Abstract
Transglutaminases are calcium-dependent enzymes that catalyze a post-translational modification of proteins through the formation of ɛ-(γ-glutamyl)lysine bonds. Although specific roles for transglutaminases have been described, recent findings have provided evidence that dysregulation of transglutaminases may contribute to many pathological processes including celiac disease and neurodegenerative diseases. A crucial step in the elucidation of biological and pathological roles of transglutaminases requires the identification of protein substrates. A strategy based on a functional proteomic analysis was set up using two well-characterized biotinylated transglutaminase substrates as affinity probes: 5–(biotinamido)pentylamine and the synthetic biotinylated peptide TVQQEL, the amino- and acyl-donor probes, respectively. A pool of known tissue type transglutaminase protein substrates was selected in order to test the procedure. Results obtained in this paper indicate that the whole strategy can be successfully applied in order to identify transglutaminases protein substrates as well as the amino acid site sensitive toward enzyme activity.
Keywords: Tandem mass spectrometry, protein identification, functional proteomics, affinity probe, transglutaminase
Transglutaminases (TGs) constitute a family of Ca2+-dependent enzymes catalyzing the cross-link formation between protein-bound glutamine residues (Q-donor) and the ɛ-amino group of protein-bound lysine residues (K-donor). Amines, diamines, and polyamines present in cells can act as amine donors (Folk and Finlayson 1977; Lorand and Conrad 1984). So far, several intracellular and extracellular TGs, showing different structural, kinetic, and immunological properties, were identified (Aeschlimann and Paulsson 1994). Most of them have well-defined functions as blood clotting (plasma factor XIIIa), keratin formation (keratinocyte TG), epidermal function (epidermal TG), and seminal vesicle coagulation (prostate TG; Aeschlimann and Paulsson 1994). The function of tissue transglutaminase (tTG), a bifunctional enzyme possessing TG and GTPase activity, remains an open question despite the fact that it has been studied extensively. tTG seems involved in many biological processes, such as proliferation and differentiation, apoptosis, formation of extracellular matrix, tissue repair, and signal transduction through α1-adrenergic receptor (Aeschlimann and Thomazy 2000; Akimov and Belkin 2001). Recently, tTG was found to be involved in many pathological processes, such as celiac disease, autoimmune diseases, inflammation, fibrosis, cancer metastases, CAG-repeat neurodegenerative disorders, for example, Huntington’s disease (Cooper et al. 2002; Kim et al. 2002). A crucial step in the elucidation of tTG effective biological functions requires the identification of protein substrates.
Most approaches to protein identification combine protein gel electrophoresis separation with mass spectrometry (MS)-based analyses. The development of tandem mass spectrometry (MS/MS) procedures based on data-dependent analyses led to the introduction of new proteomic strategies widely applicable to the identification of proteins from mixtures without performing a preliminary gel electrophoresis separation (Gygi et al. 1999Zhou et al. 2002).
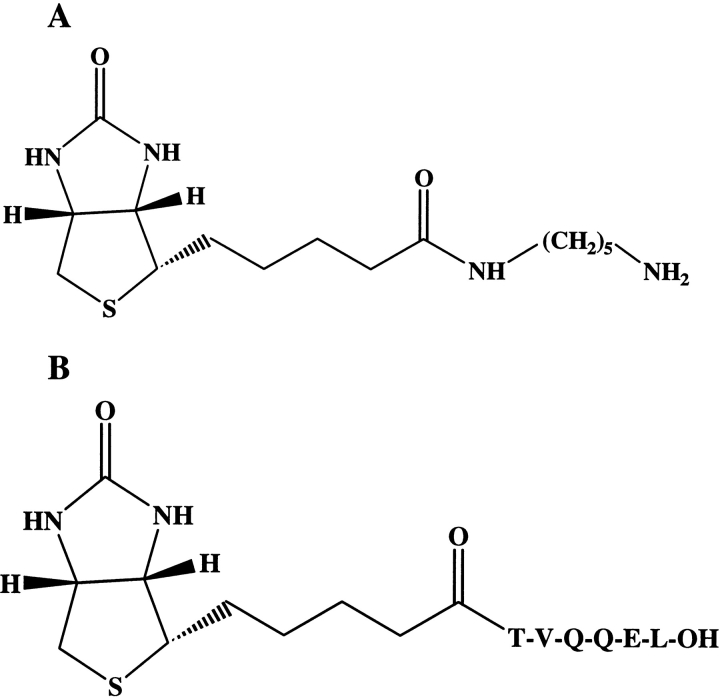
To define a functional protein map of TG substrates, we set up a procedure based on the use of two different affinity probes, shown in Figure 1 ▶. 5–(biotinamido)pentylamine (BPNH2) is a commercially available reagent, which is able to label Q-donor proteins in the TG-catalyzed reaction. The peptide biotinyl-TVQQEL (A25 peptide) was synthetized with the aim of labeling K-donor proteins in the TG-catalyzed reaction, taking into consideration the fact that this peptide had already been reported as an efficient tTG substrate (Groenen et al. 1994). The two probes share few structural features: (1) Both bear a biotin group as a tag, meaning that the TG substrate species can be selectively purified by means of avidin affinity chromatography; (2) neither are susceptible to cleavage by the most common proteolytic enzymes, thus leaving an intact biotin-labeled probe on the modified TG protein substrates when submitted to proteolysis.
Figure 1.
Biotinylated reagent structures. (A) 5–(biotinamido)pentylamine; (B) A25 peptide.
A first application of the strategy on a well-characterized set of tTG protein substrates is reported here. The pool of proteins consisted of: vasoactive intestinal peptide (VIP), a 28-amino acid long polypeptide homologous with secretin, glucagons, and other hormones (Carlquist et al. 1982; Campbell and Scanes 1992; Rawlings 1994); rat seminal vesicle protein IV (SV-IV), a 90-amino acid protein extracted from rat seminal vesicle epithelium (Ostrowski et al. 1979; Esposito et al. 2002b); bovine β casein, a very important food protein used as an additive in numerous food products and that has been used in a variety of pharmaceutical applications (Southward 1989).
Results
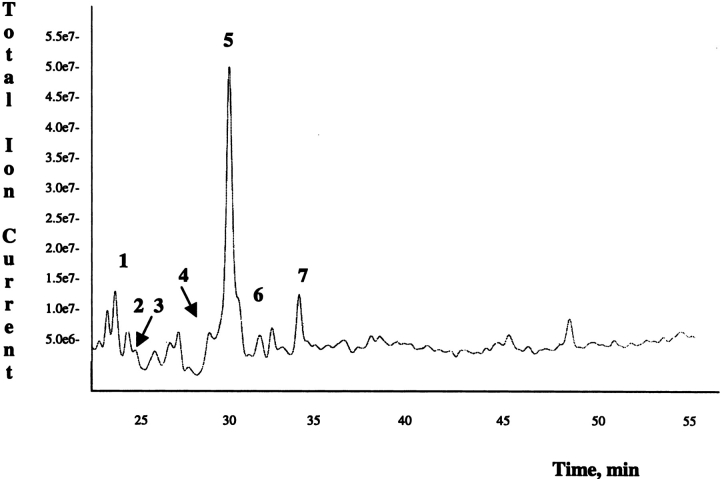
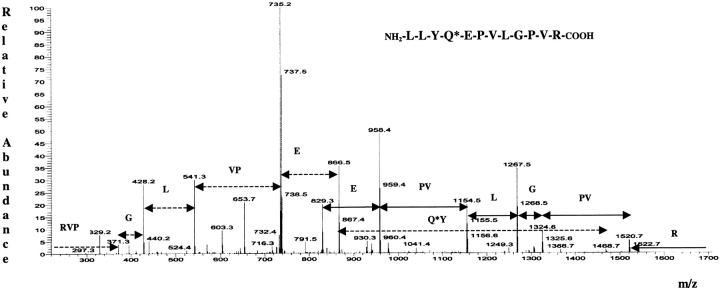
An equimolar mixture of VIP, SV-IV, and β casein was incubated with BPNH2 in the presence of tTG, as described below. The protein mixture was purified from the excess of the biotinylated substrate by performing a fast-desalting RP-HPLC. The protein mixture, eluted in a single HPLC fraction, was lyophilized, resuspended in 50 mM ammonium bicarbonate, and submitted to a double proteolytic digestion, using trypsin and chymotrypsin. Tagged and untagged peptides were then loaded onto a prepacked monomeric avidin affinity column. Untagged tryptic fragments were eluted with PBS as the unbound fraction, whereas biotinylated peptides were specifically eluted with 0.3% formic acid (Gygi et al. 1999). Six biotinylated peptide-containing fractions were collected, pooled, and concentrated to about 50 μL. Biotinylated peptides were then analyzed by LCMS/MS. Figure 2 ▶ shows the recorded total ion current (TIC) chromatogram. Every peak, corresponding to a biotinylated peptide ion, is highlighted by a number code. The data-dependent acquisition mode allowed fragmentation of the most intense ion present in each full scan spectrum. As an example, Figure 3 ▶ shows the tandem mass spectrum originated by the most intense ion eluted in peak 7 and the peptide sequence giving rise to it. The signal at m/z 1695.0 in the related full-scan spectrum is a monocharged ion. The fragmentation process generated a complex set of fragment ions mainly belonging to the b and y series. Several related patterns of b and y ions could be distinguished. The signal at m/z 1520.7 was interpreted as a bn-1 ion generated by the loss of the hydroxylic end and of an R residue as C-terminal amino acid. The signal at m/z 1324.6 was 196 Da lower than the bn-1 ion; such a mass difference could not be related to a single amino acid but could be justified by the sequence PV or VP. The signals at m/z 1324.6 and 1267.5 differed by a G residue while the signal at m/z 1154.5 could have its origin in ion 1267.5 because of the loss of a L/I residue. The whole set of mass data (molecular weight of the investigated peptide and of its fragments) was put into the query page of two Web-available softwares (see Materials and Methods) taking into account that the molecular mass of the peptide is modified by the presence of BPNH2. The mass data used for the NCBI protein database search were reduced by 311.4 Da, according to the mass difference introduced by the affinity probe used. The results obtained by the two independent Web-available softwares were in perfect agreement in identifying the sequence LLYQEPVLGPVR, a highly conserved stretch in mammalian β caseins. The assignment led to the definition of the only Q residue present in the sequence as a tTG substrate.
Figure 2.
LCMS/MS analysis of the biotinylated peptide mixture obtained from tryptic and chymotryptic hydrolysis of the protein mixture (VIP, SV-IV, and β-casein) incubated with BPNH2 in the presence of tTG. (For further details, see Materials and Methods.)
Figure 3.
Tandem mass spectrum derived by collision-induced dissociation of the MH+ precursor at m/z 1695.0. The one-letter code for encountered amino acids is shown.
Table 1 summarizes the results obtained by the LCMS/MS analysis.
Table 1.
Mascot and MS-Seq program sequence identification of the components of the protein mixture incubated with BPNH2
| Protein | Peptide | Sequence | MH+ | Fraction |
| VIP | 15–20 | KQ*MAVK | 1015.7 | 1 |
| SV-IV | 5–16 | RKTKEKYSQ*SEEVVSE | 1724.8 | 2 |
| 84–90 | FAQ*DVLN | 1117.7 | 5 | |
| 5–32/V31 | EKYSQ*-------DELVR | 3288.4 | 5 | |
| 5–32/M31 | EKYSQ*------DELMR | 3320.6 | 5 | |
| β Casein | 177–183 | AVPYPQ*R | 1141.9 | 3 |
| 194–202 | Q*EPVLGPVR | 1306.1 | 4 | |
| 192–202 | LYQ*EPVLGPVR | 1581.9 | 6 | |
| 191–202 | LLYQ*EPVLGPVR | 1695.0 | 7 |
Q* stands for a labeled glutamine residue. SV-IV protein is present as an equimolar mixture of two isoforms differentiated by a V or M residue at position 31. Numbered fractions refer to Figure 2 ▶.
The analysis allowed the identification of peptide 15–20 of the VIP sequence, revealing that the only Q residue, Q16, is a tTG substrate.
The purified SV-IV protein was shown to consist of an equimolar mixture of two isoforms differing in position 31: one isoform bearing a V residue, the other carrying an M residue (Ferranti et al. 1997). MS/MS results showed that the two Q residues present in the primary structure of SV-IV are both reactive toward tTG; labeled Q9 was identified in peptide 5–32 (5–32/V31 and 5–32/M31) and in the aspecifically cleaved fragment 5–16, whereas modified Q86 was detected in the C-terminal fragment 84–90. The presence of two signals related to peptide 5–32 in our tandem mass analysis confirmed the coexistence of the two isoforms (see Table 1).
Two Q residues, Q182 and Q194, out of 20 present in the β-casein sequence are recognized by tTG and modified by the BPNH2. Table 1 shows the peptides identified by MS/MS analysis containing modified Q residues.
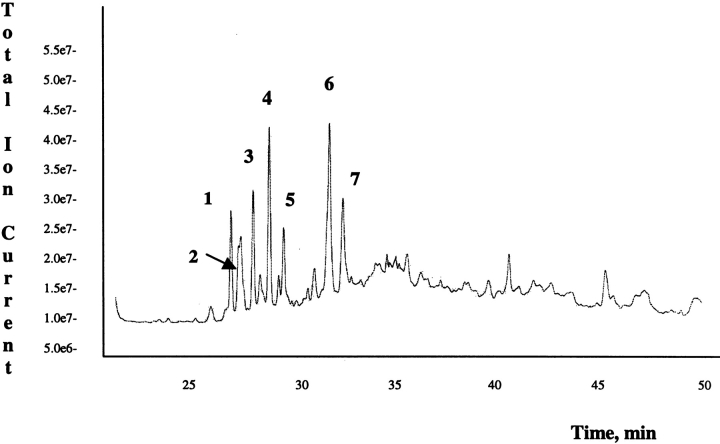
The procedure described above was applied when the mixture of proteins was incubated in the presence of a Q-donor affinity probe, A25 peptide. Biotinylated peptides were analyzed by LCMS/MS; Figure 4 ▶ shows the recorded TIC chromatogram. Few main differences were taken into account when interpreting the LCMS/MS data. Signals detected in the MS/MS data-dependent analysis must be the result of peptides carrying an A25 peptide molecule (expected mass increment 926.4 Da). Location of the modified K residues in peptides containing more than one K was unambiguously achieved taking advantage of the peptide sequence data obtained by MS/MS analysis and considering that modified K residues are not trypsin substrates. The observed fragmentation of the A25 peptide did not complicate the assignment of MS/MS data to peptide sequences. The data obtained are reported in Table 2.
Figure 4.
LCMS/MS analysis of the biotinylated peptide mixture obtained from tryptic and chymotryptic hydrolysis of the protein mixture (VIP, SV-IV and β-casein) incubated with A25 peptide in the presence of tTG. For further details, see Materials and Methods.
Table 2.
Mascot and MS-Seq program sequence identification of the components of the protein mixture incubated with A25 peptide
| Protein | Peptide | Sequence | MH+ | Fraction |
| VIP | 16–21 | QMAVK*K | 1629.8 | 5 |
| 21–23 | K*YL | 1348.7 | 3 | |
| A25 peptide | Biot-TVQQEL | 943.5 | 4 | |
| SV IV | 1–4 | RK*TK | 1457.8 | 1 |
| 3–6 | TK*EK | 1430.7 | 2 | |
| 79–80 | K*K | 1200.7 | 2 | |
| 80–81 | K*R | 1228.7 | 2 | |
| 79–81 | KK*R | 1356.8 | 2 | |
| 58–63 | SK*HISR | 1652.9 | 3 | |
| 57–63 | RSK*HISR | 1808.1 | 6 | |
| 77–83 | AK*KKRSR | 1800.8 | 7 |
K* stands for a labeled lysine residue. Numbered fractions refer to Figure 4 ▶.
The analysis showed the presence of VIP peptides 16–21 and 21–23, carrying modified K20 and K21, respectively.
Six K residues, K2, K4, K59, K78, K79, and K80, out of nine within the SV-IV primary structure, were sensitive toward tTG activity. Table 2 shows the peptides identified by MS/MS analysis.
No peptides related to the β-casein sequence were detected in the LCMS/MS analysis, suggesting the absence of NH2-donor residues sensitive to tTG action in the presence of A25 peptide.
Discussion
Proteomics is an emerging postgenomic research area that deals with the global analysis of expressed proteins using a variety of techniques (Griffin et al. 2001). Difficulties in resolving proteins have led to the development of methods based on the digestion of unfractionated protein mixtures and identification of the resulting peptides (Peng and Gygi 2001). The extreme complexity of such digests limits the number of proteins that can be identified during a single experiment. Therefore, several strategies have been used to reduce the complexity of these mixtures and thereby detect more and lower abundant proteins. One of such strategies involves the biotinylation of proteins at specific residues (Gygi et al. 1999). Following biotinylation and digestion, this procedure relies on the capture and recovery of biotinylated peptides by affinity chromatography on monomeric avidin agarose for subsequent analysis by LCMS/MS. The MS/MS spectra are then submitted to a database searching program. The whole procedure allows one not only to identify proteins but also to locate the amino acid site of the chemical modification.
Keeping in mind these considerations, we set up a procedure aimed at defining a functional protein map of TG substrates. Two biotinylated affinity probes, reported in Figure 1 ▶, were selected: BPNH2 for labeling Q-donor proteins and A25 peptide for labeling K-donor species. It must be considered that BPNH2 is a commercially available reagent thus favoring the identification of Q residues sensitive to TG action. Based on this, it is not surprising that there are many data on proteins that are reported to be Q donors in the tTG catalyzed reaction (Lee et al. 1992; Ikura et al. 1998; Madi et al. 2001). Conversely, A25 peptide was expressly synthesized with the aim of identifying K residues sensitive to tTG action. The smaller availability of efficient biotinylated probes, which can act as a Q donor in the reaction catalyzed by tTG, prevented the definition of a global map of proteins, which act as K donors in the tTG-catalyzed reaction (Lorand et al. 1992). The use of this affinity probe can then shed light on the definition of a complete functional map of tTG substrates.
A pool of well-characterized tTG protein substrates, VIP, SV-IV, and β-casein, was chosen and independently incubated with each of the two biotinylated affinity probes. The results presented in this paper showed that the whole procedure led to the identification of Q and K residues sensitive to tTG activity. Q16, K20, and K21 were found to be reactive within the VIP sequence. Published data reported Q16 and K21 as tTG substrates (Esposito et al. 1999), while no indications were available about K20. The exploitation of state-of-the-art technology in conjunction with classical biochemical methods led the authors to identify K20 as a new NH2-donor substrate for tTG within the VIP sequence.
The functional proteomic analysis showed that Q9, Q86, and K2, K4, K59, K78, K79, and K80 are tTG substrates within the SV-IV sequence. Previously published data (Porta et al. 1991) reported that K2, K4, K59, K78, K79, and K80 are involved in intramolecular polymerization with Q9 and Q86. The present data highlighted the fact that the same K residues are reactive toward an external Q-donor molecule in the presence of tTG. Intramolecular tTG-catalyzed isopeptide bonds escaped affinity capture in the monomeric avidin column and were therefore not identified in this study.
The results reported in this paper showed that β casein is a Q-donor but not a K-donor substrate of tTG. It was discovered that Q182 and Q194, along the β-casein sequence, were modified by BPNH2, while no signals related to β-casein peptides covalently linked to the A25 peptide via the ɛ-NH2 group of a K residue were detected. A previous report indicated that several Q residues present in the β-casein sequence could be considered tTG substrates: Q54, Q56, Q79, Q167, Q175 and Q182, and Q194 (Yan et al. 1984). However, it must be highlighted that the identification of modified Q54, Q56, Q79, Q167, Q175, and Q182 was very ambiguous because of difficulties in the procedure used. The same authors claimed that only Q194 was surely identified as a tTG substrate. The procedure reported here, based on up-to-date protein characterization techniques, demonstrates that Q182 and Q194 are definitively sensitive toward tTG activity. Finally, the results obtained in this study showed that β casein is not a NH2-donor protein in the reaction catalyzed by tTG toward an external Q-donor molecule. To the best of our knowledge, the only data reporting a possible involvement of β-casein K residues in tTG-catalyzed reaction is related to β-casein polymerization (Liu and Damodaran 1999) where the authors induce the formation of a polymer of β casein in the presence of tTG, without identifying K residues involved in such a reaction.
The affinity probes described in this study select proteins in a mixture according to a specific function of the chosen enzyme (i.e., TG) and not according to chemical reactivity as occurs in the isotope-coded affinity tag (ICAT) strategy developed by Gygi and coworkers (1999). The yield of an enzymatic reaction (based on recognition and binding affinity) is expected to be smaller than that of a chemical reaction, thus producing a protein mixture quantitatively different from that obtained using the ICAT strategy mentioned above.
A proper linker containing stable isotopes can be inserted in the chemical structure of the two affinity probes used in this study thus leading the development of a new generation of ICAT reagents based on a protein function and not on an amino acid-specific reactivity. Proteomic research increasingly demands quantitative rather than only qualitative results and the stable isotope methods provide an easy way to quantify proteins based on the comparison of two mass spectrometric peak areas.
The results obtained in this paper indicate that the whole strategy can be successfully applied in order to identify TG protein substrates as well as the amino acid site sensitive toward enzyme activity. Future developments will be suitable for a quantitative estimation of protein substrates of tTG, thus allowing a better understanding of tTG function in several physiological processes where it plays a key role (Aeschlimann and Thomazy 2000; Akimov and Belkin 2001). Considering the recent demonstration of the involvement of tTG in some pathological states, such as celiac disease and CAG-repeat neurodegenerative disorders (Sollid 2000; Esposito et al. 2002a), the proposed strategy could reveal significant changes in the amount of protein substrates as well as in recognizing reactive amino acids between abnormal and normal cells.
The further improvement obtained from these new reagents, along with a growing technological development, will help scientists in identifying and quantifying more and more low abundant proteins (present in small amount), and, in more general terms, will help to gain a deeper insight toward understanding the dynamic behavior of the proteins under investigation within complex biological systems.
Materials and methods
Bovine milk β casein, guinea pig liver TG (tTG), TPCK-trypsin, and chymotrypsin were purchased from Sigma. 5-(biotinamido) pentylamine (BPNH2) and the prepacked monomeric avidin column kit were from Pierce. Purified SV-IV was kindly provided by Professor Metafora (CNR International Institute of Genetics and Biophysics). VIP was obtained from Calbiochem. Reverse-phase HPLC columns C4 (25 × 0.46 cm) and C18 (25 × 0.21 cm) were purchased from Vydac (The Separation Group). All the other reagents and solvents were of the highest purity available from Carlo Erba.
Synthesis of biotin-labeled A25 peptide
The A25 peptide, TVQQEL, was synthesized manually, in a stepwise fashion, via the solid-phase method, using conventional Fmoc chemistry. Protected amino acids were added stepwise to Fmoc-Val-PEG-PS resin (0.70 g, substitution level 0.24 mmole/g); each coupling reaction was accomplished using a fivefold excess of amino acid with equimolar HBTU and HOBt in the presence of NMM (sixfold excess), for 1 h. The Nα-Fmoc protecting groups were removed after each coupling cycle by treating the protected peptide resin with a 20% solution of piperidine in DMF (1 × 1.5 min and 1 × 10 min). After deprotection of the N-terminal residue, biotin was added using HBTU/HOBt/NMM activation, as described above. Finally, the peptide was released from the resin with TFA/TIPS/H2O (90:5:5) for 75 min. The resin was removed by filtration and the crude peptide was recovered by precipitation with cold anhydrous ethyl ether to give a white powder which was liophylized. Crude yield: 75%. Purification was achieved by RP-HPLC on a semi-preparative C18 column (Jupiter 5u C18 300A, size 250 × 4.60 mm) using a gradient of CH3CN in 0.1% aqueous TFA (from 5% to 89% in 28 min) at a flow rate of 1.0 mL/min. The final product was obtained by lyophilization of the appropriate fractions after removal of the CH3CN by rotary evaporation. Analytical RP-HPLC indicated a purity >98% and molecular weights were confirmed by MS.
Incubation of the protein mixture with biotin-labeled probes
Modification of Q-donor proteins was performed by incubating a protein mixture, consisting of 9.8 μg SV-IV, 3.3 μg VIP, and 23.5 μg β casein, with 0.12 μg BPNH2 (molar ratio protein/BPNH2 = 1/20) in the presence of 2 μg tTG (molar ratio protein/tTG = 100/3) in 125 mM TRIS-HCl, 10 mM DTT, 2.5 mM CaCl2, pH 8.1. The final volume of the incubation was 100 μL.
To label K-donor proteins, the same protein mixture was incubated with 471.5 μg A25 peptide (molar ratio protein/A25 = 1/500) in the presence of 2 μg tTG (molar ratio protein/tTG = 100/3) as described above.
Both reactions were carried out for 18 h at 37°C. The reactions were then stopped by directly analyzing the mixtures by RP-HPLC, using a C4 Vydac column (25 × 0.46 cm). The eluting system consisted of 0.1% TFA (eluent A) and 0.1% TFA in acetonitrile (eluent B). The proteins were eluted in a single fraction by means of a linear gradient of eluent B in A, from 5% to 95% in 40 min at a flow rate of 1 mL/min elution was monitored at 220 nm. The purified proteins were lyophilized.
Enzymatic hydrolysis
HPLC-purified labeled protein mixtures were dissolved in 50 mM ammonium bicarbonate at pH 8.5. The mixtures were then hydrolyzed using trypsin (E/S = 1/50, w/w) at 37°C for 18 h. The reactions were stopped by boiling the sample for 5 min. Chymotrypsin was then added to the mixture (E/S = 1/50, w/w) and the reaction was kept at 37°C for 18 h. Hydrolysis was stopped by adding 5 μL 20% TFA; the sample was lyophilized and stored at −20°C.
Purification of peptides
The peptides were dissolved in 2 mL PBS and then submitted to an affinity chromatography step using a prepacked monomeric avidin column. The column was washed, saturated, and equilibrated according to the manufacturer’s instructions. After PBS conditioning, each peptide mixture was loaded onto the affinity column. The unbound fraction was eluted in PBS, whereas biotin-labeled peptides were selectively eluted with 0.3% formic acid (Gygi et al. 1999). Six biotinylated peptide-containing fractions were collected, pooled, and concentrated to about 50 μL.
The biotin-labeled tryptic peptides were fractionated by performing a RP-HPLC analysis on a C18 Vydac column (25 × 0.21 cm). The eluting system consisted of 5% formic acid, 0.05% TFA (eluent A) and 5% formic acid, and 0.05% TFA in acetonitrile (eluent B). The peptides were eluted by means of a linear gradient of eluent B in A, from 5% to 60% in 60 min at a flow rate of 200 μL/min. The elution was monitored on a LCQ mass spectrometer, without any splitting device.
Mass spectrometry analysis
Peptides eluted from the RP-HPLC C18 column were submitted on-line to MS/MS analysis by coupling the chromatographic system to an LCQ mass spectrometer (Finnigan). To determine the amino acid sequence of the peptides, the LCQ mass spectrometer was set up in a data-dependent MS/MS mode where a full-scan spectrum (m/z range from 500 to 2000 Da/e) was followed by a tandem mass spectrum; the precursor ion was selected as the most intense peak of the previous scan. The source was kept at 300°C. The mass instrument was calibrated in the whole mass range with the ultramark solution provided by the manufacturer.
Database searching
Proteins from the mixture were identified on the basis of both the molecular weights and the internal amino acid sequences of the selected fragmented peptides. Two Web-available programs (Mascot at http://www.matrixscience.com and MS-Seq at http://prospector.ucsf.edu) were searched using tandem mass spectra data and taking advantage of the specificity of the proteolytic systems used. The proteins were identified by selecting the following item: nonredundant NCBI protein database; mammals as taxonomic category; 1 Da error both for the average parent mass and for the fragments; no protein molecular weight was considered for the Matrixscience program whereas the wider molecular-weight range was used for the Protein Prospector Web site; double enzymatic digestion; up to five missed cleavages; cysteines as unmodified as well as N- and C-terminal ends; methionines both as unmodified and as oxidized.
Acknowledgments
This work was supported by CNR grant CNR G0072CF, Progetto giovani-Agenzia 2000.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
A25 peptide, Biotin-TVQQEL
BPNH2, 5(biotinamido) pentylamine
2D-PAGE, two-dimensional polyacrilamide gel electrophoresis
Fmoc, 9-fluorenylmethoxycarbonyl
HBTU, 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
HOBt, N-hydroxybenzotriazole
LCMS/MS, liquid chromatography tandem mass spectrometry
MS, mass spectrometry
MS/MS, tandem mass spectrometry
NMM: N-methyl-morpholine
PBS, phosphate buffer saline
PEG-PS: polyethylenglycol-polystyrene
RP-HPLC, reversed-phase high performance liquid chromatography
SV-IV, seminal vesicle protein IV
TFA, trifluoroacetic acid
TG, transglutaminase
tTG, guinea pig liver transglutaminase or tissue transglutaminase
TIPS: tri-isopropyl-sylane
VIP, vasoactive intestinal peptide
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0239103.
References
- Aeschlimann, D., and Paulsson, M. 1994. Transglutaminases: Protein cross-linking enzymes in body fluids. Thromb. Haemost. 71 402–415. [PubMed] [Google Scholar]
- Aeschlimann, D. and Thomazy, V. 2000. Protein cross-linking in assembly and remodeling of extra cellular matrices: The role of transglutaminases. Connect. Tissue Res. 41 1–27. [DOI] [PubMed] [Google Scholar]
- Akimov, S.S. and Belkin, A.M. 2001. Cell surface tissue transglutaminase is involved in adhesion and migration of monocytic cells on fibronectin. Blood 98 1567–1575. [DOI] [PubMed] [Google Scholar]
- Campbell, R.M. and Scanes, C.G. 1992. Evolution of the growth hormone-releasing factor (GRF) family of peptides. Growth Regul. 2 175–191. [PubMed] [Google Scholar]
- Carlquist, M., McDonald, T.J., Go, V.L., Bataille, D., Johansson, C., and Mutt, V. 1982. Isolation and amino acid composition of vasoactive intestinal peptide. Horm. Metab. Res. 14 28–29. [DOI] [PubMed] [Google Scholar]
- Cooper, A.J., Jeitner, T.M., Gentile, V., and Blass, J.P. 2002. Cross linking of polyglutamine domains catalyzed by tissue transglutaminase is greatly favored with pathological-length repeats: Does transglutaminase activity play a role in (CAG)(n)/Q(n)-expansion diseases? Neurochem. Int. 40 53–67. [DOI] [PubMed] [Google Scholar]
- Esposito, C., Cozzolino, A., Mariniello, L., Stiuso, P., De Maria, S., Metafora, S., Ferranti, P., and Carten|f9-Farina, M. 1999. Enzymatic synthesis of vasoactive intestinal peptide analogs by transglutaminase. J. Peptide Res. 53 626–632. [DOI] [PubMed] [Google Scholar]
- Esposito, C., Caputo, I., Paparo, F., Porta, R., Salvati, V.M., Mazzarella, G., Auricchio, S., and Troncone, R. 2002a. Expression and enzymatic activity of small intestinal tissue transglutaminase in coeliac disease. J. Pediatr. Gastr. Nutr. 34 473. [DOI] [PubMed] [Google Scholar]
- Esposito, C., Cozzolino, A., Porta, R., Mariniello, L., Buommino, E., Morelli, F., Metafora, V., and Metafora, S. 2002b. Protein SV-IV promotes nitric oxide production not associated with apoptosis in murine macrophages. Eur. J. Cell Biol. 81 185–196. [DOI] [PubMed] [Google Scholar]
- Ferranti, P., Mamone, G., Malorni, A., Guardiola, J., Stiuso, P., and Metafora S. 1997. Structural heterogeneity, post-translational modifications, and biological activities of SV-IV, a major protein secreted from the rat seminal vesicle epithelium. Rapid. Commun. Mass Spectrom. 11 1007–1014. [DOI] [PubMed] [Google Scholar]
- Folk, J.E. and Finlayson, J.S. 1977. The ɛ-(γ-glutamyl)lysine cross-link and the catalytic role of transglutaminases. Adv. Protein Chem. 31 1–133. [DOI] [PubMed] [Google Scholar]
- Griffin, T.J., Goodlett, D.R., and Aebersold, R. 2001. Advances in proteome analysis by mass spectrometry. Curr. Opin. Biotechnol. 12 607–612. [DOI] [PubMed] [Google Scholar]
- Groenen, P.J., Grootjans, J.J., Lubsen, N.H., Bloemendal, H., and de Jong, W.W. 1994. Lys-17 is the amine-donor substrate site for transglutaminase in β A3-crystallin. J. Biol. Chem. 269 831–833. [PubMed] [Google Scholar]
- Gygi, S.P., Rist, B., Gerber, S.A., Turecek, F., Gelb, M.H., and Aebersold, R. 1999. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17 994–999. [DOI] [PubMed] [Google Scholar]
- Ikura, K., Kita, K., Fujita, I., Hashimoto, H., and Kawabata, N. 1998. Identification of amine acceptor protein substrates of transglutaminase in liver extracts: Use of 5–(biotinamido) pentylamine as a probe. Arch. Biochem. Biophys. 356 280–286. [DOI] [PubMed] [Google Scholar]
- Kim, S.Y., Jeitner, T.M., and Steinert, P.M. 2002. Transglutaminases in disease. Neurochem. Int. 40 85–103. [DOI] [PubMed] [Google Scholar]
- Lee, K.N., Maxwell, M.D., Patterson, Jr., M.K., Birckbichler, P.J., and Conway, E. 1992. Identification of transglutaminase substrates in HT29 colon cancer cells: Use of 5–(biotinamido)pentylamine as a transglutaminase-specific probe. BBA 1136 12–16. [DOI] [PubMed] [Google Scholar]
- Liu, M. and Damodaran, S. 1999. Effect of transglutaminase-catalysed polymerasation of β-casein on its emulsifying properties. J. Agric. Food Chem. 47 1514–1519. [DOI] [PubMed] [Google Scholar]
- Lorand, L. and Conrad, S.M. 1984. Transglutaminases. Mol. Cell. Biochem. 58 9–35. [DOI] [PubMed] [Google Scholar]
- Lorand, L., Parameswaran, N., Velasco, P.T., and Murthy, S.N.P. 1992. Biotinylated peptides containing a factor XIIIa or a tissue transglutaminase-reactive glutaminyl residue that block protein cross-linking phenomena by becoming incorporated into amine donor sites. Bioconjug. Chem. 3 37–41. [DOI] [PubMed] [Google Scholar]
- Madi, A., Kele, Z., Janaky, T., Punyiczki, M., and Fésus, L. 2001. Identification of protein substrates for transglutaminase in Caernorhabditis elegans. Biochem. Biophys. Res. Commun. 283 964–968. [DOI] [PubMed] [Google Scholar]
- Ostrowski, M.C., Kistker, M.K., and Kistler, W.S. 1979. Purification and cell-free synthesis of a major protein from rat seminal vesicle secretion. A potential marker for androgen action. J. Biol. Chem. 254 3759–3765. [PubMed] [Google Scholar]
- Peng, J. and Gygi, S.P. 2001. Proteomics: The move to mixtures. J. Mass Spectrom. 36 1083–1091. [DOI] [PubMed] [Google Scholar]
- Porta, R., Esposito, C., Metafora, S., Malorni, A., Pucci, P., Siciliano, R., and Marino, G. 1991. Mass spectrometric identification of the amino donor and acceptor sites in a transglutaminase protein substrate secreted from rat seminal vesicles. Biochemistry 30 3114–3120. [DOI] [PubMed] [Google Scholar]
- Rawlings, S.R. 1994. PACAP, PACAP receptors, and intracellular signalling. Mol. Cell Endocrinol. 101 C5–C9. [DOI] [PubMed] [Google Scholar]
- Sollid, L.M. 2000. Molecular basis of celiac disease. Annu. Rev. Immunol. 18 53–81. [DOI] [PubMed] [Google Scholar]
- Southward, C.R. 1989. Uses of casein and caseinated. In Developments in diary chemistry-4 (ed. P.F. Fox), Elsevier Applied Science, London.
- Yan, S.B. and Wold, F. 1984. Neoglycoproteins: In vitro introduction of glycosyl units at glutamines in β-casein using transglutaminase. Biochemistry 23 3759–3765. [DOI] [PubMed] [Google Scholar]