Abstract
Under specified UV-MALDI conditions, the 4-dimethylaminophenylazophenyl-4‘-maleimide (DABMI) derivative of cysteine-containing peptides undergoes prompt fragmentation to produce a characteristic mass spectral pattern. As reported previously by others, derivatization with chromophoric DABMI allows cysteine-containing peptides and proteins to be tracked during HPLC by absorbance at upper UV and visible wavelengths. Here, we describe methodology by which these same peptide derivatives can be recognized by their distinctive MALDI mass spectral fragmentation pattern, then mass mapped, allowing for easy, rapid identification of cysteine-containing peptides.
Keywords: DABMI, prompt fragmentation, MALDI
Cysteine residues play critical roles in the structure and activity of proteins. Whether involved in disulfide bonds (Anfinsen 1973), metal cofactor binding (Romero-Isart and Vasak 2002), or redox control (Moran et al. 2001; Wilcox et al. 2001), the oxidation status of cysteinyl thiol groups often dictates the level of protein activity. Analytical schemes for characterizing the cysteine status in proteins frequently involve proteolytic digestion followed by analysis of the resulting peptides. Identification of the cysteine-containing peptides among the more numerous other proteolytic peptides can be tedious and time consuming, especially if the protease has low specificity.
In working with the hydrophobic maleimide 4-dimethylaminophenylazophenyl-4‘-maleimide (DABMI) derivative of cysteine-containing peptides, we have discovered a unique and useful property, evident under specific UV-MALDI conditions, that allows for the identification of these peptides through a mass spectral pattern of ions arising from prompt fragmentation of the DABMI moiety. A valuable feature of DABMI (common to all maleimides), for analyses done in the presence of disulfide bonds, is its high specificity and reactivity with sulfhydryl groups at mildly acidic pH; this provides efficient sulfhydryl labeling, while minimizing disulfide bond scrambling. Because the maleimide only reacts with reduced cysteine residues (free thiols), the presence of a DABMI-labeled peptide in a processed sample is indicative of free cysteine in the original peptide (or in the protein, if processing involved digestion).
DABMI has been used primarily in fluorescence resonance energy transfer studies. The synthesis and use of DABMI as a tool for the selective isolation of cysteine-containing peptides was first described by Chang et al (1983). In that work, both DABMI and N-[4-(4-dimethylaminophenylazo)phenyl]iodoacetamide (DABIA) were used to label free thiol-containing peptides and proteins, such that after digestion with endoproteases, the resulting peptides could be separated by RP-HPLC and selectively detected by monitoring their absorbance of visible light. Here, we describe a powerful tool for tracking, isolating, and identifying cysteinyl peptides on the basis of the capacity to detect DABMI-labeled peptides by absorbance at wavelengths not absorbed by unlabeled peptides, combined with the ability to recognize DABMI-labeled peptides by their signature MALDI mass spectrum.
Materials and methods
Materials and methods
DABMI, porcine gastric pepsin, α-cyano-4-hydroxycinnamic acid (CHCA), sinapinic acid, 2,5-dihydroxybenzoic acid (DHB), 2-mercaptobenzothiazole (MBT), and bumetanide were obtained from Sigma-Aldrich. Insulin-like growth Factor I residues 57–70 (IGF; H2N-ALLETYCATPAKSE-COOH ) was purchased from American Peptide Company. The 6-Amino-2-mercaptobenzothiazole (AMBT) and 2-mercaptobenzoxazole (MBO) were purchased from TCI. Tyrosine hydroxylase (TH) was donated by Prof. Donald M. Kuhn of Wayne State University. All other materials were of the highest purity available.
DABMI labeling of cysteine-containing peptides
IGF was dissolved in a 6-M guanidine HCl/0.1 M Citrate buffer (pH 5.0), (Gua/Cit5) that had been saturated with acetonitrile saturated with DABMI (∼290-μM DABMI in 33% ACN/67% Gua/Cit5 [v/v]). Peptides derived from peptic digestion of TH were derivatized by adding 165 μL of the above solution to 330 μL of the digestion mixture (which was in 10 mM HCl). The reaction was allowed to take place for 1 h in the dark at room temperature.
Absorbance spectra of DABMI and derivatives
Absorbance spectra of DABMI, DABMI-IGF, and base-hydrolyzed DABMI-IGF (hydDABMI-IGF) were obtained by infusing the microbore flow cell of a properly calibrated Waters 2487 dual wavelength HPLC absorbance detector with a solution (∼30 μM) of the analyte that had been prepurified by HPLC and was dissolved in 65/35 water/acetonitrile containing 0.1% trifluoroacetic acid (v/v). An absorbance spectrum was then obtained in scan mode from 190 to 700 nm, subtracting the absorbance spectrum of pure solvent as background.
Analysis by MALDI-MS
Unless otherwise indicated, samples were prepared by spotting 0.5 μL analyte + matrix onto a gold-plated or stainless-steel well-less sample plate with CHCA as the matrix, using the modified thin-layer technique of Cadene and Chait (2000). MALDI mass spectra were acquired on a Voyager DE-STR time-of-flight (TOF) mass spectrometer (Perkin-Elmer Biosystems Inc.) equipped with a 337-nm nitrogen laser. For measurements made in linear (or reflector) mode, the accelerating voltage was set to 20,000 V (20,000 V) with grid voltage at 95% (76%), guide wire at 0.05% (0.02%), and extraction delay time at 150 nsec (150 nsec). Time-of-flight to mass conversion was achieved with the use of external standards of bradykinin (monoisotopic calculated mass for MH+ = 1060.57 Da; average mass for MH+ = 1061.22 Da), and bovine pancreatic insulin (average calculated mass for MH+ = 5734.56 Da; average calculated mass to charge ratio for [M+2H]2+ = 2867.78).
Results and Discussion
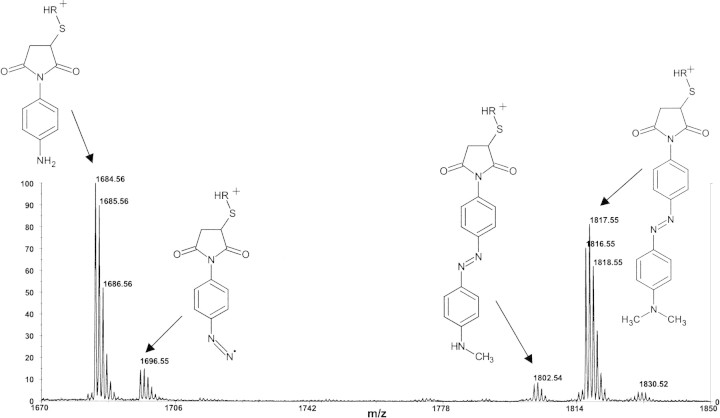
The characteristic fragmentation pattern of a DABMI-labeled peptide obtained during MALDI using CHCA as a matrix is shown in Figure 1▶. The major peaks at m/z 1684.56, m/z 1696.55, and m/z 1802.54 in Figure 1▶ (beside that at m/z 1816.55 for the protonated, intact DABMI-peptide) represent prompt fragments that, based on mass, correspond to the proposed structures shown, resulting from the loss of 132.0, 120.0, or 14.0 Da, respectively, from the protonated DABMI-peptide. Occasionally, a peak corresponding to a +14 Da mass shift can be observed. The identities of the fragments are not known with certainty, but structures are proposed in Figure 1▶. When two DABMI derivatives are present on one peptide molecule, the prompt fragmentation pattern is the same as if only one derivative were present. The fragments listed above are produced only when CHCA is used as the matrix. MALDI using sinapinic acid, bumetanide (Lavine and Allison 1999), MBT (Xu et al. 1997), AMBT (Xu et al. 1997), and MBO yield, at best, trace amounts of the fragment resulting from loss of 132 Da from MH+; under these conditions, the peaks for the other fragment ions described above are too weak to be useful. MALDI using DHB yields a substantial quantity of the −132 Da fragment, but it does not produce the −120 Da nor −14 Da fragments and, therefore, less specific information is provided. The DABMI derivative of cysteinyl peptides is chemically stable in the presence of CHCA outside of the mass spectrometer; solid-phase extraction of DABMI-IGF after being mixed and dried with CHCA and subsequent analysis by MALDI-MS with sinapinic acid as the MALDI matrix gave no indication of the prompt fragments. The thin-layer spotting technique (Cadene and Chait 2000) allows for the greatest sensitivity, but the prompt fragmentation was observed during MALDI after using other sample spotting techniques, provided CHCA was the matrix. The fragments can also be observed in negative ion mode (with CHCA as the matrix).
Figure 1.
Positive ion MALDI mass spectrum taken in reflector mode of DABMI-IGF with CHCA as the matrix. Structures are proposed for the represented ions; the charge is assumed to be located on the peptide (represented by R).
In our hands, electrospray mass spectrometry of DABMI-labeled peptides followed by low-energy collision induced dissociation in an ion trap does not produce any unusual fragmentations (none analogous to the prompt fragmentation described above); the DABMI-labeled cysteine residue behaves as any other residue (C. Borges and J. Watson, unpubl.). Viner et al (1999) have reported that under fast atom bombardment-MS/MS and increased cone-voltage electrospray conditions, a protonated fragment consisting of DABMI plus the cysteinyl sulfur atom is quite prevalent and may be useful as a marker for DABMI-labeled peptides.
Data describing the optimal wavelengths for monitoring DABMI-derivatized peptides were not available previously, and were therefore gathered by obtaining absorbance spectra of DABMI-IGF and base-hydrolyzed DABMI-IGF (hydDABMI-IGF) to determine upper UV1 and visible wavelength λmax values (Table 1). When maleimide-peptide derivatives are exposed to basic conditions, the maleimide moiety can undergo ring-opening hydrolysis or cyclization with an adjacent amine to yield a cross-linked product (Wu and Yarbrough 1976; Ishii and Lehrer 1986). (We have not observed the latter.) Thus, absorbance data on hydDABMI-IGF are included. The same prompt fragmentation products arise from base-hydrolyzed DABMI derivatives as from derivatives containing an intact maleimide ring.
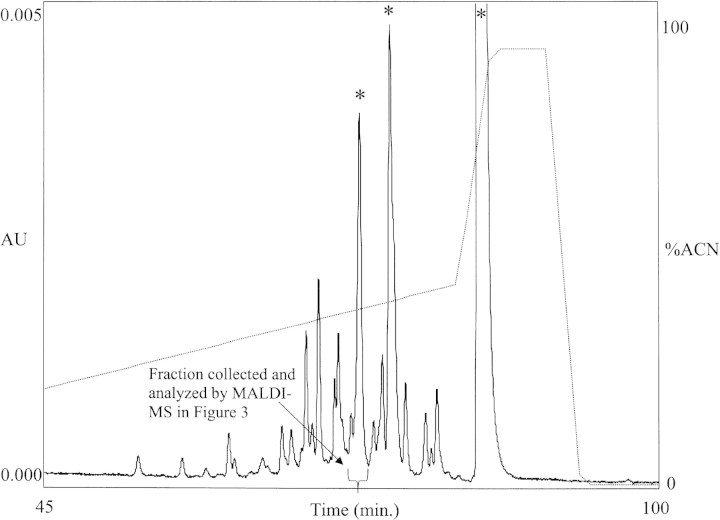
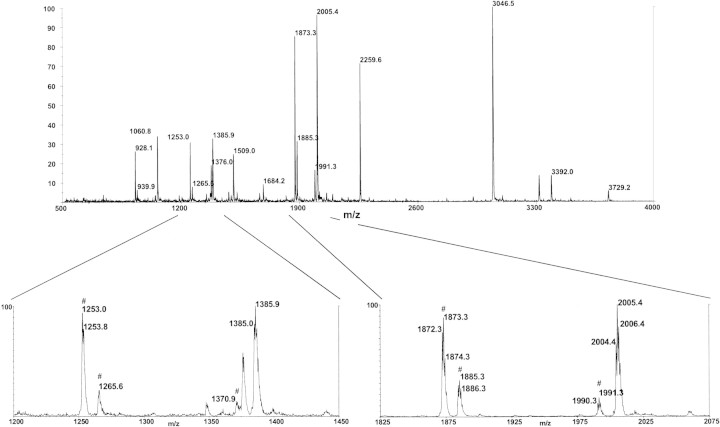
Tracking DABMI-labeled peptides by HPLC is already useful, but now cysteine-containing peptides can be recognized readily by their prompt fragmentation pattern in a MALDI mass spectrum when CHCA is used as matrix. Figure 2▶ shows an HPLC chromatogram of a peptic digest of native TH (a 498-residue protein with seven free cysteine residues) that was subsequently labeled with DABMI. (Pepsin is not a specific protease [Keil 1992], causing each cysteine residue to be distributed among 4–5 different peptic peptides.) Figure 3▶ is a MALDI mass spectrum of a chromatographic fraction collected at the time indicated in Figure 2▶, but during an analogous chromatographic run of the same sample (except ∼4.5-fold more material injected). On the basis of prompt fragmentation patterns, it is readily evident in the MALDI mass spectrum (Fig. 3▶) that there are four different cysteine-containing DABMI-labeled peptides present in the sample; their MH+ ions are represented by peaks at m/z 1060.8, m/z 1385.0, m/z 1509.0, and m/z 2004.4. Two examples are highlighted (see insets in Fig. 3▶); the peak at m/z 1385.0 represents the monoisotopic version of a DABMI-labeled MH+, whereas the peak at m/z 1253.0 represents the corresponding [MH - 132]+. The peak at m/z 2004.4 represents the monoisotopic version of another MH+, whereas the peak at m/z 1872.3 represents its corresponding [MH - 132]+ (the less abundant [MH - 120]+ and [MH - 14]+ fragment ions are also represented in the mass spectrum). (As verification that certain peaks represent fragmentation products of DABMI-labeled peptides, a MALDI mass spectrum could be taken of the sample cocrystallized with sinapinic acid instead of CHCA; in such a case, fragmentation products will not be observed.) Other peaks in Figure 3▶ represent MH+ of coeluting peptides that lack cysteine; for example, none of these other peaks is accompanied by a fragment occurring 132 u lower in the spectrum. If it were to coelute with a peptide of interest, the DABMI reagent would not interfere in the mass spectrum because its mass (320 Da) is below the low-m/z cutoff.
Figure 2.
RP-HPLC chromatogram of a DABMI-labeled peptic digest of native TH (∼60 pmole total protein). Peptides were eluted with a gradient (shown) of acetonitrile containing 0.1% trifluoroacetic acid and monitored by absorbance at 508 nm. In chromatograms of samples containing no peptides, the three largest peaks (indicated by * here) represent DABMI reagent and DABMI-related degradation products.
Figure 3.
Positive ion linear mode MALDI mass spectrum of a fraction collected between 72.25 and 73.65 min during a chromatographic run analogous to that shown in Fig. 2▶, but with a ∼4.5-fold greater quantity of the sample. Evidence for two DABMI-labeled (cysteine-containing) peptides is highlighted. One MH+ ion of a DABMI-labeled peptide is represented by a peak at m/z 1385.0, and the other by a peak at m/z 2004.4. The peaks for the accompanying fragmentation products are indicated with # in the inset spectra.
The power of using the MALDI fragmentation pattern to recognize DABMI-labeled cysteine-containing peptides can be appreciated by considering the following example of mass mapping peptic fragments of TH. As a proteolytic agent, pepsin is not specific (Keil 1992). This leads to the conundrum in results from the conventional approach that detection of a mass spectral peak for MH+ gives a molecular weight that corresponds to a dozen or more different candidate peptides. However, when DABMI fragmentation is detected as described above, the number of candidate sequences that must be considered for the associated molecular weight decreases dramatically. For example, without defining specific cleavage sites for pepsin and using a generous 0.1% error margin for the m/z measurement, the peak at m/z 2004.4 in Figure 3▶ can be mass mapped to 16 different candidate peptides originating from TH as listed in Table 2▶. However, as described earlier, the MALDI-MS fragmentation data indicate that a DABMI derivative is present, and thus, a cysteine must be present in the sequence of the peptide. Given this constraint, only 1 of the 16 candidate peptic peptides from TH fits the experimental mass value as follows: (L) KGLYATHACDABMIREHLEG ([F]; calculated monoisotopic MH+ mass = 2005.0 Da); thus the matched peptide contains Cys249. Also in Figure 3▶, the peak at m/z 1385.0 (accompanied by the peaks at m/z 1253.0, m/z 1265.6, and m/z 1370.9, constituting the DABMI signature) can be mass mapped to 18 different candidate peptides. When this list is narrowed on the basis of cysteine content requirement, two candidates remain; thus, whereas the cysteine-containing peptide remains unidentified in this case, the complexity of the assignment of the peak at m/z 1385.0 is reduced substantially.
Table 2.
Calculated masses for MH+ of candidate sequences that may be represented by the peak at m/z 2004.4 in Figure 3.
| Observed m/z | MH+ Mass Matched | Starting Residue no. | Ending Residue no. | Peptide Sequence | Cysteine Residue no. |
|---|---|---|---|---|---|
| 2004.39 | 2002.94 | 61 | 79 | (V) ASSEPGNPLEAVVFEERDG (N) | — |
| 2002.97 | 395 | 412 | (L) SSYGELLHSLSEEPEVRA (F) | — | |
| 2002.00 | 152 | 168 | (R) VSDDVRSAREDKVPWFP (R) | — | |
| 2003.03 | 20 | 37 | (S) EQDAKQAEAVTSPRFIGR (R) | — | |
| 2003.09 | 135 | 153 | (R) FEVPSGDLAALLSSVRRVS (D) | — | |
| 2003.10 | 76 | 93 | (E) ERDGNAVLNLLFSLRGTK (P) | — | |
| 2003.14 | 290 | 308 | (R) PVAGLLSARDFLASLAFRV (F) | — | |
| 2003.98 | 433 | 449 | (Y) FVSESFNDAKDKLRNYA (S) | — | |
| 2004.04 | 480 | 498 | (S) LEGVQDELHTLAHALSAIS (—) | — | |
| 2004.04 | 479 | 497 | (R) SLEGVQDELHTLAHALSAI (S) | — | |
| 2004.05 | 75 | 92 | (F) EERDGNAVLNLLFSLRGT (K) | — | |
| 2004.11 | 283 | 300 | (E) RTGFQLRPVAGLLSARDF (L) | — | |
| 2004.15 | 276 | 292 | (D) VSRFLKERTGFQLRPVA (G) | — | |
| 2004.95 | 241 | 255 | (L) KGLYATHACDABMIREHLEG (F) | 249 | |
| 2005.02 | 123 | 140 | (P) LAGSPHLEYFVRFEVPSG (D) | — | |
| 2005.13 | 197 | 212 | (S) DQVYRQRRKLIAEIAF (Q) | — |
The mass spectral fragmentation pattern indicates the presence of a DABMI-labeled cysteine, which rules out all candidates except the cysteine-containing peptide (bold).
If proteolytic cleavage and mass mapping had been carried out without derivatization by DABMI, the otherwise DABMI-labeled, cysteine-containing peptides discussed above would have been shifted down in mass by 320.1 Da, meaning that the cysteine-containing peptide would have been grouped with an entirely different list of mass-mapped peptic peptide candidates. In this case, there would have been 15 candidates [including (L) KGLYATHACREHLEG (F)] for MH+ corresponding to a theoretical peak at m/z (2004.4 − 320.1 = ) 1684.3 and five candidates for a theoretical peak at m/z (1385.0 − 320.1 = ) 1064.9. There would have been no indication, however, of cysteinyl presence and, thus, no way of distinguishing among the candidate peptides.
Conclusions
The methodology described here allows the recognition of DABMI-labeled cysteine-containing peptides based on UV or visible light absorbance and by the unique fragmentation pattern produced by the DABMI moiety during analysis by MALDI-MS. Once the MH+ ion of the DABMI derivative of a cysteine-containing peptide has been recognized through its characteristic mass spectral pattern, a simple mass-mapping procedure frequently provides the identity of the DABMI-labeled peptide. The facile recognition of cysteine-containing peptides imparted by this methodology can be particularly useful when mass-mapping peptides in a digest carried out with proteases lacking significant specificity (e.g., pepsin). During analysis of DABMI-labeled peptides, it is readily obvious when a cysteine-containing peptide is represented in the MALDI mass spectrum. When the ion represented in the mass spectrum is known to contain a cysteine residue, the number of candidate mass-matched peptides becomes greatly reduced—often allowing for identification of the peptide. Under more specific proteolytic conditions, verification of cysteinyl peptides, by way of the MALDI mass spectrum, can add a substantial degree of certainty to a mass-mapping assignment.
Table 1.
Molecular weight and λmax for UV and visible absorbance for DABMI, DABMI-IGF, and hydDABMI-IGF.
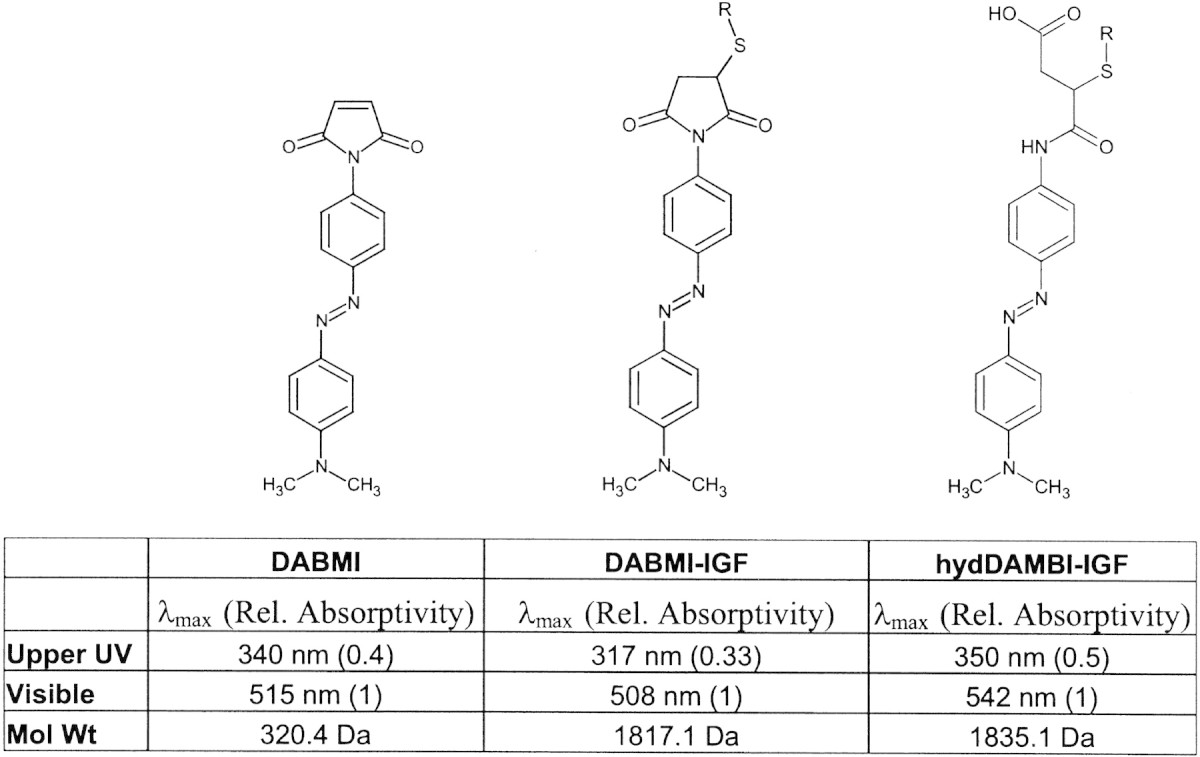
Relative absorptivity values are in relation to the visible λmax in a given spectrum.
Acknowledgments
This work was supported by NIH grant GMS R01-60576.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0301403.
Footnotes
Some absorbance detectors, such as ours, cannot simultaneously monitor both a UV and a visible wavelength due to second order interference effects. Upper UV absorbance data were therefore included as an alternative to visible wavelength absorbance detection.
References
- Anfinsen, C.B. 1973. Principles that govern the folding of protein chains. Science 181 223–230. [DOI] [PubMed] [Google Scholar]
- Cadene, M. and Chait, B.T. 2000. A robust, detergent-friendly method for mass spectrometric analysis of integral membrane proteins. Anal. Chem. 72 5655–5658. [DOI] [PubMed] [Google Scholar]
- Chang, J.Y., Knecht, R., and Braun, D.G. 1983. A new method for the selective isolation of cysteine-containing peptides. Biochem. J. 211 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, Y.and Lehrer, S.S. 1986. Effects of the state of the succinimido-ring on the fluorescence and structural properties of pyrene maleimide-labeled α α-tropomyosin. Biophys. J. 50 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil, B. 1992. Specificity of proteolysis, pp. ix, 336. Springer-Verlag, Berlin.
- Lavine, G. and Allison, J. 1999. Evaluation of bumetanide as a matrix for prompt fragmentation matrix-assisted laser desorption/ionization and demonstration of prompt fragmentation/post-source decay matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 34 741–748. [DOI] [PubMed] [Google Scholar]
- Moran, L.K., Gutteridge, J.M., and Quinlan, G.J. 2001. Thiols in cellular redox signalling and control. Curr. Med. Chem. 8 763–772. [DOI] [PubMed] [Google Scholar]
- Romero-Isart, N. and Vasak, M. 2002. Advances in the structure and chemistry of metallothioneins. J. Inorg. Biochem. 88 388–396. [DOI] [PubMed] [Google Scholar]
- Viner, R.I., Williams, T.D., and Schoneich, C. 1999. Peroxynitrite modification of protein thiols: Oxidation, nitrosylation, and S-glutathiolation of functionally important cysteine residue(s) in the sarcoplasmic reticulum Ca-ATPase. Biochemistry 38 12408–12415. [DOI] [PubMed] [Google Scholar]
- Wilcox, D.E., Schenk, A.D., Feldman, B.M., and Xu, Y. 2001. Oxidation of zinc-binding cysteine residues in transcription factor proteins. Antioxid. Redox. Signal 3 549–564. [DOI] [PubMed] [Google Scholar]
- Wu, C.W. and Yarbrough, L.R. 1976. N-(1-pyrene)maleimide: A fluorescent cross-linking reagent. Biochemistry 15 2863–2868. [DOI] [PubMed] [Google Scholar]
- Xu, N., Huang, Z.H., Watson, J.T., and Gage, D.A. 1997. Mercaptobenzothiazoles: A new class of matrices for laser desorption ionization mass spectrometry. J. Am. Soc. Mass Spectrom 8 116–124. [Google Scholar]