Abstract
The role of epigenetics in modulating gene expression in the development of organs and tissues and in disease states is becoming increasingly evident. Epigenetics refers to the several mechanisms modulating inheritable changes in gene expression that are independent of modifications of the primary DNA sequence and include post-translational modifications of nucleosomal histones, changes in DNA methylation, and the role of microRNA. This review focuses on the epigenetic regulation of gene expression in oligodendroglial lineage cells. The biological effects that post-translational modifications of critical residues in the N-terminal tails of nucleosomal histones have on oligodendroglial cells are reviewed, and the implications for disease and repair are critically discussed.
Keywords: Chromatin, Development, Epigenetics, Brain, Myelin, Differentiation, Glia
Introduction
Epigenetic regulation of gene expression has been recently shown to play a critical role in the development of organs and tissues, including the brain. It refers to the inheritable pattern of gene expression that is transmitted to daughter cells and that is independent of changes or mutations in the DNA sequence (Gottschling 2006; Bird 2007). Several factors are known to contribute to epigenetic regulation of gene expression during neural development, including microRNA (Cheng et al. 2005; Cao et al. 2006; Mehler and Mattick 2006, 2007), DNA methylation (Feng et al. 2007), ATP-remodeling complexes, changes in histone variants and post-translational modifications of nucleosomal histones (Fukuda and Taga 2005; Feng et al. 2007). This review will focus on the role of post-translational modifications of nucleosomal histones (Fig. 1).
Figure 1.
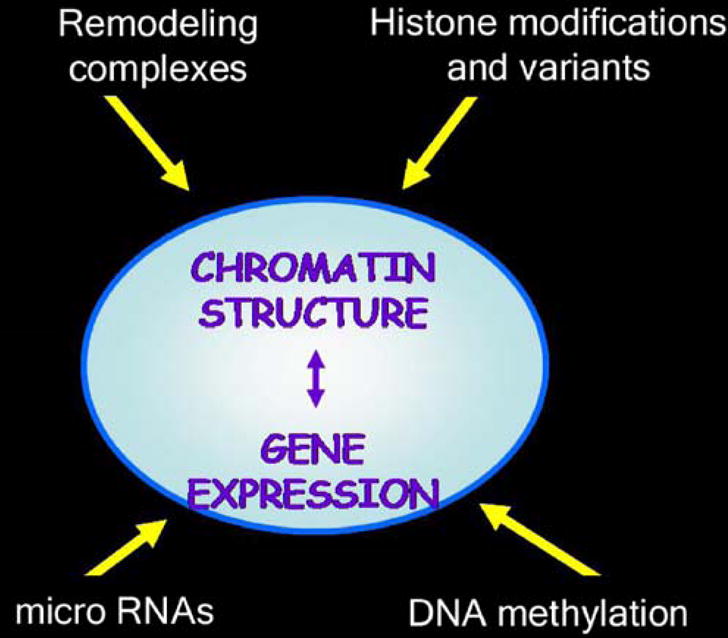
Epigenetic modulation of gene expression. The four main areas of investigation in the field of epigenetics are presented in this diagram
Nucleosomes are the basic unit of chromatin. They are formed by 150 bp of DNA wrapped around octamers of nuclear proteins called histones. At least four major nucleosomal histones have been identified: H2A, H2B, H3, and H4. Additional histone proteins (i.e., H1) are distributed in the intervening regions called linker regions.
Structural studies of nucleosomal histones have revealed that they are arranged as globular core with the N-terminal tails projecting outward (Dutnall and Ramakrishnan 1997). Because of these structural features, it becomes evident that the amino acids on the tails of these histones are the most easily accessible targets to enzymatic activities downstream of multiple signaling pathways.
Extracellular signals can communicate with the machinery responsible for the epigenetic regulation of gene expression by inducing phosphorylation on specific serine residues, transferring or removing acetyl or methyl groups on lysine residues, and methylating or deiminating arginine residues (Magnaghi-Jaulin et al. 1999; Thomson et al. 1999; Spencer and Davie 2000).
Post-Translational Modifications of Lysine Residues on Histone Tails: Acetylation and Methylation
Acetylation is the transfer of acetyl groups to the epsilon position of lysine residues in the tail of nucleosomal histones. It is important to distinguish this modification from N-alpha acetylation of eukaryotic structural proteins that occurs on alanine, serine, and methionine residues, and that is catalyzed by N-acetyl transferases. Histone acetylation is catalyzed by a family of enzymes called histone acetyltransferases (HAT), and it is functionally correlated with transcriptionally competent chromatin. The removal of acetyl groups is catalyzed by a family of enzymes called histone deacetylases (HDAC or HD) and is functionally correlated with transcriptionally inactive chromatin (Vidali et al. 1988; Wolffe 1996; Grunstein 1997; Kuo and Allis 1998; Struhl 1998; Spencer and Davie 1999; Tyler and Kadonaga 1999; Cheung et al. 2000; Turner 2000; de Ruijter et al. 2003). Hence, the ratio of HDAC to HAT activity determines the level of histone acetylation.
Histone methylation is a process of adding one, two, or three methyl groups to certain amino acids on histone tails (Ashraf and Ip 1998; Bird and Wolffe 1999; Spencer and Davie 1999; Rice and Allis 2001; Zhang and Reinberg 2001; Bannister et al. 2002; Berger 2002; Goll and Bestor 2002; Kouzarides 2002; Lachner and Jenuwein 2002; Richards 2002). Depending on the specific lysine, methylation may result in gene activation, such as methylation on lysine 4 residue on histone H3 (Me-H3K4) or in repression, such as methylation on lysine 9 or 27 (Fig. 2). It was originally thought that histone methylation was a permanent modification. This theory prevailed until the recent discovery of lysine-specific demethylase 1, which specifically removes mono- and di- but not trimethyl groups from lysine residues, and Jumonji domain-containing, which can demethylate all three status of methylated lysine residue (Kubicek and Jenuwein 2004; Bannister and Kouzarides 2005; Wysocka et al. 2005; Takeuchi et al. 2006; Anand and Marmorstein 2007; Klose and Zhang 2007; Stavropoulos and Hoelz 2007; Wilson 2007). Therefore, similar to histone acetylation, histone methylation is also a regulated dynamic process.
Figure 2.
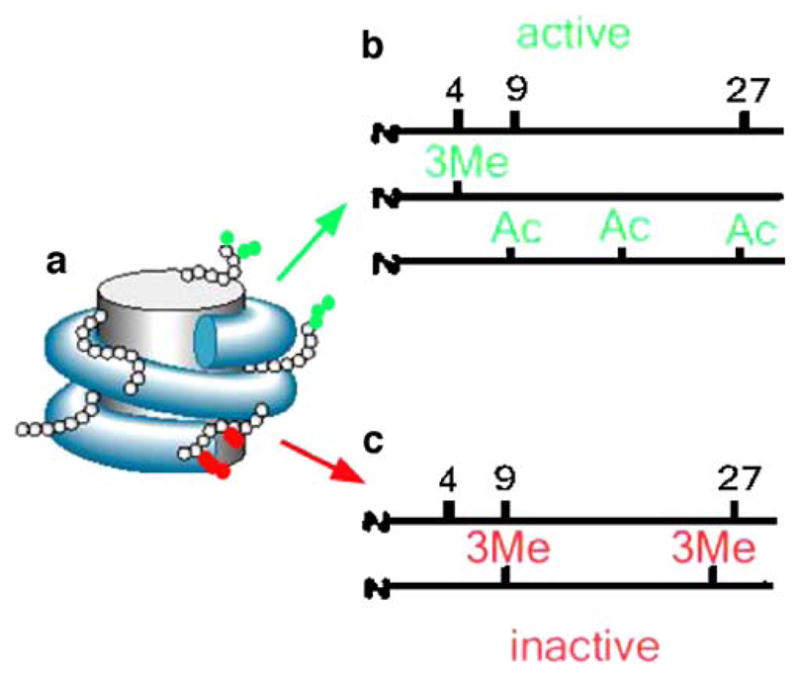
The histone code. a Schematic illustration of the nucleosome. The histones are represented by a gray cylinder and the nucleosomal histone tails are shown as small beads irradiating out of the cylinder. The post-translational modifications of the lysine residues in the histone tails are shown by red and green circles. b Post-translational modifications of lysine residues in the tail of nucleosomal histone H3, associated with active transcription (green). Note the inclusion of trimethyl lysine K4 and the acetylation of several residues. c Repressive modifications of lysine residues in position 9 and 27
Post-Translational Modifications of Arginine Residues: Methylation and Citrullination
Methylation of arginine residues in histone tails by protein arginine methyltransferases (PRMT) also plays a critical role in dynamic gene regulation (Davie and Dent 2002; Bedford and Richard 2005; Boisvert et al. 2005; Denman 2005; Pahlich et al. 2006; Thompson and Fast 2006; Wysocka et al. 2006). In mammals, PRMT1- and coactivator-associated arginine methyltransferase 1 catalyze the asymmetric methylation of arginine 3 on histone H4 (H4R3) and of arginine 17 on histone H3 (H3R17), respectively, and result in gene activation (Chen et al. 1999; Strahl et al. 2001; Wang et al. 2001a; Bauer et al. 2002). In contrast, the enzyme PRMT5 catalyzes the symmetric methylation of arginine 8 histone H3 (H3R8) and arginine 3 histone H4 (H4R3) (Pal et al. 2004), and this is associated with gene repression (Wysocka et al. 2006). Arginine methylation is also reversible and can be removed by specific deiminases, such as human peptidylarginine deiminase 4 (PAD4), which convert arginine into citrulline (Wang et al. 2004).
Modulation of Oligodendrocyte Lineage-Specific Pathways by HDAC
Model 1 Differentiation Consequent to Derepression of Differentiation Genes
Several studies have contributed to the concept of differentiation consequent to derepression of genes characteristic of the differentiated state. This model implies that neural stem cells or multipotential precursors, which have the potential to differentiate into neurons, astrocytes, and oligodendrocytes, are maintained in an undifferentiated state because of epigenetic silencing of specific lineage markers.
In embryonic neural precursor cells, for instance, the expression of the lineage specific gene glial fibrillary acidic protein (GFAP) is maintained silenced by DNA methylation of CpG islands in the promoter region (Feng et al. 2005). This render inaccessible the binding sites for the transcription factor known as signal transducers and activator of transcription (STAT), which is critical for astroglienesis (Bonni et al. 1997). As development proceeds, the progressive demethylation of the GFAP promoter correlates with the progressive increase of transcript levels (Teter et al. 1994; Takizawa et al. 2001). Besides DNA methylation, the amount of GFAP expression during differentiation is also regulated by histone acetylation and methylation because of the recruitment of a multi-protein complex containing STAT, SMAD, and the HAT p300 (Nakashima et al. 1999; Asklund et al. 2004; Song and Ghosh 2004).
Similarly, in the neuronal lineage, the expression of neural genes is maintained repressed by the activity of multiprotein complexes containing corepressors, HDAC, and the specific transcription factor known as repressor element-silencing transcription (REST) that is responsible for recognizing specific cis-element on the promoters (Huang et al. 1999; Roopra et al. 2000; Ballas et al. 2001). REST is a powerful repressor. The N terminus of the molecule has the ability to recruit HDAC1/2 complexes and Sin3A, whereas its C terminus has the ability to form complexes with HDAC1/2 and with Co-REST (Andres et al. 1999; Huang et al. 1999; Naruse et al. 1999; Roopra et al. 2000). Accordingly, neuronal differentiation of embryonic or adult neural progenitors is initiated by the removal component of these large repressive complexes. Consistent with this model, treatment of multipotential neural progenitors with pharmacological inhibitors of HDAC results in the upregulation of neurogenic marker genes, such as NeuroD, and increased neurogenesis (Hao et al. 2004; Hsieh et al. 2004).
By analogy with other lineages, it is likely, although not yet proven, that progressive acetylation of nucleosomal histones and relaxed chromatin structure occurs during the differentiation of precursors into myelinating oligodendrocytes. A number of studies have reported the presence of histone deacetylases (i.e., HDAC1 and 2) in the proximal promoter region of myelin genes in oligodendrocyte progenitors (OPC). For example, HDAC1 has been shown to be recruited by transcriptional factors Nkx2.2 (Wei et al. 2005) and also by Hes5 (Wei et al. 2005; Liu et al. 2006) to the promoter of myelin basic protein (MBP). Similarly, HDAC1/2 heterodimers, in association with Sin3B, were detected in complex with Myt-1 on the proximal region of the proteolipid protein (PLP) promoter (Romm et al. 2005). These findings suggested that the expression of myelin genes in progenitor cells was inhibited by HDAC and predicted that pharmacological inhibition of these enzymes should favor myelin gene expression and promote oligodendrocyte differentiation.
In contrast to the prediction, however, the administration of HDAC inhibitors, such as trichostatin A (TSA) or valproic acid, prevented myelin gene expression in vivo (Shen et al. 2005) and in vitro (Marin-Husstege et al. 2002; Liu et al. 2003). These data suggested at least two possible models of myelin gene activation mediated by chromatin compaction. One possibility suggests that the effect of pharmacological inhibitors of HDAC on myelin gene expression is direct. According to this possibility, activation requires the compaction of silencers caused by the remodeling of the nucleosomal tails in the chromatin regions containing negative regulatory elements. The presence of HDAC inhibitors would prevent the inactivation of these silencers by halting the formation of zones of compaction. The second possibility is that myelin gene expression results from a balance between positive and negative signals of differentiation. In this case, the post-translational modification of nucleosomes would occur in the promoter of inhibitory molecules, and myelin gene expression would result from a balance tilted in favor of the activators. Future studies providing a careful mapping of histone modifications in the regulatory regions of myelin genes will contribute to the elucidation of the model.
Model 2 Differentiation Consequent to HDAC-mediated Repression of Alternative Lineage Choices
The role of epigenetics in modulating the lineage progression of OPC and the fate choice of multipotential neural precursors have received multiple independent validations.
The first report of the negative effect of HDAC inhibitors on oligodendrocyte differentiation was in 2002 (Marin-Husstege et al. 2002). However, it was not determined whether the cells that were unable to differentiate into oligodendrocytes were redirected to other lineages.
Using adult hippocampus neural progenitor cells, Hsieh and Gage suggested that administration of HDAC inhibitors promoted neurogenesis at the expense of other lineages, possibly because of the increased neuronal gene transcripts, such as NeuroD, in response to high levels of histone acetylation (Hsieh et al. 2004). Therefore, it was conceivable that OPC were redirected to other lineages when unable to differentiate into myelin-forming cells. This concept was recently shown in a series of in vitro and in vivo experiments that clearly indicated that the probability of OPC to generate functional neurons or astrocytes (Bleached et al. 2003; Aguirre et al. 2004; Nishiyama 2007) is dependent on HDAC activity (Liu et al. 2007). These conclusions were further validated by transplantation studies of GFP-labeled OPC in recipients that were either untreated or treated with HDACi. Also in this case, HDACi treatment prevented the ability of GFP transplanted cells to generate mature oligodendrocytes, while redirecting their lineage into astrocytes or neurons (Liu et al. 2007).
These findings could be interpreted in terms of trans-differentiation event or in terms of reversion to a stem-cell-like condition, followed by differentiation along an alternate pathway. It is important to mention that the high probability for OPC to generate functional neurons was only detected in vivo in animals receiving systemic administration of HDAC1 or in vitro when cells pretreated with HDACi were cultured in neurogenic conditions. These results suggested that the differentiation of progenitors into alternative lineages was a two-step event that required, first, the “reversion” to a multipotential stage and then the “differentiation” into another cell type. The ability of OPC to revert to a multipotential stem-cell-like stage had been suggested by Kondo and Raff (2000c), who reported that only extensive culturing methods in fibroblast growth factor-2 (FGF2) would allow this reversion step (Kondo and Raff 2000c). A molecular explanation for the reversion step was provided by the same authors few years later, when Sox2 was proposed as the transcription factor responsible for the maintenance of a multipotential stage (Kondo and Raff 2004). Sox2 is the SRY-related, HMG-box-containing transcription factor that is expressed early in the developing neural tube, is essential for maintaining neural stem cells in undifferentiated state (Graham et al. 2003). It was found to be expressed in neural stem cells but not in lineage-committed progenitors because of epigenetic regulation of its promoter (Graham et al. 2003; Kondo and Raff 2004). It was reported that the reversion of OPC to neural stem cells was associated with the reactivation of the sox2 gene because of activating post-transcriptional modifications of Lys 4 and Lys 9 residues in the tail of histone H3 (Kondo and Raff 2004).
Recent findings from an independent group using a very different approach validated the model of epigenetic modulation of oligodendrocyte differentiation modulated by histone deacetylation (Lyssiotis et al. 2007). Using a line of immortalized OPC-expressing GFP from the Sox2 promoter to screen for compounds that were able to revert progenitors to multipotential neural stem cells, the authors identified several HDAC inhibitors. The compounds were able to reactivate the stem cell marker gene Sox2; however, complete reversion was rendered possible only by culturing the cells in the presence of FGF2 (Lyssiotis et al. 2007). Therefore, reactivation of Sox2 by increased histone acetylation is a prerequisite for fate reversion from a lineage-restricted OPC to a neural stem cell. However, Sox2 by itself is insufficient for the reversion, and it requires additional signals, such as FGF to favor the multipotential state (Lyssiotis et al. 2007). Together, these data suggest a model of OPC plasticity that is mediated by histone modifications and is affected by upstream signaling events.
Model 3 Differentiation Consequent to HDAC-Mediated Repression of Transcriptional Inhibitors of Differentiation
The concept of oligodendrocyte lineage progression as sequential steps of repression of inhibitory events was suggested by the observation that oligodendrogliogenesis, at least in vitro, proceeds by default. Oligodendrocytes differentiate from progenitor cells when mitogens are withdrawn from the culture medium (Noble and Murray 1984; Temple and Raff 1985; Gard and Pfeiffer 1993; Bansal and Pfeiffer 1997). This suggested that the oligodendrocytic differentiation process results from the removal of negative signals provided by mitogens. Because growth factors affect the proliferative state of the cell, it was initially proposed that differentiation could be initiated by cell cycle exit (Casaccia-Bonnefil et al. 1997; Durand et al. 1998). In this respect, multiple studies focused on the identification of the molecular regulators of the cell cycle that are responsible for the regulation of the G1 phase of the cell cycle (Durand et al. 1998; Tikoo et al. 1998; Casaccia-Bonnefil et al. 1999; Ghiani et al. 1999; Durand and Raff 2000; Tokumoto et al. 2002). The importance of cell-cycle inhibitors in oligodendrocyte differentiation was further suggested by reports of alternative biological functions for these molecules. P27Kip, for instance, was identified as part of a transcriptional complex activating the MBP promoter (Miskimins et al. 2002), as well as part of a complex with specific RNA-binding proteins like Qki (Larocque et al. 2005). Despite these exciting findings, overexpression of cell-cycle inhibitors per se was not sufficient to induce differentiation (Tikoo et al. 1998; Tang et al. 1999) and suggested that additional events must be responsible for the differentiation of progenitors into mature oligodendrocytes. Our studies using inhibitors of HDAC activity in cultured cells (Marin-Husstege et al. 2002) and in vivo (Shen et al. 2005) provided some clues regarding the additional events responsible for this transition. We proposed that after cell-cycle withdrawal, progenitors might require HDAC activity to down-regulate inhibitory molecules that halt the differentiation into myelinating oligodendrocytes. This model was in agreement with previous studies on the role of transcriptional inhibitors of oligodendrocyte differentiation (Kondo and Raff 2000b, a; Wang et al., 2001b) and with gene expression profiling studies reporting a progressive decrease of inhibitors concomitant with differentiation (Kuhlbrodt et al. 1998; Sohn et al. 2006; Dugas et al. 2007). The concept of derepression of inhibitors as potential explanation for default oligodendroglial differentiation was further supported by the evidence that inhibitors of the Id and Hes family can inhibit differentiation of oligodendroglial progenitors in multiple ways, including recruitment of HDAC (Liu et al. 2006), sequestration of activators (Samanta and Kessler 2004; Liu et al. 2006), and transcriptional inhibition of critical regulators (Guillemot 1995; Sakamoto et al. 2003; Kageyama et al. 2005; Liu et al. 2006).
The recent identification of the transcription factor Ying Yang-1 (YY-1) as a recruiter of HDAC1 on the promoter of transcriptional inhibitors of OPC differentiation and the dramatic hypomyelinating phenotype in mice with conditional ablation of YY1 in the oligodendroglial lineage provided support to the importance of chromatin modifications for the initiation of oligodendrocyte differentiation in progenitors after they have exited the cell cycle (He et al. 2007). The model of HDAC-dependent repression of differentiation inhibitors was reinforced by the fact that this is a highly conserved event among species. Indeed, it was shown that zebrafish hdac1 mutants are unable to express oligodendroglial markers, such as plp, sox10, and olig2, while they continue to express sox2 and pax6 (Cunliffe and Casaccia-Bonnefil 2006).
Therefore, together these studies support a model of oligodendrocyte differentiation characterized by the progressive epigenetic repression of inhibitory molecules that is initially dependent on histone deacetylation.
Cytoplasmic HDAC and Non-Histone Substrates: A Role in Morphological Differentiation of Oligodendrocyte Development
The mammalian sirtuin (SIRT) family of proteins, which are related to the silent information regulator 2 (Sir2) of yeast, belong to a special class of HDAC that are regulated by NAD/NADH levels (Tanny et al. 1999; Imai et al. 2000). SIRT have been reported to modulate the morphological changes associated with oligodendrocyte differentiation and myelin assembly. Nuclear SIRT1 is expressed in cultured embryonic neurospheres and in nestin cells at subventricular zone (SVZ) of adult mouse brains. Administration of SIRT1 inhibitors, nicotinamide or sirtinol, potently blocked the differentiation of neurospheres into neurons or oligodendrocytes (Horio et al. 2003).
SIRT2 is cytoplasmically localized, and its expression in the central nervous system (CNS) is oligodendrocyte-specific (Li et al. 2007; Southwood et al. 2007). Indeed, SIRT2 was reported to be a myelin component few years ago, when it was listed as one of the multiple proteins identified in proteomic screens of myelin components (Taylor et al. 2004; Vanrobaeys et al. 2005). In the developing CNS, the appearance and expression pattern of SIRT2 in the cerebrum, cerebellum, and spinal cord have been correlated with that of the myelin protein, CNPase (Li et al. 2007). However, the levels of SIRT2 are not affected by genetic deletion of cnp in mice. At cellular levels, both SIRT2 and CNPase are coexpressed in the processes but not in the cell body of oligodendrocytes, suggesting that SIRT2 may be involved in the morphological changes occurring during oligodendrocyte differentiation and myelin sheath formation (Li et al. 2007). Ultrastructural studies have further supported the localization of SIRT2 to the non-compact outer and terminal loops of myelin sheath (Li et al. 2007; Southwood et al. 2007), suggesting that its function could be important for process outgrowth and membrane expansion of oligodendrocytes.
Recent reports have uncovered an intriguing association between SIRT2 levels and the myelin protein PLP (Werner et al. 2007). It was discovered that the incorporation of SIRT2 into the myelin sheath is myelin protein PLP/DM20-dependent; as in the plp null mice, SIRT2 expression is completely lost (Werner et al. 2007).
An Integrated View of Oligodendrocyte Development
Based on the review of the current literature, we propose a model of OPC differentiation that requires global repressive events that are initiated by histone deacetylation and possibly associated with additional modifications of the chromatin structure. This is in agreement with early ultrastructural studies by electron microscopy on the developing corpus callosum of neonatal rats, reporting the progressive increase of chromatin compaction in the nuclei of OPC during their differentiation into mature oligodendrocytes (Kozik 1976). We propose that a critical functional role for massive chromatin compaction is the downregulation of molecules that preclude differentiation, including transcriptional inhibitors of myelin genes (Liu et al. 2006; Marin-Husstege et al. 2006), as well as cytoskeletal depolymerizing molecules (Liu et al. 2003; Liu et al. 2005). We further propose that the chromatin compaction initiated by histone deactylation is a critical event to prevent other lineage choice of progenitor cells and possibly prevent precocious myelin gene expression. The analysis of HDAC-mediated histone deacetylation in affecting the timing of oligodendrocyte differentiation and myelination in the developing brain revealed the specific requirement for HDAC activity during a specific temporal window of postnatal development, corresponding to the activation of a transcriptional program of differentiation after progenitors have exited the cell cycle (Marin-Husstege et al. 2002; Shen et al. 2005; He et al. 2007). At later developmental stages, however, histone deacetylation subsides and is replaced by repressive histone methylation and the establishment of a compact chromatin structure, characteristic of the differentiated oligodendrocyte phenotype (Shen et al. 2005). Together these data suggest that HDAC-mediated histone deacetylation is required for the initial stages of OPC differentiation, when the intracellular environment of the cell is “inhibitory.” However, it is dispensable later during the maturation process when the majority of the “obstacles” have been eliminated. For this reason, the role of HDAC is confined to a narrow temporal window, defined by the interface between cell-cycle exit and the beginning of differentiation. This window is characterized by the progressive decline of differentiation inhibitors and is likely to be overcome by signals that tilt the equilibrium in favor of differentiation.
Histone Modifications in the Aging Corpus Callosum and Perspective on the Potential Role of Chromatin Modifications in Neurological Diseases Affecting Myelinating Oligodendrocytes
Since histone deacetylation and methylation are critical epigenetic modulators of gene expression during oligodendrocyte differentiation and myelin formation, they serve the purpose of establishing a “molecular memory,” which is stored in the nuclei of oligodendroglial cells. This memory is responsible for the downregulation and stable repression of inhibitory molecules in differentiated oligodendrocytes (Marin-Husstege et al. 2006). In an attempt to understand the potential mechanisms underlying age-dependent decline of repair function, Shen et al. (2007) reported the progressive loss of this “epigenetic memory” in oligodendrocytes during normal aging. Decreased histone deacetylation and repressive methylation of histone H3 tails were detected in older mice. The direct result of these epigenetic changes was the increased expression of inhibitory molecules, including Hes5, Ids, and Sox2 (Shen et al. 2007). This partially explains why the responsiveness of oligodendrocytes to extracellular factors is decreased with aging and further suggests that chromatin modifications and changes in the “epigenetic memory” of oligodendrocytes may have profound effects on therapeutic outcomes.
Although not formally proven, these findings also suggest that similar epigenetic changes might affect the ability of progenitors in older brains to efficiently repair demyelinating lesions. Previous studies had suggested that remyelination failure was consequent to increased levels of inhibitory extracellular signals (John et al. 2002) or insufficient levels of oligodendrogliogenic signals (Mastronardi et al. 2003). However, it was shown that altering the levels of a single extracellular factor or signaling molecule was not sufficient per se to promote successful remyelination (Fancy et al. 2004; Stidworthy et al. 2004). Taken together, these results support a model of remyelination as a complex interplay between intrinsic changes and extracellular signaling cues.
An additional line of experimental evidence suggested the importance of aberrant modifications of nucleosomal histones in oligodendrocytes as part of the pathogenetic process leading to demyelinating disorders. Increased histone citrullination on arginine residues was reported to be increased in the normal-appearing white matter of MS patients and in animal models of demyelination (Mastronardi et al. 2006). Citrullination of arginine residues on histone tails is catalyzed by peptidylarginine deiminase 4 (PAD4). Under normal conditions, PAD4 is cytosolically localized. However, under pathological conditions, such as in the presence of abnormally increased level of tumor necrosis factor α (TNFα), PAD4 is translocated to the nucleus, and its increased nuclear expression and activity catalyzes the citrullination of nucleosomal histones (Mastronardi et al. 2006). Importantly, these molecular changes occur before the onset of symptoms in animal models of demyelination (Mastronardi et al. 2006), indicating that TNFα-induced PAD4 nuclear localization and subsequent histone citrullination may be part of the etiopathogenesis of the disease.
The potential role of histone modifications in demyelinating disorders has also led to therapeutic attempts with histone-modifying drugs, but the results, at least in the EAE models, have led to controversial results. Administration of the HDAC inhibitor TSA to C57BL/6 mice with active immunization of MOG35–55-induced EAE had favorable effects on spinal cord inflammation, demyelination volume, and axonal loss, possibly because of increased expression levels of anti-oxidants (i.e., glutathione peroxidase), glutamate transporters (i.e., excitatory amino acid transporter 2, EAAT2), and neuroprotective factors (i.e., insulin-like growth factor 2; Camelo et al. 2005). However, also the administration of curcumin, a selective HAT (CBP/p300) inhibitor that decreases the total histone acetylation levels, had positive effects on EAE. When curcumin was administered to SJL/J mice with EAE induced by adoptive transfer of MBP-immunized immune cells, it inhibited the induction phase of the disease, possibly, by blocking IL-12 signaling pathway in T lymphocytes (Natarajan and Bright 2002).
Together, these results reveal the need for the development of more specific therapies aimed at targeting epigenetic modulators only in specific cell populations.
Acknowledgments
Partially supported by grants from NIH-NINDS (RO1NS042925 and RO1 052738) and from the National Multiple Sclerosis Society (NMSS3957) to P.C.B. We thank Dr. Juan Sandoval for the help with the graphic illustrations.
References
- Aguirre AA, Chittajallu R, Belachew S, Gallo V. NG2-expressing cells in the subventricular zone are type C-like cells and contribute to interneuron generation in the postnatal hippocampus. Journal of Cell Biology. 2004;165:575–589. doi: 10.1083/jcb.200311141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand R, Marmorstein R. Structure and mechanism of lysine specific demethylase enzymes. Journal of Biological Chemistry. 2007 doi: 10.1074/jbc.R700027200. in press. [DOI] [PubMed] [Google Scholar]
- Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J. CoREST: A functional corepressor required for regulation of neural-specific gene expression. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, Ip YT. Transcriptional control: Repression by local chromatin modification. Current Biology. 1998;8:R683–686. doi: 10.1016/s0960-9822(98)70435-x. [DOI] [PubMed] [Google Scholar]
- Asklund T, Appelskog IB, Ammerpohl O, Ekstrom TJ, Almqvist PM. Histone deacetylase inhibitor 4-phenyl-butyrate modulates glial fibrillary acidic protein and connexin 43 expression, and enhances gap-junction communication, in human glioblastoma cells. European Journal of Cancer. 2004;40:1073–1081. doi: 10.1016/j.ejca.2003.11.034. [DOI] [PubMed] [Google Scholar]
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Reversing histone methylation. Nature. 2005;436:1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Kouzarides T. Histone methylation: Dynamic or static? Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- Bansal R, Pfeiffer SE. FGF-2 converts mature oligodendrocytes to a novel phenotype. Journal of Neuroscience Research. 1997;50:215–228. doi: 10.1002/(SICI)1097-4547(19971015)50:2<215::AID-JNR10>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Reports. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Molecular Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Belachew S, Chittajallu R, Aguirre AA, Yuan X, Kirby M, Anderson S, et al. Postnatal NG2 proteoglycan-expressing progenitor cells are intrinsically multipotent and generate functional neurons. Journal of Cell Biology. 2003;161:169–186. doi: 10.1083/jcb.200210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. Histone modifications in transcriptional regulation. Current Opinion in G0enetics & Development. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Chenard CA, Richard S. Protein interfaces in signaling regulated by arginine methylation. Sci STKE, 2005. 2005;(271):re2. doi: 10.1126/stke.2712005re2. [DOI] [PubMed] [Google Scholar]
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, et al. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- Camelo S, Iglesias AH, Hwang D, Due B, Ryu H, Smith K, et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. Journal of Neuroimmunology. 2005;164:10–21. doi: 10.1016/j.jneuroim.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annual Review of Neuroscience. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Hardy RJ, Teng KK, Levine JM, Koff A, Chao MV. Loss of p27Kip1 function results in increased proliferative capacity of oligodendrocyte progenitors but unaltered timing of differentiation. Development. 1999;126:4027–4037. doi: 10.1242/dev.126.18.4027. [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Tikoo R, Kiyokawa H, Friedrich V, Jr, Chao MV, Koff A. Oligodendrocyte precursor differentiation is perturbed in the absence of the cyclin-dependent kinase inhibitor p27Kip1. Genes & Development. 1997;11:2335–2346. doi: 10.1101/gad.11.18.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, et al. Regulation of transcription by a protein methyl-transferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Tavazoie M, Doetsch F. Stem cells: From epigenetics to microRNAs. Neuron. 2005;46:363–367. doi: 10.1016/j.neuron.2005.04.027. [DOI] [PubMed] [Google Scholar]
- Cheung WL, Briggs SD, Allis CD. Acetylation and chromosomal functions. Current Opinion in Cell Biology. 2000;12:326–333. doi: 10.1016/s0955-0674(00)00096-x. [DOI] [PubMed] [Google Scholar]
- Cunliffe VT, Casaccia-Bonnefil P. Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mechanisms of Development. 2006;123:24–30. doi: 10.1016/j.mod.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Davie JK, Dent SY. Transcriptional control: an activating role for arginine methylation. Current Biology. 2002;12:R59–R61. doi: 10.1016/s0960-9822(01)00674-1. [DOI] [PubMed] [Google Scholar]
- de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochemical Journal. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denman RB. PAD: the smoking gun behind arginine methylation signaling? Bioessays. 2005;27:242–246. doi: 10.1002/bies.20205. [DOI] [PubMed] [Google Scholar]
- Dugas JC, Ibrahim A, Barres BA. A crucial role for p57(Kip2) in the intracellular timer that controls oligodendrocyte differentiation. Journal of Neuroscience. 2007;27:6185–6196. doi: 10.1523/JNEUROSCI.0628-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B, Fero ML, Roberts JM, Raff MC. p27Kip1 alters the response of cells to mitogen and is part of a cell-intrinsic timer that arrests the cell cycle and initiates differentiation. Current Biology. 1998;8:431–440. doi: 10.1016/s0960-9822(98)70177-0. [DOI] [PubMed] [Google Scholar]
- Durand B, Raff M. A cell-intrinsic timer that operates during oligodendrocyte development. Bioessays. 2000;22:64–71. doi: 10.1002/(SICI)1521-1878(200001)22:1<64::AID-BIES11>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Dutnall RN, Ramakrishnan V. Twists and turns of the nucleosome: tails without ends. Structure. 1997;5:1255–1259. doi: 10.1016/s0969-2126(97)00276-1. [DOI] [PubMed] [Google Scholar]
- Fancy SP, Zhao C, Franklin RJ. Increased expression of Nkx2.2 and Olig2 identifies reactive oligodendrocyte progenitor cells responding to demyelination in the adult CNS. Molecular and Cellular Neurosciences. 2004;27:247–254. doi: 10.1016/j.mcn.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. Journal of neuroscience research. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatric Research. 2007;61:58R–63R. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- Fukuda S, Taga T. Cell fate determination regulated by a transcriptional signal network in the developing mouse brain. Anat Sci Int. 2005;80:12–18. doi: 10.1111/j.1447-073x.2005.00097.x. [DOI] [PubMed] [Google Scholar]
- Gard AL, Pfeiffer SE. Glial cell mitogens bFGF and PDGF differentially regulate development of O4 GalC-oligo-dendrocyte progenitors. Developments in Biologicals. 1993;159:618–630. doi: 10.1006/dbio.1993.1269. [DOI] [PubMed] [Google Scholar]
- Ghiani CA, Eisen AM, Yuan X, DePinho RA, McBain CJ, Gallo V. Neurotransmitter receptor activation triggers p27(Kip1)and p21(CIP1) accumulation and G1 cell cycle arrest in oligodendrocyte progenitors. Development. 1999;126:1077–1090. doi: 10.1242/dev.126.5.1077. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Histone modification and replacement in chromatin activation. Genes & Development. 2002;16:1739–1742. doi: 10.1101/gad.1013902. [DOI] [PubMed] [Google Scholar]
- Gottschling DE. DNA repair: corrections in the golden years. Current Biology. 2006;16:R956–R958. doi: 10.1016/j.cub.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Guillemot F. Analysis of the role of basic-helix-loop-helix transcription factors in the development of neural lineages in the mouse. Biology of the Cell. 1995;84:3–6. doi: 10.1016/0248-4900(96)81312-8. [DOI] [PubMed] [Google Scholar]
- Hao Y, Creson T, Zhang L, Li P, Du F, Yuan P, et al. Mood stabilizer valproate promotes ERK pathway-dependent cortical neuronal growth and neurogenesis. Journal of Neuroscience. 2004;24:6590–6599. doi: 10.1523/JNEUROSCI.5747-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, et al. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio Y, Hisahara S, Sakamoto J. Functional analysis of SIR2. Nippon Yakurigaku Zasshi. 2003;122(Suppl):30P–32P. [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Myers SJ, Dingledine R. Transcriptional repression by REST: Recruitment of Sin3A and histone deacetylase to neuronal genes. Nature Neuroscience. 1999;2:867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- John GR, Shankar SL, Shafit-Zagardo B, Massimi A, Lee SC, Raine CS, et al. Multiple sclerosis: Re-expression of a developmental pathway that restricts oligodendrocyte maturation. Natural Medicines. 2002;8:1115–1121. doi: 10.1038/nm781. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nature Reviews. Molecular Cell Biology. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. Basic helix-loop-helix proteins and the timing of oligodendrocyte differentiation. Development. 2000a;127:2989–2998. doi: 10.1242/dev.127.14.2989. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. The Id4 HLH protein and the timing of oligodendrocyte differentiation. EMBO Journal. 2000b;19:1998–2007. doi: 10.1093/emboj/19.9.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Raff M. Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science. 2000c;289:1754–1757. doi: 10.1126/science.289.5485.1754. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes & Development. 2004;18:2963–2972. doi: 10.1101/gad.309404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Current Opinion in Genetics & Development. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Kozik MB. The electron-microscopic picture of postnatal development of oligodendroglia. Folia Histochemica et Cytochemica (Krakow) 1976;14:99–106. [PubMed] [Google Scholar]
- Kubicek S, Jenuwein T. A crack in histone lysine methylation. Cell. 2004;119:903–906. doi: 10.1016/j.cell.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Enderich J, Hermans-Borgmeyer I, Wegner M. Cooperative function of POU proteins and SOX proteins in glial cells. Journal of Biological Chemistry. 1998;273:16050–16057. doi: 10.1074/jbc.273.26.16050. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T. The many faces of histone lysine methylation. Current Opinion in Cell Biology. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Larocque D, Galarneau A, Liu HN, Scott M, Almazan G, Richard S. Protection of p27(Kip1) mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nature Neuroscience. 2005;8:27–33. doi: 10.1038/nn1359. [DOI] [PubMed] [Google Scholar]
- Li W, Zhang B, Tang J, Cao Q, Wu Y, Wu C, et al. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating alpha-tubulin. Nature Neuroscience. 2007;27:2606–2616. doi: 10.1523/JNEUROSCI.4181-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Han YR, Li J, Sun D, Ouyang M, Plummer MR, et al. The glial or neuronal fate choice of oligodendrocyte progenitors is modulated by their ability to acquire an epigenetic memory. Journal of Neuroscience. 2007;27:7339–7343. doi: 10.1523/JNEUROSCI.1226-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, et al. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO Journal. 2006;25:4833–4842. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Muggironi M, Marin-Husstege M, Casaccia-Bonnefil P. Oligodendrocyte process outgrowth in vitro is modulated by epigenetic regulation of cytoskeletal severing proteins. Glia. 2003;44:264–274. doi: 10.1002/glia.10290. [DOI] [PubMed] [Google Scholar]
- Liu A, Stadelmann C, Moscarello M, Bruck W, Sobel A, Mastronardi FG, et al. Expression of stathmin, a developmentally controlled cytoskeleton-regulating molecule, in demyelinating disorders. Journal of Neuroscience. 2005;25:737–747. doi: 10.1523/JNEUROSCI.4174-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssiotis CA, Walker J, Wu C, Kondo T, Schultz PG, Wu X. Inhibition of histone deacetylase activity induces developmental plasticity in oligodendrocyte precursor cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14982–14987. doi: 10.1073/pnas.0707044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L, Ait-Si-Ali S, Harel-Bellan A. Histone acetylation in signal transduction by growth regulatory signals. Seminars in Cell & Developmental Biology. 1999;10:197–203. doi: 10.1006/scdb.1999.0301. [DOI] [PubMed] [Google Scholar]
- Marin-Husstege M, He Y, Li J, Kondo T, Sablitzky F, Casaccia-Bonnefil P. Multiple roles of Id4 in developmental myelination: Predicted outcomes and unexpected findings. Glia. 2006;54:285–296. doi: 10.1002/glia.20385. [DOI] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. Journal of neuroscience. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronardi FG, daCruz LA, Wang H, Boggs J, Moscarello MA. The amount of sonic hedgehog in multiple sclerosis white matter is decreased and cleavage to the signaling peptide is deficient. Multiple Sclerosis. 2003;9:362–371. doi: 10.1191/1352458503ms924oa. [DOI] [PubMed] [Google Scholar]
- Mastronardi FG, Wood DD, Mei J, Raijmakers R, Tseveleki V, Dosch HM, et al. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: A role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. Journal of Neuroscience. 2006;26:11387–11396. doi: 10.1523/JNEUROSCI.3349-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler MF, Mattick JS. Non-coding RNAs in the nervous system. Journal of Physiology. 2006;575:333–341. doi: 10.1113/jphysiol.2006.113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiological Reviews. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- Miskimins R, Srinivasan R, Marin-Husstege M, Miskimins WK, Casaccia-Bonnefil P. p27(Kip1) enhances myelin basic protein gene promoter activity. Journal of Neuroscience Research. 2002;67:100–105. doi: 10.1002/jnr.10080. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, et al. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Naruse Y, Aoki T, Kojima T, Mori N. Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13691–13696. doi: 10.1073/pnas.96.24.13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan C, Bright JJ. Curcumin inhibits experimental allergic encephalomyelitis by blocking IL-12 signaling through Janus kinase-STAT pathway in T lymphocytes. Journal of Immunology. 2002;168:6506–6513. doi: 10.4049/jimmunol.168.12.6506. [DOI] [PubMed] [Google Scholar]
- Nishiyama A. Polydendrocytes: NG2 cells with many roles in development and repair of the CNS. Neuroscientist. 2007;13:62–76. doi: 10.1177/1073858406295586. [DOI] [PubMed] [Google Scholar]
- Noble M, Murray K. Purified astrocytes promote the in vitro division of a bipotential glial progenitor cell. EMBO Journal. 1984;3:2243–2247. doi: 10.1002/j.1460-2075.1984.tb02122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlich S, Zakaryan RP, Gehring H. Protein arginine methylation: Cellular functions and methods of analysis. Bio-chimica et Biophysica Bcta. 2006;1764:1890–1903. doi: 10.1016/j.bbapap.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Molecular and Cellular Biology. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Allis CD. Histone methylation versus histone acetylation: New insights into epigenetic regulation. Current Opinion in Cell Biology. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- Richards EJ. Chromatin methylation: Who’s on first? Current Biology. 2002;12:R694–695. doi: 10.1016/s0960-9822(02)01208-3. [DOI] [PubMed] [Google Scholar]
- Romm E, Nielsen JA, Kim JG, Hudson LD. Myt1 family recruits histone deacetylase to regulate neural transcription. Journal of Neurochemistry. 2005;93:1444–1453. doi: 10.1111/j.1471-4159.2005.03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopra A, Sharling L, Wood IC, Briggs T, Bachfischer U, Paquette AJ, et al. Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Molecular and Cellular Biology. 2000;20:2147–2157. doi: 10.1128/mcb.20.6.2147-2157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto M, Hirata H, Ohtsuka T, Bessho Y, Kageyama R. The basic helix-loop-helix genes Hesr1/Hey1 and Hesr2/Hey2 regulate maintenance of neural precursor cells in the brain. Journal of Biological Chemistry. 2003;278:44808–44815. doi: 10.1074/jbc.M300448200. [DOI] [PubMed] [Google Scholar]
- Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. Journal of Cell Biology. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Liu A, Li J, Wolubah C, Casaccia-Bonnefil P. Epigenetic memory loss in aging oligodendrocytes in the corpus callosum. Neurobiology of Aging. 2007 doi: 10.1016/j.neurobiolaging.2006.10.026. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J, Natale J, Chew LJ, Belachew S, Cheng Y, Aguirre A, et al. Identification of Sox17 as a transcription factor that regulates oligodendrocyte development. Journal of Neuroscience. 2006;26:9722–9735. doi: 10.1523/JNEUROSCI.1716-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MR, Ghosh A. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nature Neuroscience. 2004;7:229–235. doi: 10.1038/nn1192. [DOI] [PubMed] [Google Scholar]
- Southwood CM, Peppi M, Dryden S, Tainsky MA, Gow A. Microtubule deacetylases, SirT2 and HDAC6, in the nervous system. Neurochemical Research. 2007;32:187–195. doi: 10.1007/s11064-006-9127-6. [DOI] [PubMed] [Google Scholar]
- Spencer VA, Davie JR. Role of covalent modifications of histones in regulating gene expression. Gene. 1999;240:1–12. doi: 10.1016/s0378-1119(99)00405-9. [DOI] [PubMed] [Google Scholar]
- Spencer VA, Davie JR. Signal transduction pathways and chromatin structure in cancer cells. Journal of Cellular Biochemistry Supplement. 2000;35:27–35. doi: 10.1002/1097-4644(2000)79:35+<27::aid-jcb1123>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Stavropoulos P, Hoelz A. Lysine-specific demethylase 1 as a potential therapeutic target. Expert Opinion on Therapeutic Targets. 2007;11:809–820. doi: 10.1517/14728222.11.6.809. [DOI] [PubMed] [Google Scholar]
- Stidworthy MF, Genoud S, Li WW, Leone DP, Mantei N, Suter U, et al. Notch1 and Jagged1 are expressed after CNS demyelination, but are not a major rate-determining factor during remyelination. Brain. 2004;127:1928–1941. doi: 10.1093/brain/awh217. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Current Biology. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes & Development. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S. Roles of jumonji and jumonji family genes in chromatin regulation and development. Developmental Dynamics. 2006;235:2449–2459. doi: 10.1002/dvdy.20851. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Developmental Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Tang XM, Beesley JS, Grinspan JB, Seth P, Kamholz J, Cambi F. Cell cycle arrest induced by ectopic expression of p27 is not sufficient to promote oligodendrocyte differentiation. Journal of Cellular Biochemistry. 1999;76:270–279. doi: 10.1002/(sici)1097-4644(20000201)76:2<270::aid-jcb10>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Taylor CM, Marta CB, Claycomb RJ, Han DK, Rasband MN, Coetzee T, Pfeiffer SE. Proteomic mapping provides powerful insights into functional myelin biology. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4643–4648. doi: 10.1073/pnas.0400922101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple S, Raff MC. Differentiation of a bipotential glial progenitor cell in a single cell microculture. Nature. 1985;313:223–225. doi: 10.1038/313223a0. [DOI] [PubMed] [Google Scholar]
- Teter B, Osterburg HH, Anderson CP, Finch CE. Methylation of the rat glial fibrillary acidic protein gene shows tissue-specific domains. Journal of Neuroscience Research. 1994;39:680–693. doi: 10.1002/jnr.490390609. [DOI] [PubMed] [Google Scholar]
- Thompson PR, Fast W. Histone citrullination by protein arginine deiminase: Is arginine methylation a green light or a roadblock? ACS Chem Biol. 2006;1:433–441. doi: 10.1021/cb6002306. [DOI] [PubMed] [Google Scholar]
- Thomson S, Clayton AL, Hazzalin CA, Rose S, Barratt MJ, Mahadevan LC. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO Journal. 1999;18:4779–4793. doi: 10.1093/emboj/18.17.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikoo R, Osterhout DJ, Casaccia-Bonnefil P, Seth P, Koff A, Chao MV. Ectopic expression of p27Kip1 in oligodendrocyte progenitor cells results in cell-cycle growth arrest. Journal of Neurobiology. 1998;36:431–440. [PubMed] [Google Scholar]
- Tokumoto YM, Apperly JA, Gao FB, Raff MC. Posttranscriptional regulation of p18 and p27 Cdk inhibitor proteins and the timing of oligodendrocyte differentiation. Developments in Biologicals. 2002;245:224–234. doi: 10.1006/dbio.2002.0626. [DOI] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Tyler JK, Kadonaga JT. The “dark side” of chromatin remodeling: Repressive effects on transcription. Cell. 1999;99:443–446. doi: 10.1016/s0092-8674(00)81530-5. [DOI] [PubMed] [Google Scholar]
- Vanrobaeys F, Van Coster R, Dhondt G, Devreese B, Van Beeumen J. Profiling of myelin proteins by 2D-gel electrophoresis and multidimensional liquid chromatography coupled to MALDI TOF-TOF mass spectrometry. J Proteome Res. 2005;4:2283–2293. doi: 10.1021/pr050205c. [DOI] [PubMed] [Google Scholar]
- Vidali G, Ferrari N, Pfeffer U. Histone acetylation: A step in gene activation. Advances in Experimental Medicine and Biology. 1988;231:583–596. doi: 10.1007/978-1-4684-9042-8_49. [DOI] [PubMed] [Google Scholar]
- Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001a;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001b;29:603–614. doi: 10.1016/s0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- Wei Q, Miskimins WK, Miskimins R. Stage-specific expression of myelin basic protein in oligodendrocytes involves Nkx2.2-mediated repression that is relieved by the Sp1 transcription factor. Advances in Experimental Medicine and Biology. 2005;280:16284–16294. doi: 10.1074/jbc.M500491200. [DOI] [PubMed] [Google Scholar]
- Werner HB, Kuhlmann K, Shen S, Uecker M, Schardt A, Dimova K, et al. Proteolipid protein is required for transport of sirtuin 2 into CNS myelin. Journal of Neuroscience. 2007;27:7717–7730. doi: 10.1523/JNEUROSCI.1254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JR. Targeting the JMJD2A histone lysine demethylase. Nature Structural and Molecular Biology. 2007;14:682–684. doi: 10.1038/nsmb0807-682. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Histone deacetylase: a regulator of transcription. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Allis CD, Coonrod S. Histone arginine methylation and its dynamic regulation. Frontiers in Bioscience. 2006;11:344–355. doi: 10.2741/1802. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Milne TA, Allis CD. Taking LSD 1 to a new high. Cell. 2005;122:654–658. doi: 10.1016/j.cell.2005.08.022. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Development. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]


