its genome (70% of sequence similarity) is similar to that of E. coli K12 (Hayashi et al. 2001). However, some genes are unique and clustered in the so-called "O-islands" that possibly contains major causes of pathogenicity (Perna et al. 2001). Similarly, the K12 strain contains unique genes (K islands) that do not have homologs in 0157:H7. Furthermore, the annotation of the proteome of E. coli 0157:H7 (available at NCBI) is presently not as refined as that of E. coli K12; about 35% of the chains are still annotated as hypothetical proteins in the NCBI release. Out of this set Hunter predicts 327 new all-α, 10 new all-β membrane proteins, and 1564 new globular proteins. Table 4 lists the NCBI code and the Swiss-Prot ID of the homolog (if existing) in K12. The length, the number of predicted transmembrane strands, and the Swiss-Prot annotation of the first homolog are also shown. In 0157, the proteins classified as outer membrane all-β include three chains without annotation, three chains homologous to hypothetical proteins, and four chains homologous to a surface antigen, a probable lipoprotein, an outer membrane porin, and a chain involved in TonB-dependent transport.
Table 4.
Fishing new globular, inner, and outer membrane proteins in the E. coli 0157 genome with Hunter
| New globular proteins | 1564 |
| New inner membrane proteins | 327 |
| New outer membrane proteins | 10 |
| NCBI code | Homologa in E. coli K12 | Length | No. of predicted TM strands | No. of other homologous in Swiss-Prot | Annotation of homologs (first homolog, % identity of local and global alignments) |
| a Homolog = with an E-value ≤10−7. | |||||
| 13359635 | UP05_ECOLI | 810 | 18 | 5 | Surface antigen (D152_HAEIN: 45%; 45%) |
| 13359780 | YAGZ_ECOLI | 195 | 2 | 0 | |
| 13360600 | YMCA_ECOLI | 698 | 20 | 1 | Probable lipoprotein (YJBH_ECOLI: 65%; 64%) |
| 13361464 | OMPN_ECOLI | 123 | 4 | 24 | Outer membrane porin (OMS2_SALTI: 85%; 26%) |
| 13361566 | YDDB_ECOLI | 790 | 24 | 1 | Hypothetical protein (YDDB_HAEIN: 26%; 23%) |
| 13361895 | YDIY_ECOLI | 252 | 12 | 0 | |
| 13362260 | CIRA_ECOLI | 715 | 14 | 22 | Colicin receptor; TonB dependent transport (Y262_HAEIN: 24%; 23%) |
| 13362608 | YFAZ_ECOLI | 187 | 8 | 0 | |
| 13364489 | YJBH_ECOLI | 698 | 22 | 1 | Hypothetical protein (YMCA_ECOLI: 65%; 64%) |
| 13364675 | YTFM_ECOLI | 577 | 12 | 1 | Hypothetical protein (YTFM_HAEIN: 44%; 42%) |
What did we learn?
From this analysis we may conclude that the all-α membrane protein content (including the new proteins that we add with our procedure) of both genomes is about 25% in E. coli 0157:H7 and about 22% in E. coli K12. These figures compare well with previous estimates performed with all-α membrane protein predictors based on HMM and neural networks (Krogh et al. 2001; Liu and Rost 2001). However, what is novel is that Hunter classifies and lists together with globular and inner membrane proteins, the putative contents of all-β outer transmembrane proteins, and this may be particularly interesting in pathogenic bacteria. We found that the proteome of E. coli K12 contains about 1.7% of outer membrane proteins; the estimate is similar in E. coli 0157:H7 (see also Table 5). These values are somewhat lower than a previous estimate in E. coli 0157 done with a scale-based method, however without an estimate of false positives (Wimley 2002).
Table 5.
Predicting globular, inner, and outer membrane proteins in genomes of Gram-negative bacteria with Hunter
| Organism | Outer membrane | Inner membrane | Globular | Total |
| Escherichia coli K12 | 65 (1.6%) | 907 (21.7%) | 3201 (76.7%) | 4173 |
| Newa | 18 | 136 | 1099 | 1253 |
| Escherichia coli O157:H7 | 78 (1.5%) | 1034 (19.3%) | 4249 (79.2%) | 5361 |
| New | 10 | 327 | 1564 | 1901 |
| Chlamidia pneumoniae CWL029 | 12 (1.1%) | 290 (27.6%) | 750 (71.3%) | 1052 |
| New | 2 | 181 | 236 | 419 |
| Salmonella typhimurium LT2 | 70 (1.6%) | 1002 (22.5%) | 3379 (75.9%) | 4451 |
| New | 0 | 2 | 21 | 23 |
| Neisseria meningitidis MC58 | 34 (1.7%) | 372 (18.4%) | 1619 (80.0%) | 2025 |
| New | 6 | 176 | 662 | 844 |
| Helicobacter pylori 26695 | 36 (2.3%) | 352 (22.5%) | 1178 (75.2%) | 1566 |
| New | 10 | 141 | 445 | 596 |
| Haemophylus influentiae Rd | 23 (1.3%) | 348 (20.4%) | 1338 (78.3%) | 1709 |
| New | 5 | 121 | 430 | 556 |
| Thermotoga maritima | 18 (1.0%) | 370 (20.0%) | 1458 (79.0%) | 1846 |
| New | 11 | 203 | 559 | 773 |
| Pseudomonas aeruginosa | 131 (2.4%) | 1292 (23.2%) | 4142 (74.4%) | 5565 |
| New | 62 | 616 | 1867 | 2545 |
a The number of new proteins predicted in the class with Hunter out of the nonannotated region.
What did Hunter classify?
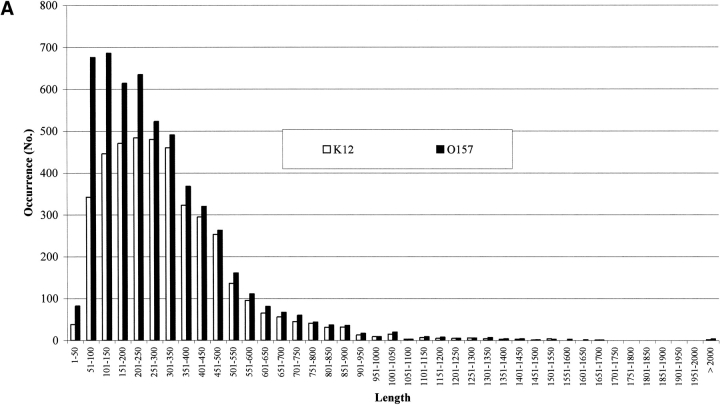
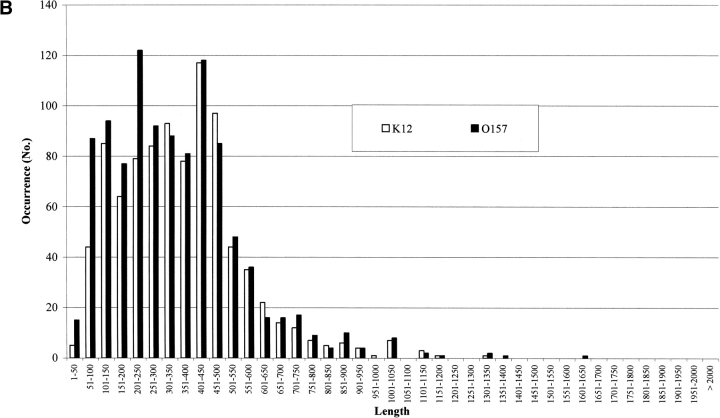
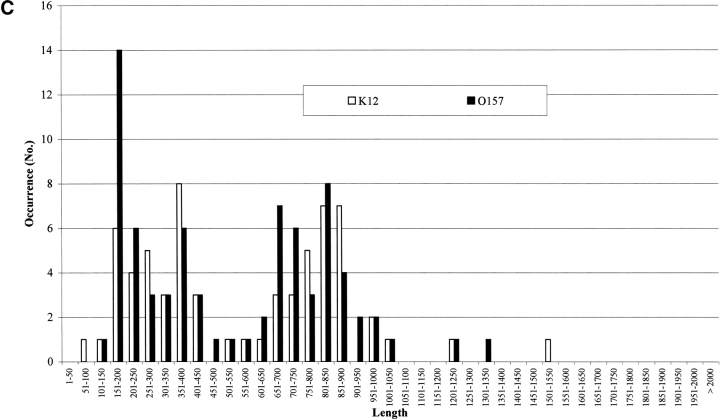
It has been estimated that about 10% of the genes in genomes are due to overannotation of too short sequences (Skovgaard et al. 2001). In Figure 2A ▶ the number of proteins in both strains is shown as a function of their length. It is evident that 9% and 14% of the genes are shorter than or include at least 100 residues, in E. coli K12 and O157:H7, respectively. However, the transmembrane protein predictors do not retain such short sequences. In Figure 2 ▶, B and C, the membrane protein chains classified by Hunter are listed as a function of their length (in residue numbers). In both K12 and 0157:H7, proteins classified membrane all-α (Fig. 2B ▶) and all-β (Fig. 2C ▶) are differently distributed according to different lengths. The membrane all-α in K12 have lengths from 1–50 to 1301–1350; in 0157, from 1–50 to 1351–1660. The percentage of short sequences (≤100 residues) is 5.4 and 9.9 in K12 and O157, respectively, and the average length of all-α transmembrane proteins is quite similar (364 residues in E. coli K12 and 342 in O157, respectively). The same observation holds also for all-β membrane proteins (Fig. 2C ▶, average length in K12 and O157, 592 and 547 residues, respectively). The percentage of short segments predicted ranges from 1.5 to 0, in K12 and O157, respectively. In conclusion, a negligible number of short sequences is classified transmembrane by the suite of predictors.
Figure 2.
(Continued on next page)
The length distribution of protein chains in E. coli K12 and O157. (A) Bar plot of the length distribution of proteins in the genomes. (B) Bar plot of the length distribution of inner membrane proteins after prediction with Hunter in both strains. (C) Bar plot of the length distribution of outer membrane proteins after prediction with Hunter in both strains.
The topography of predicted membrane proteins.
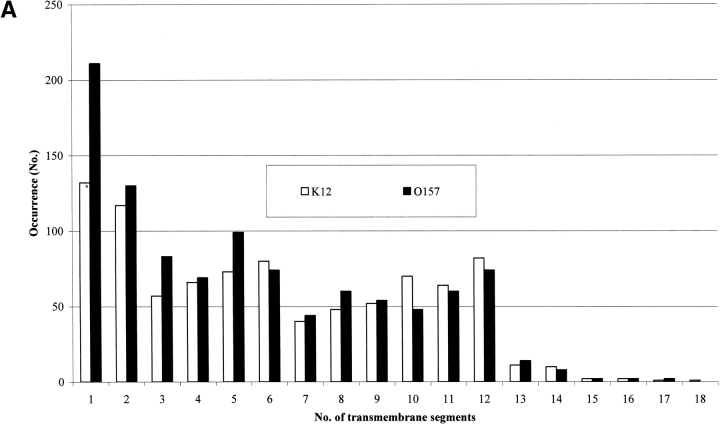
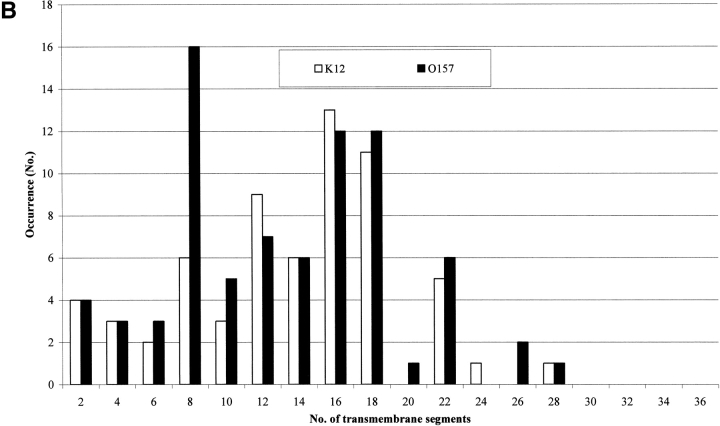
Our analysis also allows predicting the number of transmembrane segments for each protein type. The number of all-α transmembrane proteins with a given number of transmembrane segments is shown for both strains in Figure 3A ▶. Apparently, the number of monotopic all-α membrane proteins nearly doubles in 0157. For all-β membrane proteins it is worth noticing (Fig. 3B ▶) that the number of proteins with eight β-strands in the barrel is nearly threefold larger than in K12. Interestingly, the larger number of eight β-stranded barrel membrane proteins predicted in 0157 is consistent with the notion that this type of outer membrane protein, related to possible mechanisms of virulence (Vogt and Schulz 1999), may be more abundant in the pathogen compared to K12. Indeed, 10 chains belonging to this structural subset in 0157 are homologous to protein X of E. coli (OMPX_ECOLI), and belong to a family of highly conserved proteins that promote bacterial adhesion to and entry into mammalian cells (Vogt and Schulz 1999, and references therein); one is homologous to PERT-BORPE, the virulence factor P69 perctatin of Bordetella pertussis (Emsley et al. 1996), and one to OMPA_ECOLI, required for the action of colicins K and L and for the stabilization of mating aggregates in conjugation (Pautsch and Schulz 2000). The remaining are proteins found in both strains and annotated as hypothetical outer membrane proteins.
Figure 3.
Topography of transmembrane proteins in E. coli K12 and O157 as predicted with Hunter. (A) Bar plot of inner membrane proteins as a function of the number of transmembrane predicted segments in both strains. (B) Bar plot of outer membrane proteins as a function of transmembrane predicted β strands in the barrel in both strains.
On the other hand, K12 is endowed with more outer membrane proteins containing 12 β-strands (the typical architecture of phospholipase A, OMPLA, participating in secretion of colicins in E. coli; implied in virulence in pathogens; Snijder and Dijkstra 2000).
Fishing new proteins in other genomes
We also filtered other genomes of Gram-negative bacteria: one thermophile, and the other pathogenic bacteria (Table 5). We highlight what is newly labeled by Hunter in the three classes. It is evident that the number of outer membrane proteins is at least one order of magnitude lower than that of the inner membrane proteins, ranging from 1% to 2.4% in Thermotoga maritima (a thermophile bacterium) and Pseudomonas aeruginosa (another pathogen), respectively. Also, the fraction of inner membrane proteins ranges from about 18% to 28%. From these data, it may also be concluded that neither the fraction of inner nor that of outer membrane proteins seems related to the pathogenicity of the bacterium.
Conclusions
We describe the performance of Hunter, a suite of programs specifically developed for genome-wide analysis of Gram-negative bacteria, containing a predictor of signal peptide specific for this type of bacteria. The predictor is discriminative towards three protein categories of the genome: inner all-α, outer all-β membrane, and globular proteins.
We test its availability using 50% of the genome of E. coli K12 as annotated in EcoGene, and estimate the rate both of false negatives (proteins that may be missed in the class) and false positives (proteins that may be wrongly predicted in the class).
Filtering of E. coli K12 and 0157 adds new chains to inner and outer membrane proteins and globular ones. We propose Hunter for specifically fishing new inner and outer membrane proteins in genomes of Gram-negatives, and possibly highlighting new virulence factors.
Materials and methods
Databases and alignment methods
Genome annotation of E. coli K12 and 0157:H7 was taken from EcoGene (Rudd 2000) and NCBI (http://www.ncbi.nlm.nih.gov). If not specified, annotations are as in NCBI for all genomes of Gram-negative bacteria. Proteins, solved at an atomic resolution and included in training/testing sets of the predictors of membrane proteins, were taken from PDB (http://www.rcsb.org/pdb/index.html). Proteins used for implementing the signal peptide predictor were described before (Kersey et al. 2000; Menne at al. 2000). The lists of the training sets as well as the lists of the proteins predicted in a given category are available at our Web site (www.biocomp.unibo.it/hunter).
The sequence profile was derived after alignment towards the nonredundant database (July 2001) with PSI-BLAST (Altschul et al. 1997). If necessary, local and global alignments were performed with LALIGN (www.ch.embnet.org/software/LALIGN_form.html) used with default parameters.
The signal peptide predictor
We trained and tested a neural network-based predictor on 598 chains, with 301 positive examples. The network architecture includes an asymmetric input window comprising 14 residues, three neurons in the hidden layer, and one output layer. Our predictor scores similarly to SignalIP (Nielsen et al. 1999). The accuracy is 96.5% and the correlation coefficient is 0.93. When E. coli K12 was filtered with a stringent filter the accuracy per protein was 95.3%. SignalIp under the same conditions and over the same set had accuracy per protein of 95.5%. When using the predictor, a stringent threshold means that only the output values larger or equal to 0.99 are accepted. The low threshold is similarly set at 0.84.
The predictor of all-α inner membrane proteins
We trained and tested on a nonredundant set of 36 membrane proteins known with atomic resolution a neural network-based predictor. The predictor architecture includes a 17-residue long input window that uses sequence profile, 15 neurons in the hidden layer, and two output neurons. Per-residue accuracy is 86.3% and the correlation coefficient is 0.72, with an overlapping score (SOV; Zemla et al. 1999) of 86.8%. To increase the discriminative power of the network, we implemented a filter, which takes into consideration the maximal probability values characteristics of the test set. A protein is accepted only if it is predicted with at least one transmembrane region including probability values as high as 0.96. When a nonredundant set of some 800 globular proteins are predicted, the rate of false positives decreases from 26% (the majority with one α helix) to 0.5%.
The predictor of all-β membrane proteins
The neural network predicting the all-β membrane proteins has been described before (Jacoboni et al. 2001). However, in this case the rate of false positives was also decreased by using a hidden Markov model similar to that previously described (Martelli et al. 2002). It has been discussed that the transmembrane strand pattern is not as characteristic as that of alpha-transmembrane helices. When some 800 nonredundant globular proteins are presented to the neural network 30% are wrongly predicted with at least one and two transmembrane β strands. When the HMM is added on top of the network, the rate of false positive decreases down to 5%.
Hunter
Hunter is the suite of programs described above. The core tools are written in C; the parsers and the global framework are written in PERL. The suite is implemented on a Beowulf, comprising eight CPUs. The running time for a genome of medium size (about 5000 genes) is about 5 h. Most of the time is used for computing the sequence profile after sequence alignment towards the nonredundant database done with PSI-BLAST (Altschul et al. 1997).
Statistical indexes to measure the predictor efficiency have been described before (Casadio et al. 1996; Jacoboni et al. 2001; Martelli et al. 2002).
Acknowledgments
This work was partially supported by a grant of the Ministero della Università e della Ricerca Scientifica e Tecnologica (MURST) for the project "Hydrolases from Thermophiles: Structure, Function and Homologous and Heterologous Expression," a grant for a target project in Biotechnology and a project on Molecular Genetics, both of the Italian Centro Nazionale delle Ricerche (CNR), delivered to R.C. R.C. also acknowledges the EC grant Biowulf IST 1999-20232 for supporting the development of DNCBLAST, a parallelized version of PSI-BLAST for PC nets. PLM is the recipient of a fellowship from the Italian Center for National Researches (CNR) devoted to a target project of Molecular Genetics (Law No. 449–1997). We thank S. Brunak and his group for making available Signal IP.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0223603.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., Rapp, B.A., and Wheeler, D.L. 2002. GenBank. Nucleic Acids Res. 30 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadio, R., Fariselli, P., Taroni, C., and Compiani, M. 1996. A predictor of transmembrane α-helix domains of proteins based on neural networks. Eur. Biophys. J. 24 165–178. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R. 1996. Hidden Markov models. Curr. Opin. Struct. Biol. 6 361–365. [DOI] [PubMed] [Google Scholar]
- Emsley, P., Charles, I.G., Fairweather, N.F., and Isaacs, N.W. 1996. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature 381 90–92. [DOI] [PubMed] [Google Scholar]
- Fischer, D. and Eisenberg, D. 1999. Finding families for genomic ORFans. Bioinformatics 15 759–762. [DOI] [PubMed] [Google Scholar]
- Frishman, D. and Mewes, H.W. 1999. Genome-based structural biology. Prog. Biophys. Mol. Biol. 72 1–17. [DOI] [PubMed] [Google Scholar]
- Frishman, D., Albermann, K., Hani, J., Heumann, K., Metanomski, A., Zollner, A., and Mewes, H.W. 2001. Functional and structural genomics using PEDANT. Bioinformatics 17 44–57. [DOI] [PubMed] [Google Scholar]
- Gasteiger, E., Jung, E., and Bairoch, A. 2001. SWISS-PROT: Connecting biomolecular knowledge via a protein database. Curr. Issues Mol. Biol. 3 47–55. [PubMed] [Google Scholar]
- Hayashi, T., Makino, K., Ohnishi, M., Kurokawa, K., Ishii, K., Yokoyama, K., Han, C.G., Ohtsubo, E., Nakayama, K., Murata, T., et al. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8 11–22. [DOI] [PubMed] [Google Scholar]
- Iliopoulos, I., Tsoka, S., Andrade, M.A., Janssen, P., Audit, B., Tramontano, A., Valencia, A., Leroy, C., Sander, C., and Ouzounis, C.A. 2001. Genome sequences and great expectations. Genome Biol. 2 INTERACTIONS0001. [DOI] [PMC free article] [PubMed]
- Jacoboni, I., Martelli, P.L., Fariselli, P., De Pinto, V., and Casadio, R. 2001. Prediction of the transmembrane regions of β-barrel membrane proteins with a neural network-based predictor. Protein Sci. 10 779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.T. 2000. Protein structure prediction in the postgenomic era. Curr. Opin. Struct. Biol. 10 371–379. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A., MacCallum, R.M., and Sternberg, M.J. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299 499–520. [DOI] [PubMed] [Google Scholar]
- Kersey, P., Hermjakob, H., and Apweiler, R. 2000. VARSPLIC: Alternatively-spliced protein sequences derived from SWISS-PROT and TrEMBL. Bioinformatics 16 1048–1049. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305 567–580. [DOI] [PubMed] [Google Scholar]
- Liu, J. and Rost, B. 2001. Comparing function and structure between entire proteomes. Protein Sci. 10 1970–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli, P.L., Fariselli, P., Krogh, A., and Casadio, R. 2002. A sequence profile based HMM for predicting and discriminating β barrel membrane proteins. Bioinformatics 18:S1 46–53. [DOI] [PubMed] [Google Scholar]
- Menne, K.M., Hermjakob, H., and Apweiler, R. 2000. A comparison of signal sequence prediction methods using a test set of signal peptides. Bioinformatics 16 741–742. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10 1–6. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Brunak, S., and von Heijne, G. 1999. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng. 12 3–9. [DOI] [PubMed] [Google Scholar]
- Pautsch, A. and Schulz, G.E. 2000. High-resolution structure of the OmpA membrane domain. J. Mol. Biol. 298 273–282. [DOI] [PubMed] [Google Scholar]
- Perna, N.T., Plunkett III, G., Burland, V., Mau, B., Glasner, J.D., Rose, D.J., Mayhew, G.F., Evans, P.S., Gregor, J., Kirkpatrick, H.A., et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409 529–533. [DOI] [PubMed] [Google Scholar]
- Rost, B. 2002. Enzyme function less conserved than anticipated. J. Mol. Biol. 318 595–608. [DOI] [PubMed] [Google Scholar]
- Rost, B., Casadio, R., Fariselli, P., and Sander, C. 1995. Transmembrane helices predicted at 95% accuracy. Protein Sci. 4 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd, K.E. 2000. EcoGene: A genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 28 60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, G.E. 2000. β-barrel membrane proteins. Curr. Opin. Struct. Biol. 10 443–447. [DOI] [PubMed] [Google Scholar]
- Skovgaard, M., Jensen, L.J., Brunak, S., Ussery, D., and Krogh, A. 2001. On the total number of genes and their length distribution in complete microbial genomes. Trends Genet. 17 425–428. [DOI] [PubMed] [Google Scholar]
- Snijder, H.J. and Dijkstra, B.W. 2000. Bacterial phospholipase A: Structure and function of an integral membrane phospholipase. Biochim. Biophys. Acta 1488 91–101. [DOI] [PubMed] [Google Scholar]
- Thornton, J.M., Todd, A.E., Milburn, D., Borkakoti, N., and Orengo, C.A. 2000. From structure to function: Approaches and limitations. Nat. Struct. Biol. Suppl: 991–994. [DOI] [PubMed]
- Turcotte, M., Muggleton, S.H., and Sternberg, M.J. 2001. Automated discovery of structural signatures of protein fold and function. J. Mol. Biol. 306 591–605. [DOI] [PubMed] [Google Scholar]
- Vogt, J. and Schulz, G.E. 1999. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Struct. Fold. Des. 7 1301–1309. [DOI] [PubMed] [Google Scholar]
- von Heijne, G. 1999. Recent advances in the understanding of membrane protein assembly and structure. Q. Rev. Biophys. 32 285–307. [DOI] [PubMed] [Google Scholar]
- von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S.G., Fields, S., and Bork, P. 2002. Comparative assessment of large-scale data sets of protein–protein interactions. Nature 417 399–403. [DOI] [PubMed] [Google Scholar]
- Wimley, W.C. 2002. Toward genomic identification of β-barrel membrane proteins: Composition and architecture of known structures. Protein Sci. 11 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemla, A., Venclovas, C., Fidelis, K., and Rost, B. 1999. A modified definition of Sov, a segment-based measure for protein secondary structure prediction assessment. Proteins 34 220–223. [DOI] [PubMed] [Google Scholar]