Abstract
It was established previously that helical propensities of different amino acid residues in the middle of α-helix in peptides and in proteins are very similar. The statistical analysis of the protein helices from the known three-dimensional structures shows no difference in the frequency of noncharged residues in the middle and at the C terminus. Yet, experimental studies show distinctive differences for the helical propensities of noncharged residues in the middle and in the C terminus in model peptides. Is this a general effect, and is it applicable to protein helices or is it specific to the model alanine-based peptides? To answer this question, the effects of substitutions at positions 28 (middle residue) and 32 (C2 position at the C terminus) of the α-helix of ubiquitin on the stability of this protein are measured by using differential scanning calorimetry. The two data sets produce similar values for intrinsic helix propensity, leading to a conclusion that noncharged amino acid residues at the solvent-exposed positions in the middle and at the C terminus of the α-helix have the same helical propensity. This conclusion is further supported with an excellent correlation between the helix propensity scale obtained for the two positions in ubiquitin with the experimental helix propensity scale established previously and with the statistical distribution of the residues in protein helices.
Keywords: α-Helix, C-capping, stability, thermodynamic propensity, differential scanning calorimetry
The structure of the α-helix is characterized by hydrogen bonding patterns between amide hydrogen bond donors and carbonyl oxygen acceptors of residues situated four apart in sequence. This pattern of hydrogen bonding, however, implies that the four initial amide hydrogen bond donors and the last four carbonyl oxygen hydrogen bond acceptors do not have hydrogen bonding partners. The potential effect of this is fraying of the helix ends. Stereochemical (Presta and Rose 1988) and statistical (Richardson and Richardson 1988) analyses of the amino acid residues at the ends of α-helices revealed the existence of the specific capping interactions at both the N and C termini, which compensate for the unsatisfied hydrogen bonds and thus prevent ends fraying. According to Aurora and Rose (1998), amino acid residues in helices and their flanking residues can be labeled as follows:
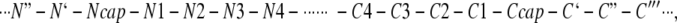 |
where numbered residues have helical backbone dihedral angles (φ = −64 ± 7 degrees; ϕ = −41 ± 7 degrees). Ncap with dihedrals φ = −94 ± 15 degrees and ϕ = 167 ± 5 degrees and Ccap with dihedrals φ = −90 ± 10 degrees and ϕ = −20 ± 15 degrees are boundary residues that belong both to the helix and to the adjacent turn (Aurora and Rose 1998). At the N termini, the capping motif consists of six residues interacting via hydrogen bonding and a hydrophobic contact (Richardson and Richardson 1988; Seale et al. 1994; Aurora and Rose 1998). Hydrogen bonding at the N termini involves residues Ncap and N2 or N3. Interactions at the C termini of α-helices have been divided into several distinct motifs (Schellman 1980; Preissner and Bork 1991; Aurora et al. 1994). Of these, the most frequent ones are those that terminate with glycine in position C`. These C-capping motifs are characterized by hydrogen bonding of the backbone amides of C` and/or C" to carbonyl oxygens of residues in C3 and/or C2, respectively. In addition, there are hydrophobic interactions between side-chains of residues in positions C" and C3 or C" and C3/C4, with Ccap frequently providing an additional hydrophobic interaction. Detailed studies of the effects of different amino acid substitutions at C`, Ccap, and C4/C" positions in the α-helix of the model protein ubiquitin, revealed the importance of different factors at each position (Thomas and Makhatadze 2000; Thomas et al. 2001; Ermolenko et al. 2002). For example, the propensity of residues at the C` position is defined by the hydration of peptide backbone (Serrano et al. 1992; Thomas et al. 2001), whereas the propensity of residues at the Ccap is largely defined by their hydrophobicity (Ermolenko et al. 2002).
It is not clear yet, however, what is the role of C2 and C1 positions in the C-capping motif. On one hand, because these residues are located within the helix, one can expect that they have helical propensities. Indeed, the statistical analysis of the helix ends does not find significant change in frequency of occurrences of different amino acid residues at the C2 and C1 positions relative to the middle of the α-helix (Aurora and Rose 1998; Kumar and Bansal 1998; Penel et al. 1999b). The exceptions are charged residues that are expected to interact with the helix dipole. These interactions will be favorable for the basic residues and unfavorable for the acidic residues (Blagdon and Goodman 1975; Wada 1976; Hol et al. 1978; Nicholson et al. 1991). On the other hand, these positions C2 and C1 are located in the last turn of the helix and might have somewhat different intrinsic helical propensities than do the residues in the middle of the helix proper. This idea is supported by the experimental data of Serrano’s group, which showed significant position dependence of the intrinsic helical propensities of the residues at the C terminus of alanine-based peptides (Petukhov et al. 2002).
Our group has a long-standing interest in deciphering the rules for helix termination and, in particular, for the interactions at the C terminus of α-helices (Thomas and Makhatadze 2000; Thomas et al. 2001; Ermolenko et al. 2002). We thus decided to directly test the helix propensity of different amino acid residues at the C terminus of an α-helix and compare it with the helix propensity in the middle of the same helix, as well as with the existing helix propensity scales (Lyu et al. 1990; O’Neil and DeGrado 1990; Horovitz et al. 1992; Blaber et al. 1993, 1994; Park et al. 1993; Chakrabartty and Baldwin 1995; Rohl et al. 1996; Myers et al. 1997a,b; Yang et al. 1997). For these experiments, we chose two solvent-exposed positions in the α-helix of ubiquitin: position 28, which is located in the middle of helix spanning residues 24–34, and position 32, which is located in the last turn of this α-helix and can be classified as C2 position of C-capping motif that includes residues 30–36. Over two dozen ubiquitin variants with substitutions at these two positions were generated, and their stabilities were measured by using differential scanning calorimetry (DSC). Analysis of the obtained thermodynamic data and their comparison with the experimental and statistical data reported previously allowed us to conclude that there is no difference in the intrinsic helical propensity of the noncharged residues at the solvent-exposed positions in the middle and in the C terminus of α-helices.
Results
α-Helix of ubiquitin and the design of substitutions
Figure 1 ▶ shows the overall structure of the ubiquitin molecule. It consists of five β-strands forming a somewhat concave surface (Vijay-Kumar et al. 1987a,b). An α-helix that includes residues 23–34 lies across the β-sheet part of the molecule. This α-helix is composed of three full turns and has both N-capping and C-capping interactions. The C-capping motif of the α-helix of ubiquitin has the backbone–backbone hydrogen bond between residues C`(G35) and C3(31) and van der Waals interactions between residues C4(I30) and C"(I36). Position 28, occupied by Ser residue in the wild-type yeast ubiquitin, is located on the solvent-exposed side of the helix. Position 32, which is C2 position of the C-capping motif, is also located on the solvent-exposed side of this helix. Thus, we used these two sites in the α-helix of ubiquitin to compare helical propensity of residues in the middle (position 28) with the helical propensities at the C terminus (position 32). Five amino acid substitutions were made at position 28, and 11 amino acid substitutions were incorporated into the position 32.
Figure 1.
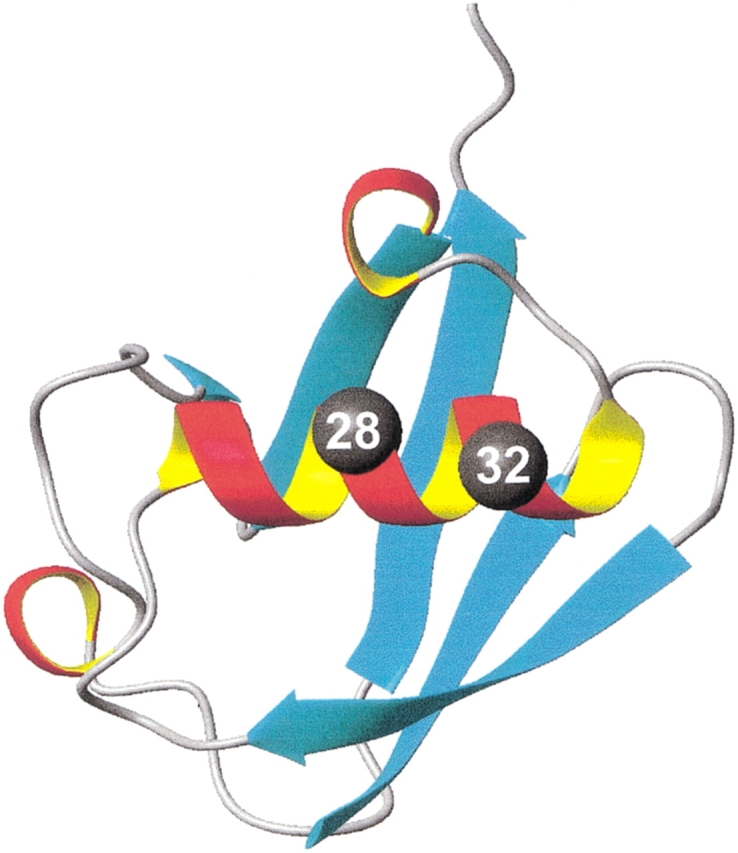
Representation of the three-dimensional structure of the ubiquitin molecule, showing the location of the position 28 (middle of α-helix) and position 32 (C2 position at the C terminus of the α-helix).
The five amino acids substituted into position 28 (G, A, S, V, I) were chosen for two reasons. First, these amino acid residues have helical propensity, as judged by well-established helical propensity scales for the residues in the middle of α-helix (see Pace and Scholtz 1998) that span the entire range, including the residue with the highest helix propensity, alanine, and the residue with the lowest (after proline) helix propensity, glycine. Second, these amino acid residues have been used by Petukhov et al. (2002) in their study of position dependence of helical propensities in the model helical peptides.
The 11 variants of ubiquitin with the amino acid substitutions at the C2 position were A, F, G, I, L, M, N, Q, S, T, and V. The ionizable amino acid residues lysine, arginine, histidine, aspartate, and glutamate were excluded because of the potential interaction between charged side-chains and the helix dipole. Indeed, from the previous extensive work of a number of groups, it is clear that positively charged side-chains at the C terminus would stabilize the helix structure through the interaction with the helix macro-dipole (Blagdon and Goodman 1975; Wada 1976; Hol et al. 1978; Nicholson et al. 1991). The effect of negatively charged residues will be just the opposite. Thus, this selected set of 11 amino acid residues is representative of different chemical nature, size, shape, and helical propensities of naturally occurring amino acid residues.
Thermodynamic stability of ubiquitin variants
Stabilities of the ubiquitin variants with the substitutions in positions 28 or 32 were evaluated by using DSC. The detailed thermodynamic data obtained from these DSC experiments are reported in three tables supplied as an electronic supplement.
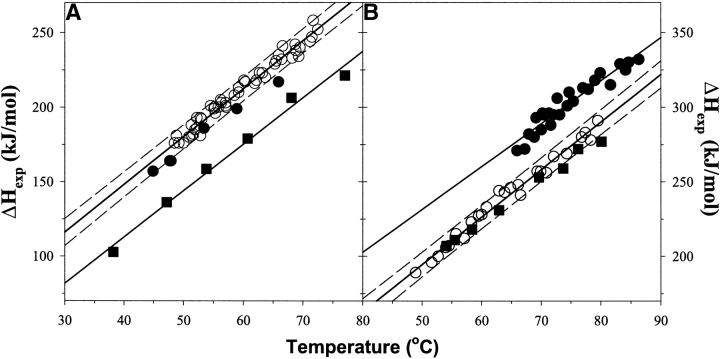
The substitutions in position 32 were generated in the background of WTAAA (K11A/K33A/E34A). These substitutions were introduced to remove potential side-chain/side-chain interactions with the neighboring sites, which can affect the intrinsic propensity. Moreover, these substitutions relieve high electrostatic potential around this site, which also can obscure intrinsic propensities. The WTAAA was previously characterized by NMR and shown to have the HSQC spectrum identical to that of the wild-type yeast ubiquitin (Ermolenko et al., 2002). The WTAAA variant is ∼13°C more thermostable than is the wild-type protein. This is expected, keeping in mind that the positions 32 and 33 are helical residues and alanine is the most helix-favoring residue. Furthermore, the amino acid preference in position 34 is defined by hydrophobic interactions, and alanine is more hydrophobic than is the wild-type glutamate (Ermolenko et al. 2002). What is less expected is a significant increase in enthalpy of unfolding. Figure 2A ▶ compares the temperature dependencies of the enthalpies of unfolding on the transition temperature. Clearly WTAAA and all the substitutions at position 32 have 35 ± 10 kJ/mole higher enthalpy of unfolding. It is also important to note that there is no change in the temperature dependencies of the enthalpy functions, indicating similar values for the heat capacity changes, ΔCp, upon unfolding. Interestingly, as can be seen from Figure 2A ▶, the G32 variant has somewhat lower enthalpies of unfolding than does the rest of position 32 substitutions; however, a definitive conclusion about it cannot be made at this time because this difference is just outside the experimental error. One possible explanation for the increase of enthalpy of unfolding for WTAAA is the observation by Luo and Baldwin (1999) that the enthalpy of helix coil transition for alanine residues is higher than that for any other residue. This explanation is further supported by the effects of the alanine substitutions in the middle of α-helix on the enthalpy of unfolding, which are discussed two paragraphs below.
Figure 2.
Temperature dependence of the enthalpy of unfolding for the studied ubiquitin variants. (A) (Squares) WT, (open circles) all but G32 substitutions in WTAAA background, and (filled circles) G32 substitution in WTAAA background. (B) (Squares) WT#, (filled circles) position 28 substitutions in the WT#4A background, and (open circles) position 28 substitutions in the WT#4AV5A background. Solid lines represent the linear fit with the slope that represents the heat capacity change upon unfolding, ΔCp = 3.2 ± 0.3 kJ/(K•mole); dashed lines show errors at the 95% confidence.
Table 1 summarizes the effects of amino acid substitutions on the Gibbs energy changes relative to alanine of 11 position 32 variants of ubiquitin. As one would expect from the known helix propensity scales, G32 is the most destabilizing substitution in the studied set of the ubiquitin variants. Addition of the side-chain leads to an increase in the stability relative to the G32 variant. The obtained rank order of the residues is G < N < T < F < S < V < I < L < M < A < Q. Higher stability of Q32 variant over A32 is a particularly interesting observation for two reasons. First, neither asparagine (similar in chemical nature to glutamine) nor leucine (similar in the overall shape to glutamine) in this position is stabilizing relative to alanine. Second, the propensity scales derived for the middle of an α-helix, in most cases, assign alanine with the highest helix propensity (see Pace and Scholtz 1998), and thus, the higher stability of Q32 variant might be specific for the C terminus of α-helix. Position 28 of ubiquitin, which is an i − 4 for residue 32, is occupied by a serine. Thus, there might be a potential side-chain/side-chain interaction of S28 and Q32. To explore this, the stabilities of ubiquitin variants with A32 and Q32 substitutions in the background of A28 were measured. In this background, the Q32/A28 variant is actually 0.4 kJ/mole less stable than the A32/A28 variant, thus confirming the side-chain/side-chain interactions between Q32 and S28. Therefore, the value of −0.4 kJ/mole for Q32 was used for further analysis.
Table 1.
Thermodynamic propensity of different amino acid residues at the helical positions
| Amino acid residue | ΔΔG(X32) | ΔΔG(X28) | ΔΔG(X28/V5A) | ΔΔG(P&S) | ΔΔG(PHD)/Pg(C2) |
| A | 0.0 ± 0.2 | 0.0 ± 0.2 | 0.0 ± 0.2 | 0.0 ± 0.5 | 0.0/1.46 |
| F | −2.4 ± 0.4 | — | — | −2.3 ± 0.5 | −1.7/0.74 |
| G | −4.3 ± 0.4 | −4.4 ± 0.3 | −4.2 ± 0.1 | −4.2 ± 0.5 | −4.1/0.28 |
| I | −0.9 ± 0.2 | −1.0 ± 0.2 | −1.0 ± 0.2 | −1.7 ± 0.5 | −0.7/1.12 |
| L | −0.6 ± 0.2 | — | — | −0.9 ± 0.2 | −0.3/1.31 |
| M | −0.4 ± 0.1 | — | — | −1.0 ± 0.3 | −0.4/1.24 |
| N | −2.8 ± 0.3 | — | — | −2.7 ± 0.4 | −2.2/0.60 |
| Q | 0.1 ± 0.1 (−0.4 ± 0.1)a | — | — | −1.6 ± 0.4 | −0.3/1.29 |
| S | −2.0 ± 0.1 | −2.7 ± 0.3 | −2.3 ± 0.1 | −2.1 ± 0.3 | −1.8/0.71 |
| T | −2.5 ± 0.1 | — | — | −2.8 ± 0.4 | −1.9/0.69 |
| V | −1.7 ± 0.4 | −2.0 ± 0.1 | −1.7 ± 01 | −2.6 ± 0.5 | −1.2/0.89 |
All ΔΔG values are relative to alanine and are in kilojoules per mole. ΔΔG(PHD) = −R • T • ln[Pg(X)/Pg(A)], where the normalized frequency for individual amino acid residues in position C2 of α-helices, Pg, was calculated from the data reported in Penel et al. (1999a,b); ΔΔG(P&S) values of helical propensity for the middle of α-helix are taken from Pace and Scholtz (1998).
a Changes in ΔΔG relative to A32 in the background of A28 variant.
The obtained propensities at position 32 can be compared with the helix propensity scales derived from the experimental studies of other systems. However, to eliminate all potential issues when comparing the results obtained by using different model systems and different experimental methods to determine the changes in stability, we measured the stabilities of selected amino acid residues at a solvent-exposed site in the middle of α-helix of ubiquitin. Five amino acid residues–G, S, V, I, and A–were incorporated to the position 28 of the ubiquitin. These amino acid residues were selected based on their established helical propensity to cover the entire range from lowest propensity (glycine) to intermediate (serine, valine, and isoleucine) to highest (alanine). To avoid any possible side-chain/side-chain interactions, the residues in positions i − 3, i − 4, i + 3, and i + 4 were substituted to alanine (i.e., D24A/N25A/Q31A/D32A in the WT# background containing stabilizing substitution R42E [Loladze et al. 2002] and designated as WT#4A). These substitutions resulted in an increase in stability of the ubiquitin molecule by ∼15°C relative to the WT# ubiquitin. However, what is more interesting (Figure 2B ▶), WT#4A and all the substitutions in the position 28 of WT#4A also resulted in an increase in the enthalpy of unfolding by 40 ± 10 kJ/mole, without noticeable change in the temperature dependence of the enthalpy function, indicating similar values for the heat capacity changes, ΔCp, upon unfolding. The similarities with the case of WTAAA background used for position 32 substitutions are obvious: Inclusion of alanine residues into α-helix leads to an increase in the enthalpy of unfolding in accord with the previously suggested effects by Luo and Baldwin (1999). These effects are currently under further investigation in the laboratory by using ubiquitin and helical peptides as model systems. The changes in the thermodynamic properties of the background in which substitutions were made might influence the relative changes in stability of different residues in position 28. To estimate whether increase in the thermostability and in the enthalpy of unfolding of the WT#4A ubiquitin background can have an effect on the intrinsic helical propensities, the effects of substitutions in position 28 on stability were also measured in the WT#4AV5A background that contains an additional V5A substitution. The V5A substitution is located on the opposite side from the position 28 of the ubiquitin molecule and was shown to lead to a decrease in stability by ∼15°C and, more importantly, to a decrease in the enthalpy of unfolding by 25±10 kJ/mole (Loladze et al. 2002). Both of these effects, decrease in the transition temperature and decrease in the enthalpy of unfolding, are well reproduced in the WT#4AV5A relative to the WT#4A (Fig. 2B ▶). The stabilities of five substitutions in position 28 in these two backgrounds relative to alanine are compared in Table 1. As it can be seen, the similarities in the effects of same amino acid substitutions in different backgrounds on the stability are remarkable and, importantly, establish the experimental error in determining the thermodynamic helix propensity.
Discussion
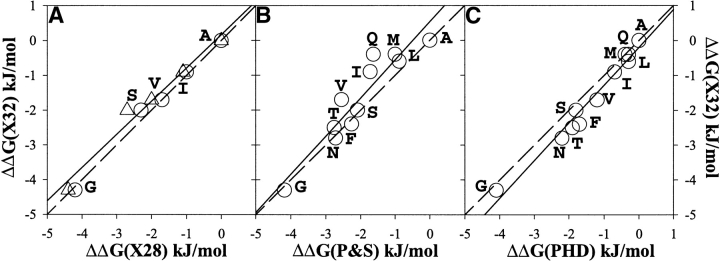
The aim of this work is to establish whether there is a difference in the intrinsic helix propensity between residues located in the middle and in the C-terminal end of the solvent-exposed positions of an α-helix in a protein. The α-helix of ubiquitin is used as a model system for these experiments. Figure 3A ▶ shows the correlation between the effects of different amino acid substitutions at positions 32 and 28 of the α-helix of ubiquitin. Position 28 is located on the solvent-exposed side in the middle of the only α-helix spanning residues from 24 to 34. Position 32 is also located on the solvent-exposed side, but in the last turn of the α-helix. The correspondence of the effects of same amino acid substitutions on the stability of ubiquitin at these two different positions is remarkable. The correlation coefficient R is 0.99 with the slope of 0.95. This result indicates that there is no difference in the intrinsic helical propensity for residues in the middle and in the C terminus of an α-helix. Further support for this conclusion comes from the comparison of the propensities in position 32 with the experimentally derived helix propensity scale for position in the middle of the α-helix by Pace and Scholtz (1998). The correlation analysis is shown in Figure 3B ▶. The correlation coefficient of 0.93 and the slope of 1.05 clearly indicate that there is no dramatic difference in the intrinsic helix propensity of residues in the middle and in the C terminus of the α-helix in proteins.
Figure 3.
Correlation between experimentally measured propensities of noncharged residues in the C2 position at the C terminus of the α-helix of ubiquitin, ΔΔG(X32). (A) With the propensity in the middle of the α-helix of ubiquitin, ΔΔG(X28) in two different backgrounds WT4A (triangles) and WT4AV5A (circles). (B) With the experimentally derived thermodynamic propensity scale of Pace and Scholtz (1998), ΔΔG(P&S). (C) With the propensity of residues at the C2 position of α-helices derived from the statistical analysis of Penel et al. (1999b), ΔΔG(PHD). The solid lines show linear fits, and the dashed lines represent the perfect correlation with the slope of one. The calculated correlation coefficients and the slopes are, respectively, 0.95 and 0.99 for (A), 0.93 and 1.05 for (B), and 0.98 and 1.05 for (C).
These experimental results are also in accord with the statistical analysis of residue distribution in protein helices (Aurora and Rose 1998; Kumar and Bansal 1998; Penel et al. 1999b). Figure 3C ▶ shows the correlation of the helical propensity in position 32 with the normalized frequencies of amino acid residues in position C2 of protein helices (Penel et al. 1999a,b). The correlation coefficient (0.98) and the slope (1.05) are similar to those for experimental helix propensity scale of Pace and Scholtz (1998).
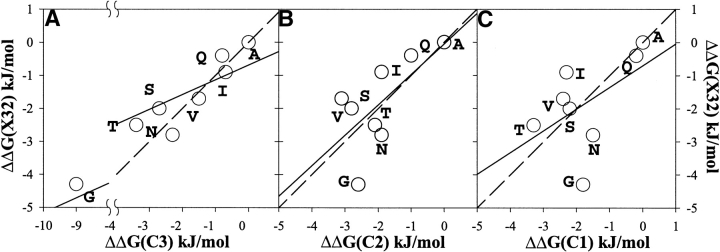
How do these results compare with the report by Petukhov et al. (2002) that there is significant positional dependence of the intrinsic helical propensity of residues? Figure 4, A–C ▶ , shows the correlation of the intrinsic helical propensity in position 32 of ubiquitin with the helical propensity at the last three positions of alanine-based helical peptide. Because, in the peptide, the end of helical segment is not defined, we compared our data with all three positions C3, C2, and C1. It should be noted that the meaning of C3, C2, and C1 in the context of helical peptides is different from their definition in protein helices. In helical peptides, C3, C2, and C1 denote the last three residues in the sequence, whereas in protein helices, C3, C2, and C1 are defined as the last three helical residues. The correlation coefficients of the propensity in position 32 with the propensities at C3, C2, and C1 positions are 0.90, 0.57, and 0.53, respectively. However, even in the case of C3 when correlation coefficient is high, the slope is only 0.44. The difference in slope is higher than is usually observed in the correlation of the effects of different amino acid substitutions in the middle of α-helix on the stability of proteins and alanine-based peptides (Pace and Scholtz 1998).
Figure 4.
Correlation between experimentally measured propensities of noncharged residues in the C2 position at the C terminus of the α-helix of ubiquitin, ΔΔG(X32), with the experimentally derived thermodynamic propensity scale at the three C-terminal residues of alanine-based peptide by Petukhov et al (2002): position C3 (A), position C2 (B), and position C1 (C). The solid lines show linear fits, and the dashed lines represent the perfect correlation with the slope of one. The calculated correlation coefficients and the slopes are, respectively, 0.82 and 0.43 for (A), 0.45 and 0.91 for (B), and 0.29 and 0.65 for (C).
What is a possible source for the differences between the experimental data on protein helices presented above and the experimental data obtained in model helical peptides of Petukhov et al. (2002)? The thermodynamic propensity scale of different amino acid residues in the middle of the helix was studied by several groups using different model systems, both peptide and protein based (Lyu et al. 1990; O’Neil and DeGrado 1990; Wojcik et al. 1990; Horovitz et al. 1992; Scholtz and Baldwin 1992; Blaber et al. 1993, 1994). However, it was not until Myers et al. (1997a,b) directly compared helix propensity in proteins and peptides by using the same 17-residue sequence, forming an α-helix in the structure of ribonuclease T1 and in isolated peptide, and showed identical propensities for a guest position in the middle of the helical segment. The conclusion that helix propensities are identical in proteins and peptides was important for justifying the use of short helical peptides as a model system for the study of thermodynamics of helix formation. It also led to a development of a "unified" intrinsic helix propensity scale by Pace and Scholtz (1998), derived from the experimental studies of different model systems. Thus, one can argue that the results obtained on model helical peptides as model systems potentially reflect the situation in the proteins, and the thermodynamic parameters are directly transferable from peptides to proteins. If this is valid for the middle of α-helix (Myers et al. 1997a,b), then by analogy, it should be valid for any position of α-helix. Conversely, one can argue that the ends of the helices in short helical peptides are subject to fraying (Aurora et al. 1994; Aurora and Rose 1998). Thus, experimental data on model peptides with the guest position at the N or C terminus cannot be used for the prediction of the helix-forming tendencies in the N or C terminus of helices within the protein structure, because the latter avoid the end-fraying effects via specific capping interactions (Aurora and Rose 1998). The results presented above support this and provide experimental evidence that the propensity scale for the C-terminal residues derived from the peptide model system are not directly applicable for the prediction of the effects of amino acid substitutions in C terminus of the protein helices on stability. Nevertheless, this finding by no means diminishes the importance of observation of the positional dependence of the helix propensities in short helical peptides. They provide important framework for predicting the fractional helicity of peptides in solution (Doig 2002) and can and are used to rationalize many important issues in protein–protein interactions (Brokx et al. 2001).
To summarize, the results presented above lead to the conclusion that the intrinsic helix propensity for the noncharged amino acid residues at the solvent-exposed positions in the middle and at the C terminus of the α-helix are the same. An important consequence of this is that the solvent-exposed C2 position of α-helix is not part of the C-capping motif.
Materials and methods
Mutagenesis, expression, and purification of the ubiquitin variants
Mutations in the codon corresponding to positions 28 and 32 in the amino acid sequence of yeast ubiquitin containing C-terminal His6-tag were introduced as described previously (Thomas and Makhatadze 2000; Thomas et al. 2001; Ermolenko et al. 2002; Loladze et al. 2002). The incorporation of mutations was confirmed by sequencing of the entire gene on an ABI PRISM 377 DNA sequencer. Overexpression of the ubiquitin variants was done in BL21(DE3) or JM109(DE3) strains of Escherichia coli. Proteins were purified to apparent homogeneity as described previously (Ermolenko et al. 2002; Loladze et al. 2002). The concentration of the ubiquitin variants was measured spectrophotometrically by using a molar extinction coefficient of ɛ280 nm = 1280. Correction for light scattering was taken into account as described previously (Ermolenko et al. 2002).
DSC
The DSC experiments were preformed on a VP-DSC (MicroCal) instrument at a scan rate of 90 deg C/h. All experiments were carried out in 30 mM glycine or sodium acetate buffers. The protein concentration in the DSC experiments varied between 1.5 and 3.5 mg/mL. Temperature-induced unfolding of all the studied ubiquitin variants was routinely checked for reversibility by recording the second scan. In all cases, it was found that the reversibility is >90% as judged by the area under the excess heat capacity function. Calorimetric profiles were analyzed according to a two-state transition model using the nonlinear regression routine NLREG and in-house written scripts (Makhatadze 1998; Lopez and Makhatadze 2002). Individual curves for a given ubiquitin variant were fit to a two-state transition model with the heat capacities of the native and the unfolded states; transition temperature, Tm; the enthalpy of unfolding at the transition temperature, ΔH(Tm); and the heat capacity of unfolding, ΔCp, as independent variables (Makhatadze 1998; Lopez and Makhatadze 2002). The standard thermodynamic functions under reference conditions were calculated as
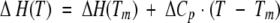 |
(1) |
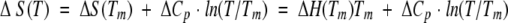 |
(2) |
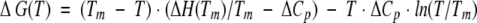 |
(3) |
where ΔS(T) and ΔG(T) are the entropy and Gibbs energy functions of a protein, respectively. The value of ΔCp for all ubiquitin variants was found to be 3.2 ± 0.5 kJ/(K•mole).
Acknowledgments
This work was supported by a grant from the NIH (RO1-GM54537).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0304303.
Supplemental material: See www.proteinscience.org.
References
- Aurora, R. and Rose, G.D. 1998. Helix capping. Protein Sci. 7 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurora, R., Srinivasan, R., and Rose, G.D. 1994. Rules for α-helix termination by glycine. Science 264 1126–1130. [DOI] [PubMed] [Google Scholar]
- Blaber, M., Zhang, X.J., and Matthews, B.W. 1993. Structural basis of amino acid α helix propensity. Science 260 1637–1640. [DOI] [PubMed] [Google Scholar]
- Blaber, M., Zhang, X.J., Lindstrom, J.D., Pepiot, S.D., Baase, W.A., and Matthews, B.W. 1994. Determination of α-helix propensity within the context of a folded protein: Sites 44 and 131 in bacteriophage T4 lysozyme. J. Mol. Biol. 235 600–624. [DOI] [PubMed] [Google Scholar]
- Blagdon, D.E. and Goodman, M. 1975. Mechanisms of protein and polypeptide helix initiation. Biopolymers 14 241–245. [DOI] [PubMed] [Google Scholar]
- Brokx, R.D., Lopez, M.M., Vogel, H.J., and Makhatadze, G.I. 2001. Energetics of target peptide binding by calmodulin reveals different modes of binding. J. Biol. Chem. 276 14083–14091. [DOI] [PubMed] [Google Scholar]
- Chakrabartty, A. and Baldwin, R.L. 1995. Stability of α-helices. Adv. Protein Chem. 46 141–176. [PubMed] [Google Scholar]
- Doig, A.J. 2002. Recent advances in helix-coil theory. Biophys. Chem. 101–102 281–293. [DOI] [PubMed] [Google Scholar]
- Ermolenko, D.N., Thomas, S.T., Aurora, R., Gronenborn, A.M., and Makhatadze, G.I. 2002. Hydrophobic interactions at the Ccap position of the C-capping motif of α-helices. J. Mol. Biol. 322 123–135. [DOI] [PubMed] [Google Scholar]
- Hol, W.G., van Duijnen, P.T., and Berendsen, H.J. 1978. The α-helix dipole and the properties of proteins. Nature 273 443–446. [DOI] [PubMed] [Google Scholar]
- Horovitz, A., Matthews, J.M., and Fersht, A.R. 1992. α-Helix stability in proteins, II: Factors that influence stability at an internal position. J. Mol. Biol. 227 560–568. [DOI] [PubMed] [Google Scholar]
- Kumar, S. and Bansal, M. 1998. Dissecting α-helices: Position-specific analysis of α-helices in globular proteins. Proteins 31 460–476. [DOI] [PubMed] [Google Scholar]
- Loladze, V.V., Ermolenko, D.N., and Makhatadze, G.I. 2002. Thermodynamic consequences of burial of polar and non-polar amino acid residues in the protein interior. J. Mol. Biol. 320 343–357. [DOI] [PubMed] [Google Scholar]
- Lopez, M.M. and Makhatadze, G.I. 2002. Differential scanning calorimetry. Methods Mol. Biol. 173 113–119. [DOI] [PubMed] [Google Scholar]
- Luo, P. and Baldwin, R.L. 1999. Interaction between water and polar groups of the helix backbone: An important determinant of helix propensities. Proc. Natl. Acad. Sci. 96 4930–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu, P.C., Liff, M.I., Marky, L.A., and Kallenbach, N.R. 1990. Side chain contributions to the stability of α-helical structure in peptides. Science 250 669–673. [DOI] [PubMed] [Google Scholar]
- Makhatadze, G.I. 1998. Measuring protein thermostability by differential scanning calorimetry. In Current protocols in protein chemistry (eds. J.E. Coligan et al.), pp. 7.9.1–7.9.14. John Wiley & Sons, New York. [DOI] [PubMed]
- Myers, J.K., Pace, C.N., and Scholtz, J.M. 1997a. A direct comparison of helix propensity in proteins and peptides. Proc. Natl. Acad. Sci. 94 2833–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1997b. Helix propensities are identical in proteins and peptides. Biochemistry 36 10923–10929. [DOI] [PubMed] [Google Scholar]
- Nicholson, H., Anderson, D.E., Dao-pin, S., and Matthews, B.W. 1991. Analysis of the interaction between charged side chains and the α-helix dipole using designed thermostable mutants of phage T4 lysozyme. Biochemistry 30 9816–9828. [DOI] [PubMed] [Google Scholar]
- O’Neil, K.T. and DeGrado, W.F. 1990. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science 250 646–651. [DOI] [PubMed] [Google Scholar]
- Pace, C.N. and Scholtz, J.M. 1998. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 75 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.H., Shalongo, W., and Stellwagen, E. 1993. Residue helix parameters obtained from dichroic analysis of peptides of defined sequence. Biochemistry 32 7048–7053. [DOI] [PubMed] [Google Scholar]
- Penel, S., Hughes, E., and Doig, A.J. 1999a. Side-chain structures in the first turn of the α-helix. J. Mol. Biol. 287 127–143. [DOI] [PubMed] [Google Scholar]
- Penel, S., Morrison, R.G., Mortishire-Smith, R.J., and Doig, A.J. 1999b. Periodicity in α-helix lengths and C-capping preferences. J. Mol. Biol. 293 1211–1219. [DOI] [PubMed] [Google Scholar]
- Petukhov, M., Uegaki, K., Yumoto, N., and Serrano, L. 2002. Amino acid intrinsic α-helical propensities, III: Positional dependence at several positions of C terminus. Protein Sci. 11 766–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissner, R. and Bork, P. 1991. On α-helices terminated by glycine, 1: Identification of common structural features. Biochem. Biophys. Res. Commun. 180 660–665. [DOI] [PubMed] [Google Scholar]
- Presta, L.G. and Rose, G.D. 1988. Helix signals in proteins. Science 240 1632–1641. [DOI] [PubMed] [Google Scholar]
- Richardson, J.S. and Richardson, D.C. 1988. Amino acid preferences for specific locations at the ends of α-helices. Science 240 1648–1652. [DOI] [PubMed] [Google Scholar]
- Rohl, C.A., Chakrabartty, A., and Baldwin, R.L. 1996. Helix propagation and N-cap propensities of the amino acids measured in alanine-based peptides in 40 volume percent trifluoroethanol. Protein Sci. 5 2623–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellman, C. 1980. The α-L conformation at the ends of helices, pp. 53–61. Elsevier, New York.
- Scholtz, J.M. and Baldwin, R.L. 1992. The mechanism of α-helix formation by peptides. Annu. Rev. Biophys. Biomol. Struct. 21 95–118. [DOI] [PubMed] [Google Scholar]
- Seale, J.W., Srinivasan, R., and Rose, G.D. 1994. Sequence determinants of the capping box, a stabilizing motif at the N- termini of α-helices. Protein Sci. 3 1741–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano, L., Sancho, J., Hirshberg, M., and Fersht, A.R. 1992. α-Helix stability in proteins, I: Empirical correlations concerning substitution of side-chains at the N and C-caps and the replacement of alanine by glycine or serine at solvent-exposed surfaces. J. Mol. Biol. 227 544–559. [DOI] [PubMed] [Google Scholar]
- Thomas, S.T. and Makhatadze, G.I. 2000. Contribution of the 30/36 hydrophobic contact at the C-terminus of the α-helix to the stability of the ubiquitin molecule. Biochemistry 39 10275–10283. [DOI] [PubMed] [Google Scholar]
- Thomas, S.T., Loladze, V.V., and Makhatadze, G.I. 2001. Hydration of the peptide backbone largely defines the thermodynamic propensity scale of residues at the C` position of the C-capping box of α-helices. Proc. Natl. Acad. Sci. 98 10670–10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar, S., Bugg, C.E., and Cook, W.J. 1987a. Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol. 194 531–544. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar, S., Bugg, C.E., Wilkinson, K.D., Vierstra, R.D., Hatfield, P.M., and Cook, W.J. 1987b. Comparison of the three-dimensional structures of human, yeast, and oat ubiquitin. J. Biol. Chem. 262 6396–6399. [PubMed] [Google Scholar]
- Wada, A. 1976. The α-helix as an electric macro-dipole. Adv. Biophys. 9 1–63. [PubMed] [Google Scholar]
- Wojcik, J., Altmann, K.H., and Scheraga, H.A. 1990. Helix-coil stability constants for the naturally occurring amino acids in water, XXIV: Half-cystein parameters from random poly (hydroxybutylglutamine-co-l-proline). Biopolymers 30 121–134. [DOI] [PubMed] [Google Scholar]
- Yang, J., Spek, E.J., Gong, Y., Zhou, H., and Kallenbach, N.R. 1997. The role of context on α-helix stabilization: Host-guest analysis in a mixed background peptide model. Protein Sci. 6 1264–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]