Abstract
β-1,4-Galactanases hydrolyze the galactan side chains that are part of the complex carbohydrate structure of the pectin. They are assigned to family 53 of the glycoside hydrolases and display significant variations in their pH and temperature optimum and stability. Two fungal β-1,4-galactanases from Myceliophthora thermophila and Humicola insolens have been cloned and heterologously expressed, and the crystal structures of the gene products were determined. The structures are compared to the previously only known family 53 structure of the galactanase from Aspergillus aculeatus (AAGAL) showing ~56% identity. The M. thermophila and H. insolens galactanases are thermophilic enzymes and are most active at neutral to basic pH, whereas AAGAL is mesophilic and most active at acidic pH. The structure of the M. thermophila galactanase (MTGAL) was determined from crystals obtained with HEPES and TRIS buffers to 1.88 Å and 2.14 Å resolution, respectively. The structure of the H. insolens galactanase (HIGAL) was determined to 2.55 Å resolution. The thermostability of MTGAL and HIGAL correlates with increase in the protein rigidity and electrostatic interactions, stabilization of the α-helices, and a tighter packing. An inspection of the active sites in the three enzymes identifies several amino acid substitutions that could explain the variation in pH optimum. Examination of the activity as a function of pH for the D182N mutant of AAGAL and the A90S/ H91D mutant of MTGAL showed that the difference in pH optimum between AAGAL and MTGAL is at least partially associated with differences in the nature of residues at positions 182, 90, and/or 91.
Keywords: β-1,4-galactanase; family 53 glycoside hydrolase; thermostability; pH optimum; clan GH-A; thermophile; alkalophile
Pectin is a major and complex component of the plant cell wall, requiring a large battery of enzymes for its degradation (de Vries and Visser 2001). It consists of “smooth” regions of α-1,4-linked galacturonic acid (homogalacturonan) and “hairy” regions of rhamnogalacturonan (O’Neill et al. 1990). The most abundant form of rhamnogalacturonan, rhamnogalacturonan I (RG-I), consists of alternating α-1,2-linked rhamnose and α-1,4-linked galacturonic acids (Lau et al. 1985). β-1,4 galactanases hydrolyze the β-1,4 O-glycosidic bonds in galactan and arabinogalactan, which attach to the C4 position of the rhamnose residues and form the branches in the “hairy” region of pectin. We previously determined the structure of the mesophilic Aspergillus aculeatus β-1,4-galactanase (AAGAL) at 293K (PDB-ref code: 1FHL) and 100K (PDB-ref code: 1FOB) by multiple isomorphous replacement (Ryttersgaard et al. 2002). AAGAL belongs to family 53 in the glycoside hydrolases classification (Couthino and Henrissat 1999), which in turn belongs to clan GH-A (Henrissat et al. 1995; Jenkins et al. 1995), containing glycoside hydrolase families with different specificities.
All of the glycoside hydrolases of clan GH-A perform glycosidic bond cleavage via the catalytic retaining mechanism (McCarter and Withers 1994). This mechanism usually involves two glutamic acid residues, which are located at the ends of β-strands 4 (acid/base) and 7 (nucleophile). A nucleophilic attack is performed on the anomeric carbon by one of the carboxylic residues, and the glycosidic oxygen is protonated by the catalytic acid/base residue, which leads to the formation of a covalent enzyme-substrate intermediate. This intermediate is released by a second nucleophilic attack performed by a water molecule previously deprotonated by the acid/base residue. For family 53, the catalytic glutamates have been identified experimentally in P. cellulosa galactanase (Braithwaite et al. 1997).
Enzymes sharing similar structure and catalytic mechanisms can exhibit surprisingly different biochemical properties. A. aculeatus galactanase is a mesophilic enzyme with pH optimum and stability at low pHs (Christgau et al. 1995). Here, we report the isolation, characterization, and structure determination of two thermophilic fungal galactanases from Humicola insolens and Myceliophthora thermophila with pH optimum and stability in the neutral to basic range. These two galactanases have high sequence identity (about 56%) with AAGAL but some rather striking differences in amino acid composition. Given the high sequence identity, the structures were determined by molecular replacement using the AAGAL structure as a starting point.
In the last 25 years, considerable efforts have been made to explain the thermostability enhancement of proteins by comparison of the sequences and tertiary structures of thermophilic proteins to their mesophilic counterparts. Some of the most commonly invoked structural determinants of enhanced thermostability are hydrogen bonds, α-helices stabilization, improved packing, and ion pairs, as shown in several of the larger-scale comparative studies performed over the years (Argos et al. 1979; Querol et al. 1996; Vogt et al. 1997; Szilàgyi and Zàvodsky 2000). pH adaptation and stability have also stimulated considerable interest, especially the adaptation to high pH, which is important in many enzymatic industrial processes (Shirai et al. 2001). Given the high structural and sequence similarity but very different properties of AAGAL, HIGAL, and MTGAL, this represents an ideal case to study nature’s protein engineering in terms of temperature and pH adaptation.
Results and Discussion
Characterization of AAGAL, HIGAL, and MTGAL
The pH, temperature optimum, and pH range where MTGAL, HIGAL, and AAGAL maintain more than half of their optimal activity were determined and are presented in Table 1. Although AAGAL has been previously characterized (Christgau et al. 1995), we repeated the pH and temperature profile determinations to ensure that they were comparable to those of MTGAL and HIGAL. As seen from Table 1, MTGAL and HIGAL are both more active and more stable at neutral to basic pH, whereas AAGAL is most active and stable at acidic pH. MTGAL and HIGAL are also more thermophilic than AAGAL. Although this is not always true for other enzymes, in the case of fungal GH-53 galactanases, the pH optimum and stability range are close to the pI of the individual enzymes.
Table 1.
Characterization of MTGAL, HIGAL, and AAGAL
| Enzyme | MTGAL | HIGAL | AAGAL |
| pH optimum (at 37°C) | 7.0 | 8.5 | 4.0 |
| pH of crystallization | 8.5 | 6.5 | 7.5 |
| pH range where activity > 50% | 5.5–8.5 | 6.5–9.5 | 3.0–5.5 |
| Temperature optimum | 65°C (at pH 6.5) | 65°C (at pH 6.5) | 37°C (at pH 6.5) |
| 50°C (at pH 3.5) | |||
| pH stability rangea | 3.5–8.5 | 3.5–8.5 | 2.5–5.5 |
| Theoretical pIb | 6.0 | 6.7 | 4.0 |
a This is the pH interval where activity > 50% is left after incubation for 2 h at 37°C.
b Calculated in http://us.expasy.org/tools/protparam.html.
Structures of MTGAL and HIGAL
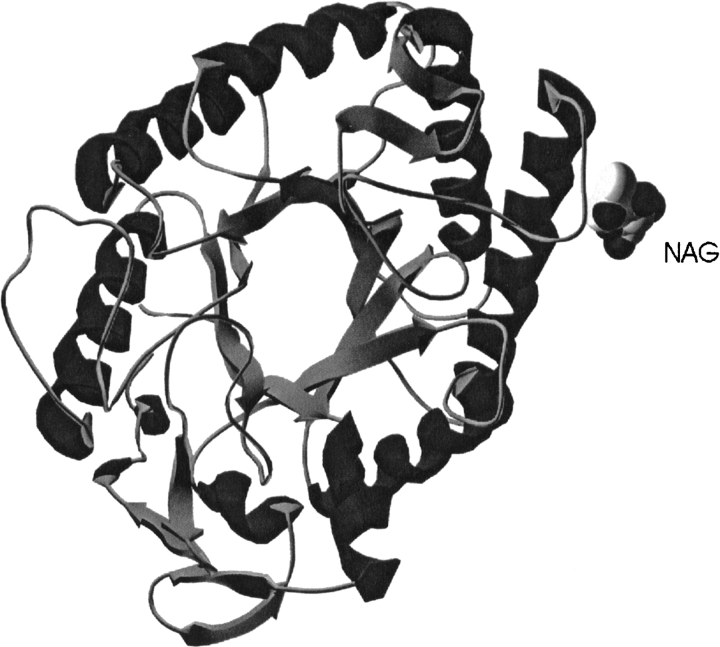
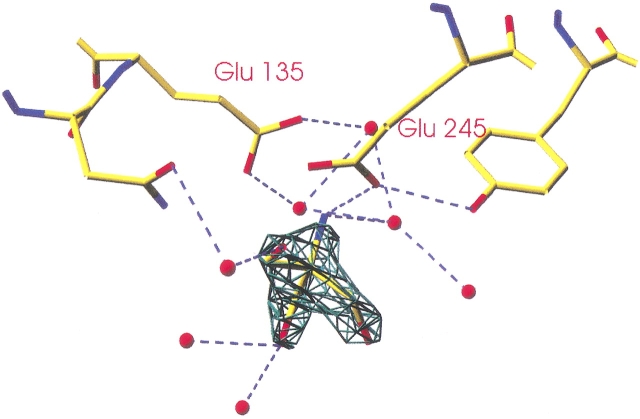
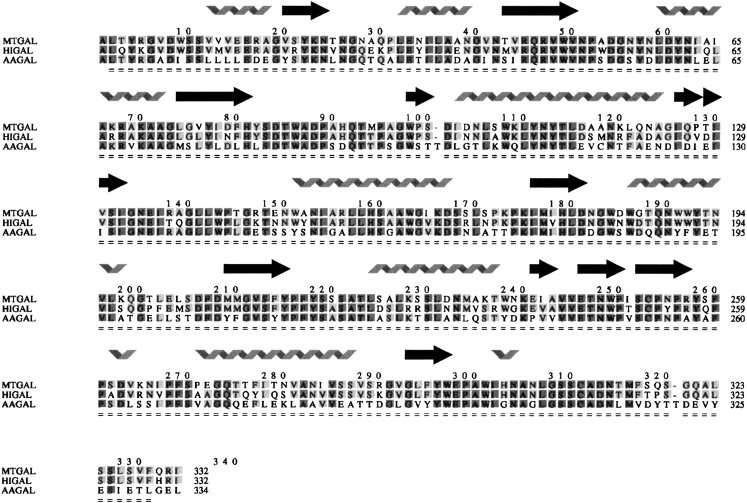
Like other enzymes belonging to clan GH-A, HIGAL (Fig. 1 ▶) and MTGAL share a (β/α)8-barrel fold (Nagano et al. 2002). The two catalytic amino acids are positioned at the ends of β-strands 4 (acid/base, E135) and 7 (nucleophile, E245) in MTGAL and HIGAL. By hydrogen bonding to the catalytic nucleophile and directly (in molecule D) or through a water molecule (in molecule A) to the conserved Asn preceding the acid/base, the TRIS molecule found in MTGAL’s structure mimics the substrate at the active site (Fig. 2 ▶). A structure-based sequence alignment was made for the three fungal galactanases (Fig. 3 ▶). The sequence identity between MTGAL and AAGAL is 56%, and that between MTGAL and HIGAL is 78%. The backbone of MTGAL superimposes well on the backbones of HIGAL and AAGAL, with root mean square deviation values of 0.4 Å and 0.7 Å, respectively, based on 332 Cα atoms. The residues in the substrate binding groove are very similar. Thus, the main differences between the structures are likely to be mostly related to thermal and pH adaptation.
Figure 1.
Illustration of the overall fold of HIGAL. Made with Swiss PDB-Viewer (Guex and Peitsch 1997).
Figure 2.
Close-up of a TRIS molecule in the MTGAL active site (molecule A). Produced with Swiss PDB-Viewer (Guex and Peitsch 1997).
Figure 3.
Structure-based sequence alignment of MTGAL, HIGAL, and AAGAL. Created using Indonesia (D. Madsen and G. Kleywegt, in prep.). Shading indicates the degree of conservation.
Analysis of amino acid composition
Despite the high sequence identity (56%) between AAGAL and MTGAL/HIGAL, some differences were observed in amino acid composition. The percentage composition of Pro in MTGAL and HIGAL is 5.1% whereas in the mesophilic AAGAL, 3.9% of the residues are proline. The number of glycine residues is slightly higher in the mesophilic AAGAL (7.2%) than in MTGAL (6.5%) and HIGAL (6.6%). The correlation between a higher number of proline residues and a thermostability enhancement of proteins has been reported (Aghajari et al. 1998). In particular, Ala→Pro and Gly→Ala substitutions were previously shown to increase thermostability by decreasing the entropy of unfolding (Matthews et al. 1987). MTGAL and HIGAL contain 3.3% and 4.5% of arginine residues, respectively, whereas AAGAL contains only 1.5%. Arginine is able to provide an additional hydrogen bond compared to other polar side chains, and thus a significantly higher percentage of this residue could contribute to an enhancement of the thermostability.
AAGAL also has a slightly higher percentage of charged residues (17.6%) than MTGAL (15.1%) and HIGAL (15.9%). This observation goes against the opposite tendency observed by Szilàgyi and Zàvodsky (2000) for mesophilic versus thermophilic proteins. MTGAL and HIGAL have similar percentages of negatively (8.0% and 8.1%, respectively) versus positively charged residues (8.6% and 9.6%, respectively). In contrast, AAGAL has 13.4% of glutamates and aspartates, resulting in its very low pI (Table 1).
Analysis of structural factors important for temperature optimum and stability
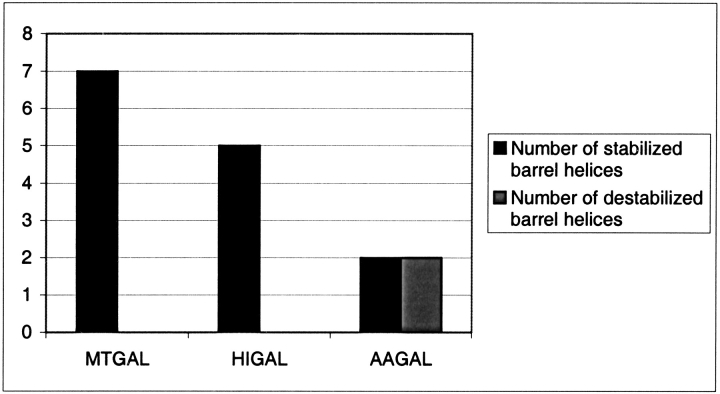
The three β-1,4-galactanases present different temperature profiles, and by inference different temperature stabilities despite high sequence and structural similarity. Figure 4 ▶ shows that the two thermostable galactanases (MTGAL, HIGAL) have a higher number of stabilized barrel helices than the mesophilic AAGAL, taking into account only helix dipole stabilization by charged side chains. A similar finding was shown for the distantly related family 10 xylanases (Lo Leggio et al. 1999; Teixeira et al. 2001).
Figure 4.
Comparison of α-helix stabilization in MTGAL, HIGAL, and AAGAL. Stabilized/destabilized helices were classified from the observation of favorable or unfavorable interactions between the α-helix dipole, the three first N-terminal residues, and the three last C-terminal residues as in Teixeira et al. (2001). Positively charged residues (Lys, Arg, His) were considered favorable at the C-terminus and unfavorable at the N-terminus. For negatively charged residues (Asp, Glu), the opposite was true. Helices where favorable interactions outnumbered unfavorable interactions were considered stabilized. Where unfavorable interactions outnumbered the favorable ones, the helix was considered destabilized. Only barrel helices were considered.
Increases in the number of salt bridges and hydrogen bonds are common features often associated with thermostability, as shown by extensive studies (Vogt et al. 1997; Szilàgyi and Zàvodsky 2000). For the GH-53 galactanases, a slight increase in ion pairs can be seen with a 4 Å cutoff (14 ion pairs for AAGAL, 16 and 15 for MTGAL and HIGAL, respectively) but not with a 6 Å cutoff (24 ion pairs for AAGAL and MTGAL and 23 for HIGAL). A slight increase in the number of hydrogen bonds can be observed in the mesophile AAGAL (1.003 Hbond/residue) compared to the thermophiles MTGAL and HIGAL (1.048 Hbond /residue and 1.024 Hbond /residue, respectively), which might be related to the increase in percentage of arginine in the latter two enzymes. It is also known that a cation-π interaction can be energetically equivalent or stronger than a hydrogen bond or a salt bridge (Gallivan and Dougherty 2000). Eight cation-π interactions can be found in HIGAL and MTGAL, whereas AAGAL contains six.
Thermophilic proteins usually have better packed hydrophobic cores than their mesophilic counterparts, and atom accessibility can provide a measure of this. The percentage of atoms accessible to a probe of 1.4 Å is 47.9% and 48.2% in MTGAL and HIGAL, respectively, and 49.3% for AAGAL. The small difference is however larger than the standard deviation (0.5%) for the eight molecules of MTGAL refined in the two crystal forms. The difference might be significant given the high similarity of the three galactanases, as it would correspond to complete burial of one/two large side chains. Evidence for better packing of HIGAL and MTGAL also comes from cavity analysis. HIGAL does not contain any cavities, MTGAL contains an average of 1.2 cavities corresponding to a total volume of 264 Å3, and AAGAL contains two cavities, which corresponds to a total volume of 358 Å3.
The AAGAL structure also has a wide B-factor distribution as opposed to the tight distribution for MTGAL (HEPES) which was determined at the same temperature and to similar resolution. The B-factor distribution is also shifted to higher B-factors. The B-factor average (Main Chain) is 16.8Å2 in MTGAL, and 26.9Å2 in AAGAL. This indicates the presence of areas of high thermal motions in the mesophile that can affect the kinetics of unfolding and thus also contribute to differences in thermostability.
Analysis of structural factors important for pH optimum and stability
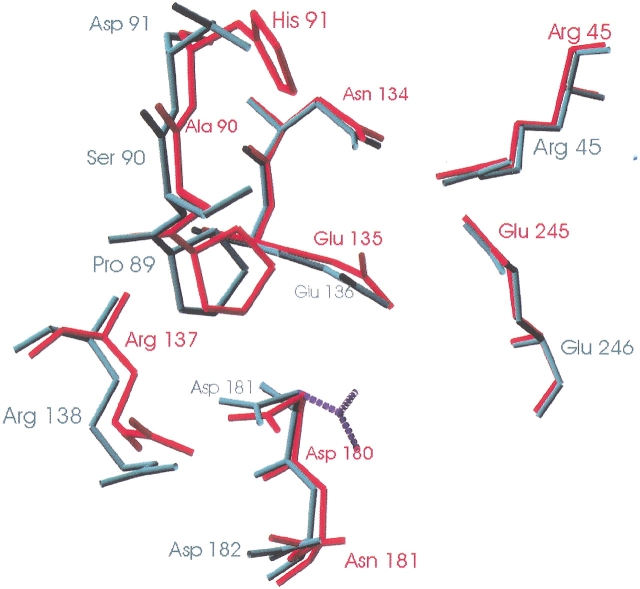
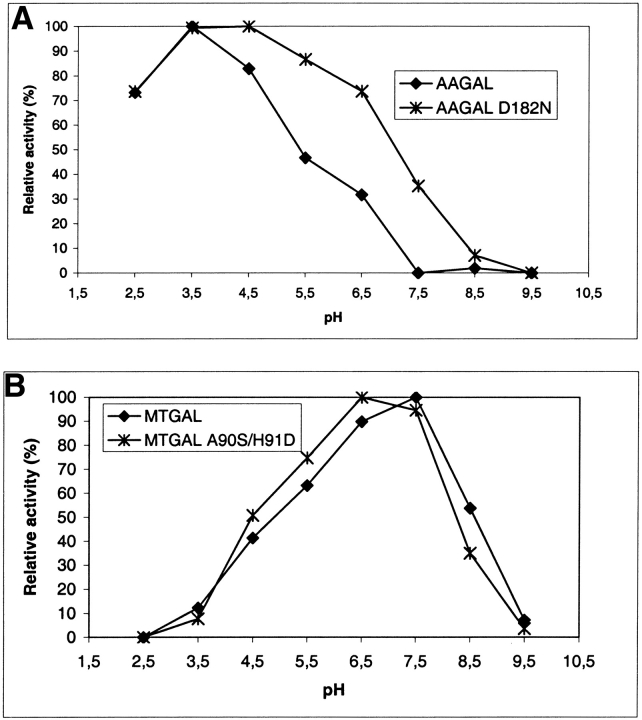
The structures of the three fungal galactanases were examined for polar or charged residues in the vicinity of the active site directly or indirectly interacting with the catalytic carboxylates and which differ between AAGAL and MTGAL/HIGAL. Three residues were identified: Ser 90 in AAGAL (Ala in MTGAL and HIGAL), Asp 91 in AAGAL (His in MTGAL and HIGAL), and Asp 182 in AAGAL (Asn 181 in MTGAL and HIGAL). These residues are shown in Figure 5 ▶. Asp 182 in AAGAL was modified to Asn, and this indeed generated a broadening of the pH profile towards higher pH by about 1 pH unit (Fig. 6A ▶). The MTGAL double mutant A90S/H91D shifted the pH profile by about 0.5 unit towards lower pH compared to the MTGAL wild type (Fig. 6B ▶). None of the mutations gave significant change in the specific activity towards AZCL-lupin galactan at their respective pH optima.
Figure 5.
Superposition of the MTGAL (in red) and AAGAL (in green) active sites. The possible position of Asp 181 at pH 4.0 is represented by dashed lines. Produced with Swiss PDB-Viewer (Guex and Peitsch 1997).
Figure 6.
(A) pH activity profile of native and D182N-substituted AAGAL. (B) pH activity profile of native and A90S/H91D-substituted MTGAL.
At the pH optimum, one would expect the acid base to be protonated and the nucleophile to be deprotonated if glycosylation is the rate-limiting step. This hypothesis is plausible because galactooligosaccharides are poor leaving groups. If we wish to shift the pH optimum towards alkaline pH, it would seem logical to shift the pKa of the acid/base up. As negative charges normally increase the pKa of their neighbors, it seems counterintuitive that the D to N mutation should increase the pH optimum. However, this type of effect is not unique to the galactanases; it is also seen in family 11 glycoside hydrolases. A very elegant study of Bacillus circulans xylanase shows that the N35D mutation increases the pKa of catalytic carboxylates Glu 78 and Glu 172, but decreases the pH optimum. To explain this phenomenon, those authors invoke reverse protonation, where hydrolysis requires that the group with higher pKa (Glu78) is deprotonated and a group with lower pKa (Asp 35) is protonated (Joshi et al. 2000, 2001). This requires an inherent catalytic efficiency of the mutant about two orders of magnitude higher than the native for it to have a similar level of activity, which is indeed the case. Joshi et al. suggested that this efficiency comes from glycosyl-enzyme transition state stabilization by a short hydrogen bond between Asp 35 and Glu 172.
The substitution of Asp 182 (AAGAL) with Asn in MTGAL (Asn 181) and in the D182N mutant has an effect on the pH optimum analogous to the effect described by Joshi and coworkers (2000, 2001), although in this case the Asp is in the native AAGAL, which has the lower pH optimum, implying that reverse protonation is occurring in a native enzyme. However, a similar explanation of higher inherent catalytic efficiency is hardly justified, due to the rather large (>10Å) distance between Asp 182 and the catalytic residues. Although the variant residue is about 10Å from the catalytic acid/base, in all of the galactanases investigated, an aspartate (Asp 181 in AAGAL and Asp 180 in MTGAL/HIGAL) is situated between the variant residue and the catalytic acid/base (Fig. 6 ▶). This aspartate is hydrogen-bonded to an arginine (Thr in HIGAL) at the pH of crystallization (Table 1). However, whereas MTGAL was crystallized near its pH optimum, for AAGAL, the pH of crystallization (pH=7.5) and the pH optimum (pH=4.0; Ryttersgaard et al. 1999) are in the basic and acidic range, respectively. Thus, we assume that in the model (pH=7.5), Asp 181 is deprotonated and negatively charged and interacts with the positive charge of Arg 138. At pH 4.0, Asp 181 might be protonated and flip (dashed line in Fig. 6 ▶), and provide a pathway for Asp 182 in AAGAL to interact with the catalytic acid/base.
As noted in the analysis of amino acid compositions, the acidophilic AAGAL has a much larger number of aspartates and glutamates than MTGAL and HIGAL. In particular, a large patch of carboxylate groups is found near the active site, which explains the low stability and activity at high pH of AAGAL. If all of these carboxylate groups become deprotonated, significant destabilization of the structure can be expected by charged repulsion. We do not observe in MTGAL and HIGAL the shift from Lys/Asp salt bridges to Arg/Asp salt bridges as seen in the alkali stable family 5 cellulases (Shirai et al. 2001), which can be rationalized by the fact that the two galactanases are only moderately alkalophilic, whereas the pH optimum of the cellulase is 9.5.
Conclusions
The genes for two new thermophilic fungal galactanases from M. thermophila and H. insolens were cloned, overexpressed in Aspergillus oryzae and the gene products characterized, showing that they are active and stable at neutral to high pHs.
The structures of the two enzymes were determined and compared to the structure of the mesoacidophilic galactanase from Aspergillus aculeatus. MTGAL and HIGAL show structural characteristics typical for thermophilic/thermostable enzymes: improved packing, increased rigidity, increased helix stability, and a slight increase in the number of electrostatic interactions. The most significant single difference between the mesophilic and thermophilic galactanases is the helix-dipole stabilization. However, the analysis confirms the general picture that has emerged since 1975 (Matthews et al. 1974): Thermostability is achieved by a combination of small structural features rather than a single one. Several residues, which might be responsible for the difference in pH optimum, were also identified. Substitution of AAGAL Asp182 to Asn (Asn 181 in MTGAL and HIGAL) confirmed experimentally that this residue is important for the acidic pH optimum of AAGAL. Similarly, substitution of MTGAL Ala90 to Ser and His91 to Asp (as in AAGAL) demonstrated that the nature of residues 90 and/or 91 also influence the pH profile.
The described Asp to Asn substitution counterintuitively shifts the pH profile of AAGAL up one pH unit, similarly to what has been seen in family 11 xylanases, suggesting that this might be a more general strategy for manipulating the pH optimum of enzymes.
Materials and methods
Cloning, expression, and purification of MTGAL and HIGAL
The cloning and expression is described in detail elsewhere (U.S. Patent 6,242,237 B1). In brief, mRNA was isolated from mycelium of Humicola insolens and Myceliophthora thermophila as described by Dalbøge and Heldt-Hansen (1994). Double-stranded cDNA was made according to standard protocols. The cDNA was digested with NotI and ligated to BstXI adaptors (Invitrogen). cDNAs larger than 0.7 Kb were selected, then cloned into BstXI-NotI-cleaved pYes 2.0 vector, and a library was constructed in E. coli DH10B cells (Bethesda Research Laboratories). This library was subdivided into pools, and each pool was transformed into S. cerevisiae W3124. Galactanase-positive clones were identified as the colonies surrounded by a halo on SC agar plates containing 0.1% AZCL galactan (Megazyme). DNA from positive clones was isolated and transformed into E. coli. The cDNA insert was excised and ligated into an Aspergillus expression vector. Aspergillus oryzae strains were transformed and tested for galactanase activity. A galactanase-positive strain was used for production of the galactanase for purification.
MTGAL was purified by the following procedure: Ammonium sulfate was added to the fermentation supernatant to give a 1.4 M final concentration, and the galactanase was applied to a Butyl Toyopearl 650S column equilibrated in 20 mM succinic acid, 1.4 M (NH4)2SO4, pH 6.0. After the column was washed with equilibration buffer, followed by elution with a linear ammonium sulfate gradient (1.4 M → 0 M). The eluted galactanase activity was dialyzed against 20 mM Tris/HCl, pH 9.0 and applied to a Q-Sepharose HP column equilibrated in the same buffer. After a washing with equilibration buffer, the column was eluted with a linear NaCl gradient (0 M → 0.2 M). Eluted fractions were checked for activity and then analyzed by SDS-PAGE, and pure fractions were pooled. The pH was adjusted to 8.0 with CH3COOH, and the pooled fractions were concentrated on an Amicon ultrafiltration cell. HIGAL was purified by a similar procedure as MTGAL, except that the galactanase activity eluting from the Butyl Toyopearl 650S column was applied to a cation exchanger (SP-Sepharose HP) equilibrated in 20 mM CH3COOH/NaOH, pH 5.0 and eluted with a linear NaCl gradient (0 M → 0.25 M).
Characterization of MTGAL and HIGAL
The activities of MTGAL and HIGAL were measured either by the release of reducing sugars from lupin galactan (Megazyme) or by the release of blue color from AZCL-lupin-galactan (Megazyme).
For the pH and temperature profiles, 4 mg/mL AZCL-lupin-galactan (Megazyme) was homogenously suspended in deionized water by stirring. Next, 500 μL AZCL galactan suspension and 500μL assay buffer (50 mM CH3COOH, 50 mM KH2PO4, 50 mM H3BO3, 1 mM CaCl2, 0.01% Triton X-100, adjusted to the relevant pH-values with HCl or NaOH) were mixed in an Eppendorf tube and placed on ice. Twenty μL galactanase sample (diluted in assay buffer) was added. An enzyme concentration range of 40–500 ng in 1 mL assay mixture was used. An Eppendorf thermomixer was used for this assay and set to the desired temperature. The tube was incubated for 15 min. The incubation was stopped by transferring the tube back to the ice bath and then centrifuged for a few minutes. OD650 was read as a measure of galactanase activity.
Site-directed mutagenesis, purification, and characterization of D182N AAGAL and MTGAL A90S/H91D
The D182N mutation was introduced in the AAGAL encoding gene by the use of the mutagenic oligonucleotide 5′- CAT TTG GAC AAC GGC TGG AGC -3′ and the mega-priming method described (Sarkar and Sommer 1990). The resulting variant gene was cloned into plasmid pHD464 (Dalbøge and Heldt-Hansen 1994), and the correct introduction of the mutation was verified by DNA sequencing.
The A90S, H91D double mutation was introduced in the MTGAL encoding gene essentially as described above by the use of the mutagenic oligonucleotide 5′- GCC GAT CCT TCT GAT CAG ACC ATG CC -3′. Proteins were expressed in and secreted from Aspergillus oryzae essentially as described by Christensen et al. (1988).
AAGAL D182N and MTGAL A90S/H91D were purified using similar protocols. Both cultures were filtered to remove the mycelia. Ammonium sulphate (1.6 M) was added, the solution was loaded onto a butyl column equilibrated with 25 mM sodium acetate, 1.6 M ammonium sulphate pH 5.0, and eluted using a linear ammonium sulphate gradient decreasing from 1.6 M to 0 M over 10 column volumes. Fractions containing galactanase activity of both variants eluted around 1 M ammonium sulphate, and were pooled and dialysed against 10 mM sodium citrate pH 3.5. The dialysate was loaded onto an S-Sepharose column equilibrated with 10 mM sodium citrate pH 3.5. Galactanase did not bind to this column, and was concentrated on an Amicon ultrafiltration device with a 10-kD cut-off filter. The concentrates of the two galactanase variants were at least 95% pure as estimated by SDS-PAGE.
pH profiles of AAGAL D182N and MTGAL A90S/H91D and the corresponding wild-type galactanases were measured as described above for MTGAL and HIGAL.
Crystallization
Crystallization was carried out in hanging drops by vapor diffusion at room temperature. MTGAL was crystallized in two nonisomorphous crystal forms with similar cell dimensions and the same space group C2 containing four molecules in the asymmetric unit. The packing of three of the molecules is similar in the two crystal forms (although orientations differ), but the packing of the fourth molecule is different.
The final structures presented here were refined against data collected on crystals grown in 32% PEG 4000, 0.2 M ammonium sulphate, 0.1 M TRIS, pH 8.5 and 30% PEG 4000, 0.2 M ammonium sulphate, 0.1 M HEPES pH 7.5, representing the two different crystal forms, respectively. The structures are referred to as MTGAL (TRIS) and MTGAL (HEPES), respectively. A droplet of 2 μL protein solution (11 mg/mL) and 4 μL of reservoir solution was mixed and equilibrated against 0.5 mL of reservoir solution. For HIGAL, a 4 μL drop containing the protein solution (18 mg/mL) and the precipitant solution in a 1:1 volume ratio was equilibrated against 1 mL of the precipitant solution. The crystals grew in 0.1 M sodium cacodylate, pH 6.5 and 1.4 M sodium acetate trihydrate. HIGAL crystallizes in the orthorhombic space group P212121 with one molecule in the asymmetric unit.
Data collection
A data set was measured for HIGAL and each of the two MTGAL crystal forms on a cryo-cooled (100K) crystal at beamline I 711 (MAXLAB; Cerenius et al. 2000). Mother liquor containing 15% glycerol was used as cryoprotectant. The data were collected with a MAR 345 imaging plate detector system. The data sets were processed using the programs DENZO and SCALEPACK (Otwinowski and Minor 1997). Details of all data collection statistics are in Table 2.
Table 2.
Data collection statistics
| Data set | HIGAL | MTGAL (TRIS) | MTGAL (HEPES) |
| Space group | P212121 | C2 | C2 |
| Cell dimensions (Å) | 45.25, 68.95, 135.16 | 107.99, 136.58, 110.88 | 108.375, 136.145, 111.01 |
| Cell angles (°) | 90, 90, 90 | 90, 118.9, 90 | 90, 119.0, 90 |
| Mol/asym.unit | 1 | 4 | 4 |
| Resolution range (Å) | 20–2.55 (2.61–2.55)a | 18–2.14 (2.22–2.14) | 20–1.88 (1.95–1.88) |
| Unique reflections | 13628 | 76436 | 114360 |
| Completeness overall (%) | 94.6 (90.8) | 99.9 (99.6) | 97.6 (89) |
| Overall I/σ(I) >2 (%) | 77 (55.9) | 90.4 (76.2) | 84.4 (61.3) |
| Overall Rsym (%) | 14.2 (44.7) | 7.6 (21.1) | 7.6 (41.7) |
a Numbers in parentheses refer to the outer resolution shell.
Structure determination and refinement
The structure of MTGAL was originally determined by molecular replacement using AMoRe (Navaza 1994) from a crystal grown using a reservoir containing 30% PEG 4000, 0.2 M ammonium sulphate, 0.1 M HEPES, pH 7.5, and using the AAGAL structure determined at room temperature (Ryttersgaard et al. 2002) as a search model. After placing of four molecules in the asymmetric unit and rigid body refinement, the correlation coefficient (F) and the R-factor were 63.2% and 36.5%, respectively. The necessary amino acid substitutions were made and the structure was refined. However, the refinement statistics obtained were not satisfactory, with notably abnormal B-factor values beyond the expected physical range. After several tests, the individual B-factors were not refined and one overall anisotropic B-factor was fixed.
Later, another crystal grown in similar conditions was found not to be isomorphous despite similar cell parameters. The structure was determined by molecular replacement using AMoRe (Navaza 1994) and the first MTGAL structure as model. The correlation coefficient (F) was 74.6% and the R-factor 27.7%. However, as data were collected only to 2.8 Å resolution, this model was not refined.
New data were therefore necessary for accurate refinement. For the two final data sets of the two MTGAL crystal forms, the starting points for refinement were the original structures determined by Molecular Replacement with AMoRe. The structure of HIGAL has been determined by molecular replacement using the program EPMR (Kissinger et al. 1999) and one molecule of MTGAL as a search model. A correlation coefficient (F) of 61.1% and an R-factor of 40.5% were obtained for the correct solution. Mutations from the model to target sequence were made in O (Jones et al. 1991), which was also used for manual rebuilding against the electron density maps.
The refinement was carried out by the CNS package version 1.1 (Brünger et al. 1998), and refinement summaries are presented in Table 3. Atomic B-factors have been refined. Restrained NCS was used except for a few side chains during MTGAL refinement. The water molecules were added automatically in CNS according to the criterion that they had a peak height above 3.0 σ in the Fobs-Fcalc map and above 1.5 σ in the 2Fobs-Fcalc electron density map, at least one hydrogen bond between water and any atoms in the protein, and a B factor below 50Å2. Six double conformations were modeled in the MTGAL (TRIS) and four in the MTGAL (HEPES) model. In the MTGAL (HEPES) model, a HEPES molecule from the crystallization conditions was also refined in one of the four molecules in the asymmetric unit (molecule A), and several diethylene glycol molecules were added throughout. A TRIS molecule from the crystallization mixture was found in the active site of MTGAL (TRIS; Fig. 2 ▶) in two of four molecules (A and D) in the asymmetric unit. The Fo-Fc difference density map revealed an N-glycosylation site at Asn 111 in the MTGAL structures in both crystal forms and in the HIGAL structure, which enabled us to insert a 2-N-acetyl-β-D-glucose unit in each molecule.
Table 3.
Refinement summary
| HIGAL | MTGAL (TRIS) | MTGAL (HEPES) | |
| Resolution range (Å) | 20–2.55 | 18–2.14 | 20–1.88 |
| R-factor | 0.202 | 0.185 | 0.194 |
| Rfreea | 0.250 | 0.209 | 0.208 |
| Protein atomsb | 2665 | 10416 | 10416 |
| Water molecules | 131 | 988 | 860 |
| Heteroatoms | |||
| Sulphate ion | 0 | 50 | 55 |
| TRIS | 0 | 16 | 0 |
| Diethylene glycol | 0 | 28 | 91 |
| GlcNAc | 14 | 56 | 56 |
| HEPES | 0 | 0 | 15 |
| RMSD | |||
| Bond lengths (Å) | 0.006 | 0.006 | 0.005 |
| Bond angles (°) | 1.3 | 1.3 | 1.3 |
| Average B-factor (Å2) | |||
| Main chain | 18.2 | 20.2 | 16.8 |
| Side chains | 18.8 | 21.2 | 18.0 |
| Water molecules | 19.0 | 22.6 | 18.2 |
| Double conformations modeled | 0 | 6 | 4 |
| Ramachandran plotc (% residues) | |||
| In most favorable regions | 86.2 | 86 | 85.9 |
| In allowed regions | 13.1 | 13.7 | 14.0 |
| In generously allowed regions | 0.7 | 0.3 | 0.1 |
a Rfree calculated using 10% of the reflections.
b Nonhydrogen protein atoms inserted in the model/asymmetric unit.
c Calculated by PROCHECK (Laskowski et al. 1993).
The structures were deposited in the PDB with codes 1hjq (HIGAL), 1hjs (MTGAL HEPES), and 1hju (MTGAL TRIS).
Structural and composition analysis
The analysis of α-helix stabilization considered only helix dipole stabilization by charged residues and was performed as described by Teixeira et al. (2001) and in the legend to Figure 4 ▶.
The PROMOTIF (Hutchinson and Thornton 1996) program was used to determine secondary structural elements, and only helices belonging to (β/α)8 barrel were considered.
The NACCESS (Hubbard and Thornton 1993) program was used with a probe of 1.4 Å to determine the number of accessible atoms (atoms with accessible surface larger than 0 Å2) in the three structures. An average value was calculated with respect to MTGAL, which contains four molecules in the asymmetric unit and has been determined in two crystal forms.
CNS version 1.1 was used to obtain lists of the interactions at less than 4 Å and 6 Å between side chain atoms of Asp and Glu and side chains of Arg, Lys, and His. The number of ion pairs was calculated for these two cut-offs.
The HBPLUS (McDonald and Thornton 1994) program was used to identify the number and the type of hydrogen bonds in the three enzymes.
The VOIDOO program http://alpha2.bmc.uu.se/~gerard/manuals/voidoo_man.html; Kleywegt et al. 2001) with a probe of 1.2 Å was used to determine the number of cavities in the three structures. The CAPTURE (Gallivan and Dougherty 1999) program was used to evaluate the number of energetically significant interactions between cationic residues (Arg and Lys) and π electrons containing residues (Trp, Tyr, and Phe). The structure-based sequence alignment was performed using the program Indonesia (D. Madsen and G. Kleywegt, in prep.), and PROCHECK (Laskowski et al. 1993) and WHATIF (Vriend 1990) were used to check the quality of the crystallographic models.
The B-factor distribution was calculated with the CCP4 program Baverage (Collaborative Computational Project, Number 4, 1994). The amino acids composition was calculated by submitting the three sequences to http://us.expasy.org/tools/protparam.html.
Acknowledgments
We thank Eva Johansson and Flemming Hansen for help with data collection, and MAX-LAB, Lund, Sweden, for provision of synchrotron time. We are grateful to Dansync and the European Union for contribution to travel expenses under the ‘Access to Research Infrastructures’ program. We also thank Michael McDonough for fruitful discussions on thermophilic adaptation. The research is funded by the Danish National Research Foundation. Initial crystallization trials of the Humicola insolens galactanase were performed by Vibeke Jorgensen as a student in the Ph.D. Course in Protein Crystallography held at the University of Copenhagen.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
RG-I, rhamnogalacturonan I
AAGAL, Aspergillus aculeatus galactanase
MTGAL, Myceliophthora thermophila galactanase
HIGAL, Humicola insolens galactanase
PEG, polyethyleneglycol
HEPES, 4-(2-hydroxyethyl)-1-1-piperazineethane sulfonic acid
TRIS, tris (hydroxymethyl) aminomethane
GlcNac, 2-N-acetyl-β-D-glucose
SC, Simmon’s citrate
AZCL, Azurine-Cross linked
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0300103.
References
- Aghajari, N., Feller, G., Gerday, C., and Haser, R. 1998. Structures of the psychrophilic Alteromonas haloplanctis α-amylase give insights into cold adaptation at a molecular level. Structure 6 1503–1516. [DOI] [PubMed] [Google Scholar]
- Argos, P., Rossmann, M., Grau, U., Zuber, H., Frank, G., and Traschin, J. 1979. Thermal stability and protein structure. Biochemistry 25 5698–5703. [DOI] [PubMed] [Google Scholar]
- Braithwaite, K.L., Barna, T., Spurway, T.D., Charnock, S.J., Black, G.W., Hughes, N., Lakey, J.H., Virden, R., Hazlewood, G.P., Henrissat, B., et al. 1997. Evidence that galactanase A from Pseudomonas fluorescens subspecies cellulosa is a retaining family 53 glycosyl hydrolase in which E161 and E270 are the catalytic residues. Biochemistry 36 15489–15500. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54 905–921. [DOI] [PubMed] [Google Scholar]
- Cerenius, Y., Ståhl, K., Svensson, A., Ursby, T., Oskarsson, Å., Albertsson, J., and Liljas, A. 2000. The crystallography beamline I711 at MAX II. J. Synchrotron Radiat. 7 203–208. [DOI] [PubMed] [Google Scholar]
- Christensen, T., Wöldike, H., Boel, E., Mortensen, S.B., Hjortshøj, K., Thim, L., and Hansen, M.T. 1988. High level expression of recombinant genes in Aspergillus oryzae. Biotechnology 6 1419–1422. [Google Scholar]
- Christgau, S., Sandal, T., Kofod, L.V., and Dalbøge, H. 1995. Expression cloning, purification and characterization of a β-1,4-galactanase from Aspergillus aculeatus. Curr. Genet. 27 135–141. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. 1994. The CCP4 Suite: Programs for protein crystallography. Acta Crystallogr. D 50 760–763. [DOI] [PubMed] [Google Scholar]
- Couthino, P.M. and Henrissat, B. 1999. Carbohydrate-active enzymes: An integrated database approach. In Recent advances in carbohydrate bioengineering (eds. H.J. Gilbert, G. Davies, et al.), pp. 3–12. The Royal Society of Chemistry, Cambridge, UK.
- Dalbøge, H. and Heldt-Hansen, H. 1994. A novel method for efficient expression cloning of fungal enzyme genes. Mol. Gen. Genet. 243 253–260. [DOI] [PubMed] [Google Scholar]
- de Vries, R.P. and Visser, J. 2001. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Rev. 65 497–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallivan, J.P. and Dougherty, D.A. 1999. Cations-π interactions in structural biology. Proc. Natl. Acad. Sci. 96 9459–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. A computational study of cation-π interactions vs. salt bridges in aqueous media: Implications for protein engineering. J. Am. Chem. Soc. 122 870–874. [Google Scholar]
- Guex, N. and Peitsch, M.C. 1997. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 18 2714–2723. [DOI] [PubMed] [Google Scholar]
- Henrissat, B., Callebaut, I., Fabrega, S., Lehn, P., Mornon, J.-P., and Davies, G. 1995. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc. Natl. Acad. Sci. 92 7090–7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard, S.J. and Thornton, J.M. 1993. “NACCESS” computer program. Department of Biochemistry and Molecular Biology, University College, London.
- Hutchinson, E.J. and Thornton, J.M. 1996. PROMOTIF—A program to identify and analyze structural motifs in proteins. Protein Sci. 5 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, J., Lo Leggio, L., Harris, G., and Pickersgill, R.W. 1995. β-Glucosidase, β-galactosidase, family A cellulases, family F xylanases and two barley glucanases form a superfamily of enzymes with 8-fold β/α architecture and with two conserved glutamates near the carboxy-terminal ends of β-strands four and seven. FEBS Lett. 362 281–285. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J., Cowan, S., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Joshi, M.D., Sidhu, G., Pot, I., Brayer, G.D., Withers, S.G., and McIntosh, L.P. 2000. Hydrogen bonding and catalysis: A novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase. J. Mol. Biol. 299 255–279. [DOI] [PubMed] [Google Scholar]
- Joshi, M.D., Sidhu, G., Nielsen, J.E., Brayer, G.D., Withers, S.G., and McIntosh, L.P. 2001. Dissecting the electrostatic interactions and pH-dependant activity of a family 11 Glycosidase. Biochemistry 40 10115–10139. [PubMed] [Google Scholar]
- Kissinger, C.R., Gehlaar, D.K., and Fogel, D.B. 1999. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D 55 484–491. [DOI] [PubMed] [Google Scholar]
- Kleywegt, G.J., Zou, J.Y., Kjeldgaard, M., and Jones, T.A. 2001. Around O. In International tables for crystallography (eds. M.G. Rossmann, E. Arnold), Vol. F, pp. 353–356, 366–367. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK—A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 24 283–291. [Google Scholar]
- Lau, J.M., Mc Neil, M., Darvill, A.G., and Albersheim, P. 1985. Structure of the backbone of rhamnogalacturonan I, a pectic polysaccharide in the primary cell walls of plants. Carbohydr. Res. 137 111–125. [Google Scholar]
- Lo Leggio, L., Kalogiannis, S., Bhat, M.K., and Pickersgill, R.W. 1999. High resolution structure and sequence of T. aurantiacus xylanase I: Implications for the evolution of thermostability in family 10 xylanases and enzymes with βα-barrel architecture. Proteins 36 295–306. [PubMed] [Google Scholar]
- Matthews, B.W., Weaver, L.H., and Kester, W.R. 1974. The conformation of thermolysin. J. Biol. Chem. 249 8030–8044. [PubMed] [Google Scholar]
- Matthews, B.W., Nicholson, H., and Becktel, W.J. 1987. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc. Natl. Acad. Sci. 84 6663–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc Carter, J.D. and Withers, S.G. 1994. Mechanisms of enzymatic glycoside hydrolysis. Curr. Opin. Struct. Biol. 4 885–892. [DOI] [PubMed] [Google Scholar]
- Mc Donald, I.K. and Thornton, J.M. 1994. Satisfying hydrogen-bonding potential in proteins. J. Mol. Biol. 238 777–793. [DOI] [PubMed] [Google Scholar]
- Nagano, N., Orengo, C.A., and Thornton, J.M. 2002. One fold with many functions: The evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 321 741–765. [DOI] [PubMed] [Google Scholar]
- O’Neill, M., Albersheim, P., and Darvill, A.G. 1990. The pectic polysaccharides of primary cell walls, pp. 415–422. Academic Press Limited, London.
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Querol, E., Perez-Pons, J.A., and Mozo-Villarias, A. 1996. Analysis of protein conformational characteristics related to thermostability. Protein Eng. 9 265–271. [DOI] [PubMed] [Google Scholar]
- Ryttersgaard, C., Lo Leggio, L., Coutinho, P.M., Henrissat, B., and Larsen, S. 2002. Aspergillus aculeatus β-1,4-galactanase: Substrate recognition and relation to other glycosides hydrolases in clan GH-A. Biochemistry 41 15135–15143. [DOI] [PubMed] [Google Scholar]
- Ryttersgaard, C., Poulsen, J.-C.N., Christgau, S., Sandhal, T., Dalbøge, H., and Larsen, S. 1999. Crystallization and preliminary X-ray studies of β1,4-galactanase from Aspergillus aculeatus. Acta Crystallogr. D 55 929–930. [DOI] [PubMed] [Google Scholar]
- Sarkar, G. and Sommer, S.S. 1990. The “Megaprimer” method of site-directed mutagenesis. BioTechniques 8 404–407. [PubMed] [Google Scholar]
- Shirai, T., Ishida, H., Noda, J.I., Yamane, T., Ozaki, K., Hakamada, Y., and Ito, S. 2001. Crystal structure of alkaline cellulase K: Insight into the alkaline adaptation of an industrial enzyme. J. Mol. Biol. 310 1079– 1087. [DOI] [PubMed] [Google Scholar]
- Szilágyi, A. and Závodszky, P. 2000. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: Results of a comprehensive survey. Structure 8 493–504. [DOI] [PubMed] [Google Scholar]
- Teixeira, S., Lo Leggio, L., Pickersgill, R.W., and Cardin, C. 2001. Anisotropic refinement of the structure of Thermoascus aurantiacus xylanase I. Acta Crystallogr. D 57 385–392. [DOI] [PubMed] [Google Scholar]
- Vogt, G., Woell, S., and Argos, P. 1997. Protein thermal stability, hydrogen bonds, and ion pairs. J. Mol. Biol. 269 631–643. [DOI] [PubMed] [Google Scholar]
- Vriend, G. 1990. A molecular modeling and drug design program. J. Mol. Graph. 8 52–56. [DOI] [PubMed] [Google Scholar]