Abstract
The fusion of a protein of interest to a large-affinity tag, such as the maltose-binding protein (MBP), thioredoxin (TRX), or glutathione-S-transferase (GST), can be advantageous in terms of increased expression, enhanced solubility, protection from proteolysis, improved folding, and protein purification via affinity chromatography. Unfortunately, crystal growth is hindered by the conformational heterogeneity induced by the fusion tag, requiring that the tag is removed by a potentially problematic cleavage step. The first three crystal structures of fusion proteins with large-affinity tags have been reported recently. All three structures used a novel strategy to rigidly fuse the protein of interest to MBP via a short three- to five-amino acid spacer. This strategy has the potential to aid structure determination of proteins that present particular experimental challenges and are not conducive to more conventional crystallization strategies (e.g., membrane proteins). Structural genomics initiatives may also benefit from this approach as a way to crystallize problematic proteins of significant interest.
Keywords: Chimera, fusion protein, protein crystallization, protein expression, membrane proteins, molecular replacement, structural genomics, X-ray crystallography
Fusion (or chimeric) proteins are utilized in the forefront of protein science research for applications as diverse as biochemical purification, immunodetection, protein therapies, vaccine development, functional genomics, analysis of protein trafficking, and analyses of protein–protein or protein–nucleic acid interactions (Beckwith 2000). In structural biology, where milligram quantities of homogeneous protein sample are usually required, the most common utility of chimeras involves the separation of the fusion protein from the cell lysate using affinity chromatography. The most common affinity tags include the hexa-histidine (His-tag; Bornhorst and Falke 2000), Escherichia coli maltose-binding protein (MBP; Sachdev and Chirgwin 2000), Schistosoma japonicum glutathione-S-transferase (GST; Smith 2000), E. coli thioredoxin (TRX; LaVallie et al. 2000), and avidin/streptavidin Strep tags (Skerra and Schmidt 2000). Several other tags have also been developed (Stevens 2000).
To grow crystals of a protein of interest for X-ray diffraction studies, large-affinity tags, such as MBP or GST, are usually removed using site-specific proteolysis in the engineered linker region, followed by purification to separate the protein of interest from the affinity tag moiety and the protease. However, particular problems may be encountered during the cleavage step, including low yield, precipitation of the target protein, tedious optimization of cleavage conditions, high cost of proteases (e.g., factor Xa and enterokinase), or failure to recover active or structurally intact protein (Baneyx 1999). The alternative is to circumvent the cleavage and repurification steps and leave the affinity tag in place for crystallization trials. Unfortunately, this brings about a new challenge, as multidomain proteins are usually (1) less conducive to forming well-ordered, diffracting crystals, presumably due to the conformational heterogeneity allowed by the flexible linker region; and (2) too large for NMR studies. These problems explain why small affinity tags, such as the His-tag, are the tags of choice in structural biology, especially for high throughput/structural genomics approaches; they do not increase the size of the protein substantially, and cleavage of small tags is often not required to grow suitable crystals (Bucher et al. 2002).
Despite the considerations mentioned above, the first three-dimensional (3D) structures of fusion protein containing large-affinity tags have recently been reported (Kobe et al. 1999; Liu et al. 2001; Ke and Wolberger 2003). The aim of this review is to compare the use of small and large-affinity tags, focusing on their use in structural biology. In particular, we highlight the factors that have contributed to the successful crystallizations of MBP fusion proteins, and to present ideas for the potential utility of large-affinity tags for problematic structural targets. The advantages of large-affinity tags in facilitating the structural studies of small peptides have been discussed previously (Zhan et al. 2001).
Expression of fusion proteins using small and large-affinity tags
Recent estimates indicate that perhaps one-third to one-half of all prokaryotic proteins cannot be overexpressed in bacteria in soluble form using a His-tag (Edwards et al. 2000; Stevens 2000). Three recent high-throughput studies have indicated that this number is higher for eukaryotic proteins (Braun et al. 2002; Hammarstrom et al. 2002; Shih et al. 2002), particularly larger multidomain proteins. If the problem of insoluble expression of the His-tagged protein in E. coli is encountered, one or more of the following options are typically explored: altering culture growth conditions, coexpressing chaperones, changing cell lines, or switching to a different affinity tag such as MBP, GST, TRX, or NusA (Stevens 2000). Single domains may be targeted after accurate mapping of the domain boundaries using limited proteolysis and fragment analysis and/or emerging bioinformatics tools (Marsden et al. 2002; Miyazaki et al. 2002). Alternatively, eukaryotic expression systems may be used. In structural genomics, the protein may initially be left behind in the pursuit of the "low-hanging fruit", to maximize the output and leave the problematic cases to be revisited when methodology improves (Edwards et al. 2000).
Apart from affinity purification, the aforementioned large-affinity tags offer several advantages. In a recent report, TRX and MBP enhanced the solubility and expression of a test set of 32 small (<20 kD) human proteins in E. coli, compared to the His-tag expression (Hammarstrom et al. 2002). For the test sets of 32 larger human proteins (17–158 kD; Braun et al. 2002) and 40 eukaryotic proteins (9–100 kD; Shih et al. 2002), the large-affinity tags MBP (40 kD), NusA (54 kD), and GST (26 kD) were demonstrated to be helpful in improving the yield of soluble protein, whereas thioredoxin (12 kD) did not provide significant improvement in solubility compared to His-tag. In an earlier study, thioredoxin and GST provided only minor or no improvement in solubility of six notoriously insoluble proteins, whereas MBP greatly enhanced the solubility of five of the proteins (Kapust and Waugh 1999). Furthermore, chaperone-like qualities have been attributed to MBP when fused at the N-terminus, assisting in correct protein folding and acquiring active proteins (Baneyx 1999; Kapust and Waugh 1999; Sachdev and Chirgwin 2000).
The use of large-affinity tags in structural biology
The considerations discussed above suggest that largeaffinity tags may offer several advantages for structural biology applications. The 3D structures of E. coli MBP in apo- (Sharff et al. 1992) and maltose-bound (Spurlino et al. 1991; Quiocho et al. 1997) forms, S. japonicum GST (McTigue et al. 1995), and oxidized and reduced forms of E. coli TRX (Katti et al. 1990; Jeng et al. 1994) have been determined. These structures can be used as search models to solve the crystallographic phase problem by molecular replacement (MR) methods. Another possible advantage may be that the crystal contacts and the crystallization conditions successful in crystallizing the affinity tag may also be explored for crystallizing the fusion protein (Carter et al. 1994). However, this particular benefit has only been demonstrated for small peptides of 5–42 residues in length fused to S. japonicum GST (Zhan et al. 2001) or Pyrococcus furiosus MBP (Bucher et al. 2002), where the short peptides occupy the void near the location of the fusion, present among neighboring GST or MBP molecules in the crystal. Larger polypeptides would not fit into the available space, reducing the described advantages. The largest detriment to successful crystallization of fusion proteins with large-affinity tags is considered to be the conformational heterogeneity introduced by the flexible linker between the affinity tag and the protein of interest.
Recently, the first crystal structures of fusion proteins containing large-affinity tags have been reported (Kobe et al. 1999; Liu et al. 2001; Ke and Wolberger 2003); all three structures contain E. coli MBP as the affinity tag. Preliminary X-ray diffraction results have also been reported for crystals of GST (Kuge et al. 1997), TRX (Stoll et al. 1998), and MBP fusion proteins (Kukimoto et al. 2000; Table 1▶).
Table 1.
Summary of crystallization data for protein fusions to large affinity tags
| Protein | Fused/total amino acids (kDa) | Cloning vector | Linker | Protein concentration | Well solution | Resolution (Å) |
|---|---|---|---|---|---|---|
| MBP/gp21 (338–425) | 88/459 (9.9/50.4) | pMAL-c2a | AAA | 18 mg/mL | 22% PEG 4000 | 2.5 |
| 0.1 M NaOAc, pH 4.7 | ||||||
| 0.2 M (NH4)SO4 | ||||||
| MBP/gp21b (338–445) | 108/479 (9.9/52.7) | pMAL-c2a | AAA | 18 mg/mL | 1) 20% PEG 10,000 | — |
| 0.1 M HEPES, pH 7.5 | ||||||
| 2) 18% PEG 8000 | ||||||
| 0.1 M cacodylate, pH 6.5 | ||||||
| 0.2 M Zn(OAc)2 | ||||||
| MBP/SarR | 115/488 (13.7/54.5) | pMAL-c2a | AAAEF | 15 mg/mL | 18–22% PEG-MME 2000 | 2.3 |
| 0.1 M NaOAc, pH 4.6 | ||||||
| 0.1 M NaCl | ||||||
| 5 mM β-mercaptoethanol | ||||||
| MBP/MATa1 (77–126) | 50/422 (6.0/46.5) | pMAL-c2a | AAAAA | 15 mg/mL | 1) 2.4 M (NH4)2SO4 | 1) 2.1 |
| 0.1 M MES, pH 5.0 | 2) 2.3 | |||||
| 2) 20% PEG 6000 | ||||||
| 0.1 M MES pH 6.0 | ||||||
| MBP/CD38c (45–300) | 256/∼635 (29.6/71.9) | pMAL-cR1 | NAe | 10 mg/mL (+1.5 mg/mL GT1b) | 10–20% PEG 20,000 | 2.4 |
| 0.1 M HEPES, pH 7.5 | ||||||
| + 0.1 M NaI (Form I) or | ||||||
| + 0.1 M glycine (Form II) | ||||||
| TRX/VanHd | 322/∼440 (35.8/∼48) | pTRxFus | NAe | 4 mg/mL | 0.8 M NaH2PO4 | 3.0 |
| 0.4 M K2HPO4 | ||||||
| 0.1 M HEPES, pH 7 | ||||||
| GST/DREF (16–115) | 100/326 | pGEX-2T | SDLVPRGS | 15 mg/mL | 5% PEG 3350 | 2.5 |
| 50 mM KH2PO4, pH 5.2 | ||||||
| 10% ethylene glycol |
a pMal-c2 was modified to introduce truncations and mutations (see Table 2▶).
b Two crystal habits were grown but were unsuitable for X-ray diffraction.
c Two polymorphs.
d Two polymorphs grown in the same drop.
e NA, information not available.
Crystallization of proteins fused to large-affinity tags
Maltose binding protein (MBP).
In the first crystallization report of an MBP-fusion protein, two fragments of the ectodomain of the human T cell leukaemia virus type 1 (HTLV-1) envelope protein gp21 were crystallized (Center et al. 1998). Crystallization of MBP fusion proteins was pursued because of the low solubility of the gp21 fragments on their own. Crystallization trials of the longer MBP/gp21 construct (residues 335–445) containing the unmodified linker between MBP and gp21 yielded no crystals. However, after extensive modification of the junction between the MBP and gp21 domains (Table 2▶), thin plate and needle crystals were obtained, albeit not suitable for structure determination. Expressing a shorter fragment (residues 338–425) with the same modifications at the fusion junction yielded 3D crystals that diffracted to 2.5 Å resolution and allowed the structure to be determined (Kobe et al. 1999). The sequence modifications in the MBP-gp21 junction included the substitution of the 25-amino acid linker with only three alanine residues, and the mutation of charged residues near the C-terminus of MBP to alanines (Table 2▶; Center et al. 1998).
Table 2.
Comparison of the fusion junctions of MBP-fusion protein structures
| Fusion protein | MBP | Linker | Protein |
|---|---|---|---|
| MBP/gp21 (338–445) | TVDEALKDAQTN | S3N10LGIEGRISEFGS | TGSMSLAS |
| MBP/gp21 (338–425) | TVDAALAAAQTN | AAA | MSLASGKS |
| MBP/SarR | TVDEALAAAQTN | AAAEF | MSKINDIND |
| MBP/MATa1 (77–126) | TVDAALAAAQT | AAAAA | ISPQARAF |
Three sections are shown, the C-terminal helix of MBP (MBP), the linker region (Linker), and the N-terminal region of the fused protein (Protein). Residues constituting and the N-terminal helix of the fused protein are underlined, with the effective linker region between these secondary structural elements highlighted in bold. Mutations are highlighted in italic. The linker sequence typical for pMAL-c2 is shown in the example of the sequence corresponding to the fusion protein of MBP/HTLV gp21 (335–445).
The extracellular domain of the cell surface antigen CD38 in complex with ganglioside GT1b, a heptasaccharide containing lipid, is another example of a protein crystallized as a fusion to MBP (Kukimoto et al. 2000). Two crystal forms were found, one of which diffracted to 2.4 Å resolution. Attempts to solve the structure using MR with MBP or a CD38 homolog as the search model have failed, and no structure has yet been reported.
The Staphylococcus accessory regulator R (SarR) from Streptococcus aureus was also crystallized as an MBP fusion protein (Liu et al. 2001), and the analysis of the sequence of the crystallized construct shows that similar modifications to those used for the successful crystallization of MBP/gp21 were employed at the fusion junction in this case (Table 2▶). Some charged residues at the C-terminus of MBP were mutated to alanine, and the linker length was shortened to the five-residue sequence AAAEF. The crystals diffracted to 2.3-Å resolution and the structure was determined using MR methods analogous to those reported for MBP/gp21.
The most recent example of a crystallized MBP fusion protein involves a fragment of the MATa1 protein (residues 77–126) from Saccharomyces cerevisiae (Ke and Wolberger 2003). The fusion junction of MBP/MATa1 was also modified to closely resemble that of MBP/gp21 (Table 2▶), with the linker sequence truncated to a penta-alanine. Two crystal forms were obtained, diffracting to 2.1 Å and 2.3 Å resolution, respectively. The structure was determined using MR, with the maltotetraose-bound MBP structure (Quiocho et al. 1997) as a search model.
The inspection of the crystallization conditions of MBP-fusion proteins (Table 1▶) shows that polyethylene glycols (PEG) and related molecules are the most successful precipitants, and the pH is generally low (Table 1▶). These observations may be of some use in devising specific crystallization screens for MBP fusion proteins (e.g., focusing on PEGs and acidic pH); however, no clear conclusions can be drawn from the small sample size presently available, and the two MBP/MATa1 crystal forms used very different conditions.
Glutathione- S-transferase (GST).
Successful crystallizations of target proteins fused to GST have been reported for the DNA-binding domain (residues 16–115) of the Drosophila DNA replication-related element-binding factor (GST/DREF; Kuge et al. 1997) and for the mouse estrogen receptor hormone binding domain (residues 281–599; GST/ERHBD; Lally et al. 1998). These reports have been discussed in the context of carrier-driven GST-peptide crystallization (Zhan et al. 2001). Of the two proteins, only GST/DREF yielded X-ray diffraction quality crystals; however, no subsequent structure has been reported. Although the thin crystals of GST/ERHBD were unsuitable for X-ray analysis, gel electrophoresis and electron microscopy confirmed the presence of the intact fusion protein.
Thioredoxin (TRX).
Vancomycin resistance protein (VanH), a d-lactate dehydrogenase from Enterococcus faecium, has been crystallized fused to thioredoxin (Stoll et al. 1998). Crystallization of the VanH-TRX fusion protein was attempted only after enterokinase cleavage failed to yield structurally intact VanH. Two crystal forms grew from the same crystallization conditions, one of which diffracted to 3.0 Å resolution (Table 1▶). No structure has yet been reported.
Crystal structures of MBP-fusion proteins
MBP/gp21.
The envelope protein gp21 is involved in the fusion of the viral and the host cell membranes during HTLV-1–mediated infection. The chimeric protein containing MBP and the HTLV-1 gp21 ectodomain has a trimeric mushroom-like structure (Fig. 1A▶) (Kobe et al. 1999). The ∼70-Å long "stalk" is comprised of the 88 residues of each gp21 monomer assembled around a threefold crystallographic symmetry axis to form a parallel coiled coil structure. The three MBP units comprise the "cap," and importantly do not hinder the formation of the trimeric complex despite the short linker between the two domains. The trimeric state is biologically relevant in vitro and in vivo (Center et al. 1998).
Figure 1.
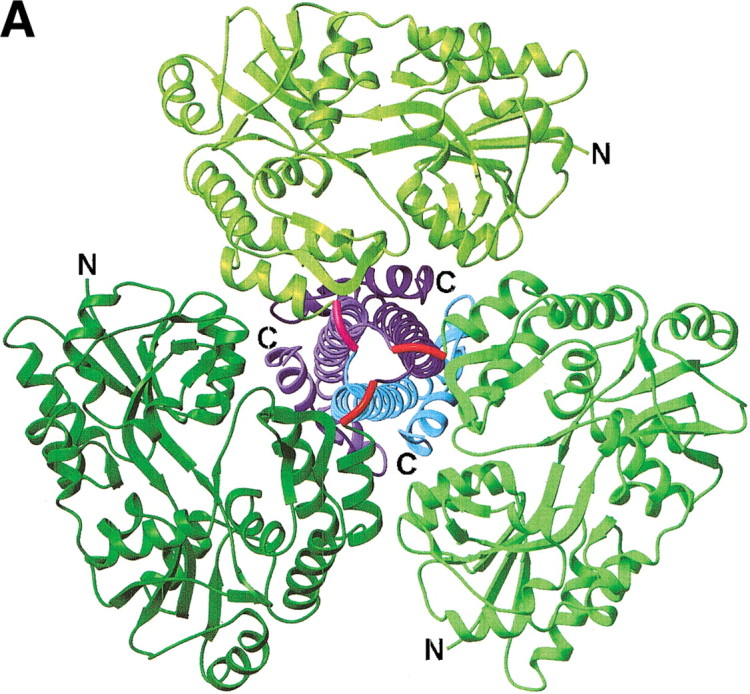
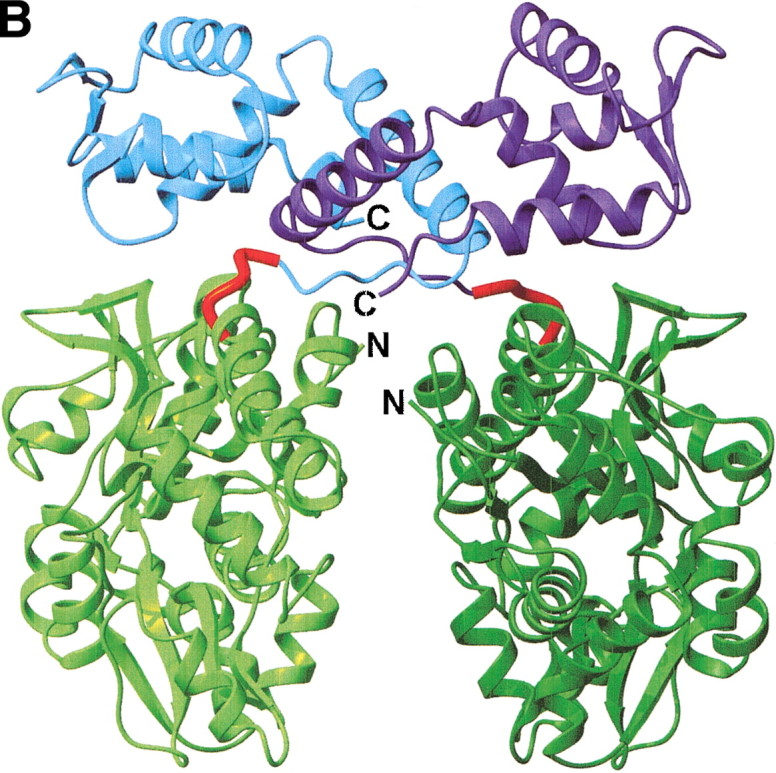
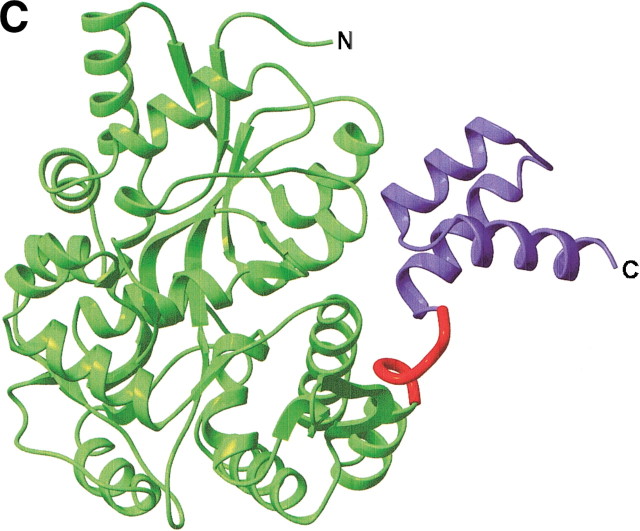
Structures of MBP fusion proteins. (A) MBP/gp21 trimer. The MBP moieties, the gp21 moieties, and the linkers are shown in different shades of green, blue, and red, respectively. (B) MBP/SarR dimer, shown as in (A). SarR moieties are shown in different shades of blue. (C) MBP/MATa1, shown as in (A). MATa1 moiety is shown in blue. The N- and C-termini are indicated. The figure was prepared with the programs MOLMOL (Koradi et al. 1996) and POV-Ray (http://www.povray.org/).
The tri-alanine linker connects the C-terminal helix of MBP with the N-terminal helix of gp21 by forming a 90° turn. This geometry buries the first 15 Å of the gp21 trimeric coiled coil in the center of the three MBP molecules, enhancing the rigid nature of the fusion. The structure also reveals that the 20 residues truncated from the C-terminus of gp21 would make little contact with the rest of the trimeric core, and could interfere with the arrangement of the MBP moieties.
MBP/SarR.
SarR from S. aureus regulates SarA expression through DNA binding. MBP/SarR forms a dimeric structure through extensive hydrophobic contacts mediated by the SarR domains (Fig. 1B▶; Liu et al. 2001). The dimer reveals a groove with appropriate dimensions and charge to bind a DNA double helix. The SarR monomers contain the typical helix-turn-helix DNA-binding domain. The MBP domains do not participate in, or hinder the dimer formation.
The first residue of the AAAEF linker represents the last residue of the C-terminal helix of MBP. The remaining four residues form part of a 10-residue loop joining the Cterminal helix of MBP to the first helix in SarR (Table 2▶; Fig. 1B▶).
MBP/MATa1.
MATa1 and MATα2 from S. cerevisiae bind DNA cooperatively to repress the transcription of haploid-specific genes. Crystallization as MBP-chimera was pursued because no crystals of free MATa1 could be obtained; the MBP/MATa1 (residues 77–126) chimera, on the other hand, produced crystals readily (one-fifth of conditions in a commercial crystallization screen produced crystals; Ke and Wolberger 2003). The structure reveals a typical homeodomain structure for the MATa1 fragment. The functional regions of MATa1 (the DNA-recognition helix and the MATα2-binding tail) are not obstructed by MBP, and the DNA-binding behavior of the chimera is the same as for the free protein.
The five-alanine linker between MBP and MATa1 adopts a turn conformation. The exact same disposition of the two moieties in the chimera is found in an alternative crystal form of the same protein. There are two residues between the poly-alanine linker and the first helix in MATa1 (Table 2▶; Fig. 1C▶).
Comparative analysis.
The most significant similarity between the three MBP fusion structures involves the short linker fusing the target proteins to MBP. The long flexible linker containing the protease cleavage site was substituted with AAA for gp21, AAAEF for SarR and AAAAA for MATa1. Each linker sequence terminates the C-terminal MBP α-helix with a turn motif immediately preceding an N-terminal α-helix from the protein of interest. The original rationale behind using a three-alanine linker in the case of MBP/gp21 was to attempt to form a rigid connection through constructing a continuous helix between the C-terminal helix of MBP and the N-terminal helix of gp21. The MBP fusion protein structures now show that instead there may be structural reasons for the formation of a 90° turn at the end of the C-terminal helix of MBP.
Significantly, no obstruction of the biologically relevant quaternary states of gp21 and SarR is induced by the close proximity of the large MBP moiety in MBP/gp21 and MBP/SarR. Only limited interactions are formed between MBP and the fused proteins, making MBP an appealing affinity tag. The structures reveal physiologically relevant multimeric states and give insight into the mechanism of biologic function.
MBP adopts distinct conformational states depending on the absence or presence of the bound maltose (Spurlino et al. 1991; Sharff et al. 1992; Boos and Shuman 1998). The MBP/gp21 and MBP/SarR structures feature MBP in the "closed" conformation, retaining a bound maltose molecule (Kd = 35 μM; Quiocho et al. 1997) from the purification stage (no maltose was added to the crystallization solution). By contrast, the two crystal forms of MBP/MATa1 have no maltose in the active site of MBP. MBP/MATa1 was purified using strong cation exchange resin, which may have facilitated the release of maltose. Importantly, the structure could still be solved by MR using a ligand-bound structure as a search model. However, the MATa1 case emphasizes that care must be taken to avoid a partial occupancy of maltose, which would result in mixed conformational states and may inhibit well-ordered crystals from forming. Extra maltose is not required to ensure conformational homogeneity of the MBP using purification and crystallization conditions similar to those employed for MBP/gp21 and MBP/SarR. However, there may be some conditions that facilitate partial maltose release producing a mixture of the two MBP conformations, in which case maltose would have to be added to the protein solution for crystallization trials.
The three protein targets in MBP/gp21, MBP/SarR, and MBP/MATa1 are small in comparison to the 368 residues of E. coli MBP to which they are fused (Table 1▶). Until further structures are determined, it is difficult to determine whether the MBP/protein ratio is significant to the success of the crystallization process. One obvious advantage of a greater MBP to protein ratio is that it facilitates the structure determination by molecular replacement methods using MBP as a search model. A further implication of having small proteins fused to MBP is the dominance of MBP in crystal lattice formation. No direct crystal contacts (other than within the oligomer) are observed between gp21, SarR, or MATa1 molecules. Instead, the crystals are assembled by the combination of MBP/MBP and MBP/protein contacts. An analysis of the crystal packing arrangements in the presently available MBP and MBP fusion protein crystal structures does not suggest any clear parallels, except that the loops protruding out furthest from the globular structure of MBP (around residues 83, 141, and 173) are most frequently involved in crystal contacts. More correlations may emerge as new structures become available, and these may be exploited to design focused crystallization protocols. Although structures of larger fusion proteins should be possible, small proteins may be more conducive to this technique by allowing the affinity tag to direct the construction of the crystal lattice.
It is possible that some of the reported crystals of fusion proteins never led to successful structure determinations because no interpretable electron density for the protein of interest could be found, as a result of the mobility of this portion of the protein in the crystals. The chance of such an outcome is minimized, in parallel with increasing the likelihood of crystallization, by a rigid connection between the affinity tag and the protein of interest.
It is also possible that a soluble fusion protein is produced, but the protein of interest is not completely folded and exists as an ensemble of conformers (Sachdev and Chirgwin 1998; Nomine et al. 2001). As such heterogeneity would be expected to impede crystallization, it is advisable to characterize the fusion protein using biophysical tools or activity assays.
Potential applications of large-affinity tags in structural biology
The use of rigid fusions of proteins to large-affinity tags may be a viable alternative to the use of small tags in structural biology, when it overcomes the problem of obtaining an active, soluble sample in sufficient quantity and concentration (particularly when there are additional problems associated with the removal of the tag). This strategy is likely to find a niche role in the determination of challenging target proteins that would otherwise prove fruitless using other more conventional strategies.
Membrane proteins
Membrane proteins represent a specific class of proteins that continue to be challenging for structural studies. Difficulties pose themselves both during recombinant expression in heterologous systems and crystallization steps. Overexpression of membrane proteins in E. coli (Grisshammer and Tate 1995; Tate 2001; Quick and Wright 2002) fused to large-affinity tags has been reported using MBP (Grisshammer et al. 1993, 1994; Chen and Gouaux 1996; Su et al. 1996; Kanamori et al. 1999; Stanasila et al. 1999; Weiss and Grisshammer 2002), GST (Panayotova-Heiermann et al. 1999; Huang et al. 2002), and thioredoxin (Therien et al. 2002).
Crystals of membrane proteins are categorized into three general architectures: 2D, type I 3D, and type II 3D crystals (Michel 1991; Abramson and Iwata 1999). A 2D crystal is constructed when membrane protein molecules are ordered side by side in a lipid bilayer representative of the native membrane structure (Fig. 2A▶). Type I 3D crystals are layers of 2D crystals built up in the third dimension. In type II crystals, detergent-solubilized protein molecules are held together by crystal contacts mediated by the hydrophilic portions of the protein (protruding from the detergent-covered, hydrophobic transmembrane regions). Medium- to low-resolution 3D structural information can be obtained from ultrathin 2D crystals (by cryoelectron microscopy; Saibil 2000), although type I and type II 3D crystals may be suitable for X-ray diffraction.
Figure 2.
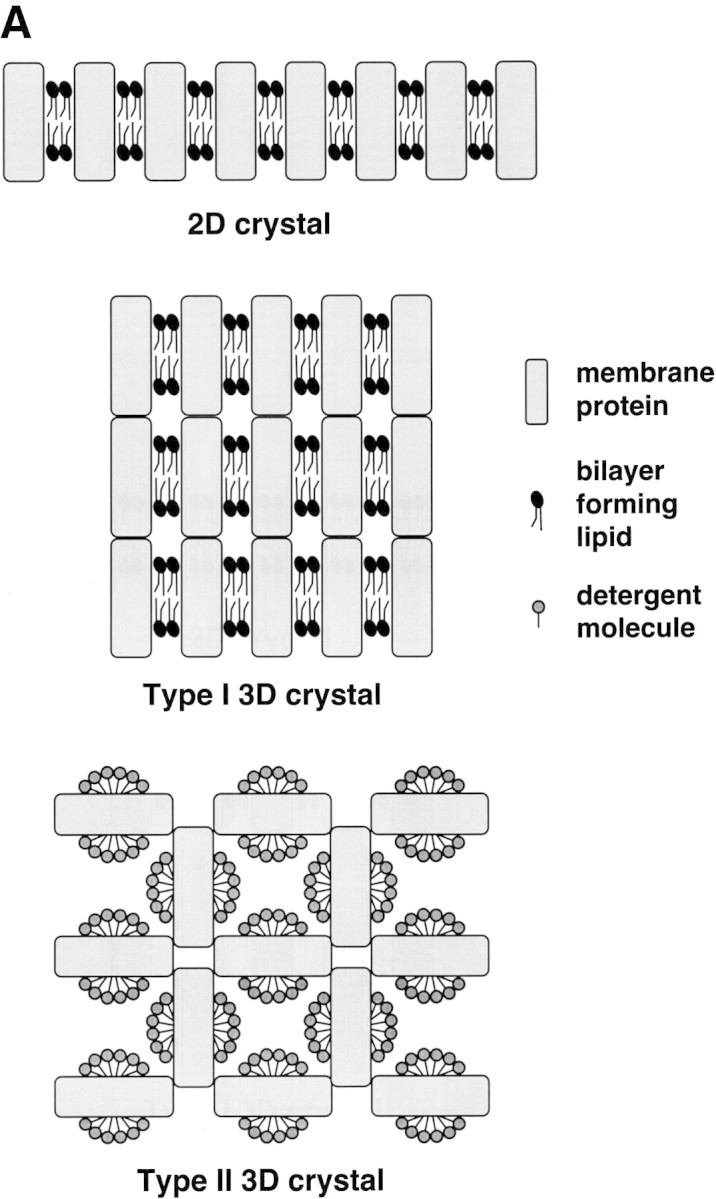
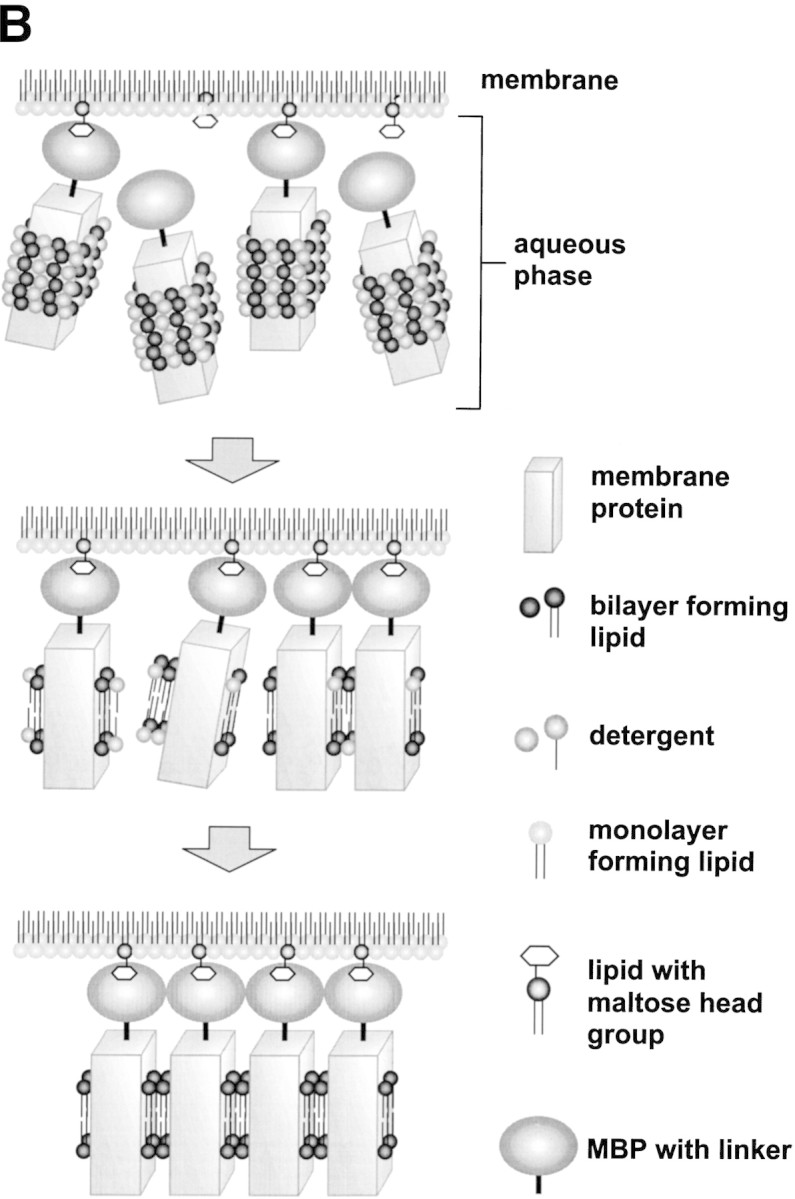
(A) Three architectures of membrane protein crystals. (B) A method for growing 2D membrane protein crystals using MBP fusion proteins.
One method of growing 2D crystals utilizes affinity of the protein for the polar head groups of natural or synthetic lipids, to allow the self-assembly of protein molecules on a planar lipid film (Fig. 2B▶; Brisson et al. 1999; Levy et al. 1999, 2001). Using this technique, the ligand lipid is diluted with a second lipid and deposited onto an aqueous solution of the protein so that the lipids spread into a monolayer at the water–air interface. Fusion proteins containing large-affinity tags may bind lipids that incorporate the ligand for the affinity tag; for example, lipids carrying a maltose polar head group may be used to bind MBP fusion proteins. Assembly into 2D crystals occurs through substitution of the detergent micelles with a lipid bilayer and the formation of specific intermolecular contacts between neighboring hydrophilic portions of molecules.
Increasing the size of the hydrophilic domain of membrane proteins through the use of antibody fragments has been demonstrated to aid crystallization (Hunte and Michel 2002). A hydrophilic domain, such as an affinity tag, may be used to play a similar role (Hunte and Michel 2002). Rigid fusion can be achieved through the short linker approach described above for soluble proteins, or by insertion of the fusion protein into one of the extramembrane loops. This latter approach has been successful in preparing 2D crystals of the fusion of cytochrome b562 to lactose permease (Prive and Kaback 1996; Zhuang et al. 1999).
Structural genomics.
In the early stages of structural genomics, challenging targets such as those that express poorly or yield insoluble protein will be put aside in the pursuit of the "lower hanging fruit" (Edwards et al. 2000). The "low-hanging fruit" usually corresponds to proteins that express as soluble His-tag fusion proteins in bacteria, but they may not always be the most desired targets (Vitkup et al. 2001). One alternative is expression in E. coli as a fusion to a large-affinity tag such as MBP, NusA, GST, thioredoxin, and others (Braun et al. 2002; Hammarstrom et al. 2002; Shih et al. 2002). Incorporation of an additional small affinity tag (such as the His-tag) greatly increases the efficiency of purification as well as minimizes the losses due to poor binding to the affinity column, a problem often encountered for MBP fusions (Baneyx 1999; Braun et al. 2002; Routzahn and Waugh 2002). The proteins expressed and purified in this way need to progress through cleavage and repurification steps before entering crystallization trials or NMR studies.
Expression of proteins rigidly fused to the MBP tag could be pursued in parallel as an alternative approach. Potentially, proteins expressed this way would retain the solubility and purification advantages observed for the longer construct (Routzahn and Waugh 2002) yet could proceed straight from purification to crystallization trials.
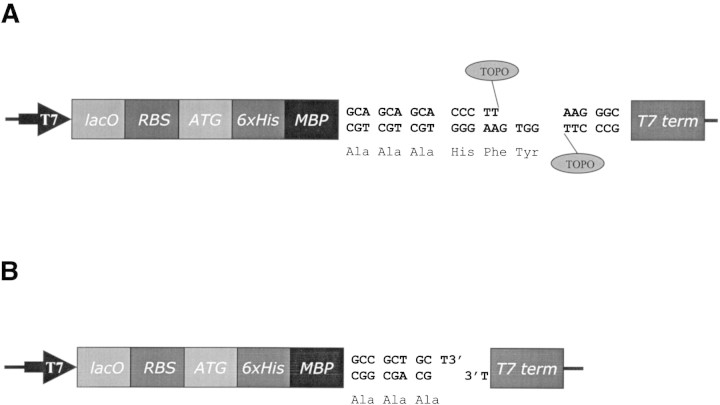
High-throughput approaches require methods for cloning and expression that can be applied simultaneously to a large number of target proteins. One efficient system with such properties is the directional TOPO cloning technology (Invitrogen Life Technologies) that requires no ligase or restriction enzymes. The technology could be adapted to the large-affinity tag-short linker system; however, the linker would have to contain three amino acids (the result of topoisomerase I action and an integral part of TOPO directional cloning), in addition to the three-alanine or similar spacer of choice (Fig. 3A▶).
Figure 3.
(A) Schematic diagram of the proposed TOPO expression vector, incorporating the MBP affinity tag, three alanines, and a TOPO directional cloning site. T7, T7 promoter; lacO, lac operon; RBS, ribosome binding site; ATG, translation initiation codon; 6xHis, His-tag; MBP, maltose binding protein; T7 term, T7 termination region; TOPO, topoisomerase I. (B) Schematic diagram of the proposed nondirectional expression vector, incorporating the MBP affinity tag and three alanines, labeled as in (A). 3’ T overhangs are indicated.
Another possibility would be to use a modification of a classic bacterial plasmid expression vector, encoding MBP followed by the sequence GCCGCTGCGCA. This sequence encodes three alanines, with the underlined nucleotides representing the recognition site for the blunt end-producing restriction enzyme FspI. The sequence could be modified to incorporate more alanine residues, if required. After a restriction digest of the vector (resulting in linearized blunt-ended DNA), 3′-terminal thymidines would be added to both ends (Fig. 3B▶), and the resulting overhangs used for an efficient ligation of the target protein PCR product (generated by thermostable polymerases producing A-overhangs). The use of this vector system would avoid the incorporation of any unwanted amino acids (as in the case of the TOPO-based strategy), as well as the use of a multiple cloning site (used in most available expression systems). However, the proposed method is a nondirectional cloning method, requiring an additional step confirming the correct orientation of cloned sequence.
An advantage of the "rigid fusion" technology is that X-ray diffraction data could potentially be collected at the home laboratory and solved using MR methods, or known binding sites for heavy atoms can be utilized for phasing (Spurlino et al. 1991; Rubin et al. 2002), as alternatives to Se-Met incorporation routinely used in structural genomics (MBP contains 7 Met residues).
Conclusions
It is clear that large-affinity tags offer an advantage over a small tag when expressing recombinant proteins, and the resulting fusion proteins have found extensive use in diverse functional studies. Although only a few successful crystallizations of proteins fused to large-affinity tags have been reported, they set an important precedent for also using the fusion proteins for structural studies. The analysis of these structures demonstrates that deliberate modification of the fusion junction will usually be required to generate high quality crystals for X-ray diffraction. A rigid connection must be established between the two domains to remove the impediment of conformational heterogeneity that exists when long flexible linkers are present. This technique may occupy a niche role in the structural biologist’s toolbox for obtaining 3D information of challenging target proteins.
Acknowledgments
We thank Ben Hankamer and Trazel Teh for useful discussions. B.K. is an NHMRC Senior Research Fellow.
Note added in proof
Chao et al. reported the crystal structure of the MBP–Saccharomyces cerevisiae ribosomal protein L30 fusion protein. The protein was crystallized in the presence of maltose, and the crystal structure was solved by molecular replacement using the structure of MBP, and refined at 2.31 Å resolution. No modifications to the MBP–protein linker were reported. (Chao, J.R., Prasad, G.S., White, S.A., Stout, D.C., and Williamson, J.R. 1999. Inherent protein structural flexibility at the RNA-binding interface of L30e. J. Mol. Biol. 326: 999–1004.)
Abbreviations
2D, two-dimensional
3D, three-dimensional
GST, glutathione-S-transferase
His-tag, hexahistidine-tag
MBP, maltose-binding protein
MR, molecular replacement
PEG, polyethylene glycol
TRX, thioredoxin
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0243403.
References
- Abramson, J. and Iwata, S. 1999. Crystallization of membrane proteins. In Protein crystallization: Techniques, strategies, and tips: A laboratory manual (ed. T.M. Bergfors), pp. 199–210. International University Line, La Jolla, CA.
- Baneyx, F. 1999. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 10 411–421. [DOI] [PubMed] [Google Scholar]
- Beckwith, J. 2000. The all purpose gene fusion. Methods Enzymol. 326 3–7. [DOI] [PubMed] [Google Scholar]
- Boos, W. and Shuman, H. 1998. Maltose/maltodextrin system of Escherichia coli: Transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornhorst, J.A. and Falke, J.J. 2000. Purification of proteins using polyhistidine affinity tags. Methods Enzymol. 326 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, P., Hu, Y.H., Shen, B.H., Halleck, A., Koundinya, M., Harlow, E., and LaBaer, J. 2002. Proteome-scale purification of human proteins from bacteria. Proc. Natl. Acad. Sci. 99 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson, A., Lambert, O., and Bergsma-Schutter, W. 1999. Two-dimensional crystallization of soluble proteins on planar lipid films. In Crystallization of nucleic acids and proteins: A practical approach, 2nd ed. (eds. A. Ducruix and R. Giegé). Oxford University Press, Oxford, UK.
- Bucher, M.H., Evdokimov, A.G., and Waugh, D.S. 2002. Differential effects of short affinity tags on the crystallization of Pyrococcus furiosus maltodextrin-binding protein. Acta Crystallogr. D58 392–397. [DOI] [PubMed] [Google Scholar]
- Carter, D.C., Rüker, F., Ho, J.X., Lim, K., Keeling, K., Gilliland, G., and Ji, X. 1994. Fusion proteins as alternative crystallization paths to difficult structure problems. Protein Pept. Lett. 1 175–178. [Google Scholar]
- Center, R.J., Kobe, B., Wilson, K.A., Teh, T., Howlett, G.J., Kemp, B.E., and Poumbourios, P. 1998. Crystallization of a trimeric human T cell leukemia virus type 1 gp21 ectodomain fragment as a chimera with maltose-binding protein. Protein Sci. 7 1612–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G.Q. and Gouaux, J.E. 1996. Overexpression of bacterio-opsin in Escherichia coli as a water-soluble fusion to maltose binding protein: Efficient regeneration of the fusion protein and selective cleavage with trypsin. Protein Sci. 5 456–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, A.M., Arrowsmith, C.H., Christendat, D., Dharamsi, A., Friesen, J.D., Greenblatt, J.F., and Vedadi, M. 2000. Protein production: Feeding the crystallographers and NMR spectroscopists. Nat. Struct. Biol. Suppl. 7 970–972. [DOI] [PubMed] [Google Scholar]
- Grisshammer, R. and Tate, C.G. 1995. Overexpression of integral membrane-proteins for structural studies. Q. Rev. Biophys. 28 315–422. [DOI] [PubMed] [Google Scholar]
- Grisshammer, R., Duckworth, R., and Henderson, R. 1993. Expression of a rat neurotensin receptor in Escherichia coli. Biochem. J. 295 571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisshammer, R., Little, J., and Aharony, D. 1994. Expression of rat Nk-2 (Neurokinin-a) receptor in Escherichia coli. Recept. Channels 2 295–302. [PubMed] [Google Scholar]
- Hammarstrom, M., Hellgren, N., Van den Berg, S., Berglund, H., and Hard, T. 2002. Rapid screening for improved solubility of small human proteins produced as fusion proteins in Escherichia coli. Protein Sci. 11 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B., Subramaniam, S., Frey, J., Loh, H., Tan, H.M., Fernandez, C.J., Kwang, J., and Chua, K.L. 2002. Vaccination of ducks with recombinant outer membrane protein (OmpA) and a 41 kDa partial protein (P45N‘) of Riemerella anatipestifer. Vet. Microbiol. 84 219–230. [DOI] [PubMed] [Google Scholar]
- Hunte, C. and Michel, H. 2002. Crystallisation of membrane proteins mediated by antibody fragments. Curr. Opin. Struct. Biol. 12 503–508. [DOI] [PubMed] [Google Scholar]
- Jeng, M.F., Campbell, A.P., Begley, T., Holmgren, A., Case, D.A., Wright, P.E., and Dyson, H.J. 1994. High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin. Structure 2 853–868. [DOI] [PubMed] [Google Scholar]
- Kanamori, M., Kamata, H., Yagisawa, H., and Hirata, H. 1999. Overexpression of the alanine carrier protein gene from thermophilic bacterium PS3 in Escherichia coli. J. Biochem. (Tokyo) 125 454–459. [DOI] [PubMed] [Google Scholar]
- Kapust, R.B. and Waugh, D.S. 1999. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 8 1668–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katti, S.K., Lemaster, D.M., and Eklund, H. 1990. Crystal structure of thioredoxin from Escherichia coli at 1.68 Å resolution. J. Mol. Biol. 212 167–184. [DOI] [PubMed] [Google Scholar]
- Ke, A. and Wolberger, C. 2003. Insights into binding cooperativity of MATa1/MATα2 from the crystal structure of a MATa1 homeodomain-maltose binding protein chimera. Protein Sci. 12 306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe, B., Center, R.J., Kemp, B.E., and Poumbourios, P. 1999. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. 96 4319–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14 51–55. [DOI] [PubMed] [Google Scholar]
- Kuge, M., Fujii, Y., Shimizu, T., Hirose, F., Matsukage, A., and Hakoshima, T. 1997. Use of a fusion protein to obtain crystals suitable for X-ray analysis: Crystallization of a GST-fused protein containing the DNA-binding domain of DNA replication-related element-binding factor, DREF. Protein Sci. 6 1783–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukimoto, M., Nureki, O., Shirouzu, M., Katada, T., Hirabayashi, Y., Sugiya, H., Furuyama, S., Yokoyama, S., and Hara-Yokoyama, M. 2000. Crystallization and preliminary X-ray diffraction analysis of the extracellular domain of the cell surface antigen CD38 complexed with ganglioside. J. Biochem. (Tokyo) 127 181–184. [DOI] [PubMed] [Google Scholar]
- Lally, J.M., Newman, R.H., Knowles, P.P., Islam, S., Coffer, A.I., Parker, M., and Freemont, P.S. 1998. Crystallization of an intact GST-estrogen receptor hormone binding domain fusion protein. Acta Crystallogr. D54 423–426. [DOI] [PubMed] [Google Scholar]
- LaVallie, E.R., Lu, Z.J., Diblasio-Smith, E.A., Collins-Racie, L.A., and McCoy, J.M. 2000. Thioredoxin as a fusion partner for production of soluble recombinant proteins in Escherichia coli. Methods Enzymol. 326 322–340. [DOI] [PubMed] [Google Scholar]
- Levy, D., Mosser, G., Lambert, O., Moeck, G.S., Bald, D., and Rigaud, J.L. 1999. Two-dimensional crystallization on lipid layer: A successful approach for membrane proteins. J. Struct. Biol. 127 44–52. [DOI] [PubMed] [Google Scholar]
- Levy, D., Chami, M., and Rigaud, J.L. 2001. Two-dimensional crystallization of membrane proteins: The lipid layer strategy. FEBS Lett. 504 187–193. [DOI] [PubMed] [Google Scholar]
- Liu, Y.F., Manna, A., Li, R.G., Martin, W.E., Murphy, R.C., Cheung, A.L., and Zhang, G.Y. 2001. Crystal structure of the SarR protein from Staphylococcus aureus. Proc. Natl. Acad. Sci. 98 6877–6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden, R.L., McGuffin, L.J., and Jones, D.T. 2002. Rapid protein domain assignment from amino acid sequence using predicted secondary structure. Protein Sci. 11 2814–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTigue, M.A., Williams, D.R., and Tainer, J.A. 1995. Crystal structures of a Schistosomal drug and vaccine target—Glutathione-S-transferase from Schistosoma japonica and its complex with the leading antischistosomal drug praziquantel. J. Mol. Biol. 246 21–27. [DOI] [PubMed] [Google Scholar]
- Michel, H. 1991. General and practical aspects of membrane protein crystallization. In Crystallization of membrane proteins (ed. H. Michel), pp. 73–88. CRC Press, Boca Raton, FL.
- Miyazaki, S., Kuroda, Y., and Yokoyama, S. 2002. Characterization and prediction of linker sequences of multi-domain proteins by a neural network. J. Struct. Funct. Genom. 2 37–51. [DOI] [PubMed] [Google Scholar]
- Nomine, Y., Ristriani, T., Laurent, C., Lefevre, J.F., Weiss, E., and Trave, G. 2001. Formation of soluble inclusion bodies by hpv e6 oncoprotein fused to maltose-binding protein. Protein Expr. Purif. 23 22–32. [DOI] [PubMed] [Google Scholar]
- Panayotova-Heiermann, M., Leung, D.W., Hirayama, B.A., and Wright, E.M. 1999. Purification and functional reconstitution of a truncated human Na+/glucose cotransporter (SGLT1) expressed in E. coli. FEBS Lett. 459 386–390. [DOI] [PubMed] [Google Scholar]
- Prive, G.G. and Kaback, H.R. 1996. Engineering the lac permease for purification and crystallization. J. Bioenerg. Biomembr. 28 29–34. [PubMed] [Google Scholar]
- Quick, M. and Wright, E.M. 2002. Employing Escherichia coli to functionally express, purify, and characterize a human transporter. Proc. Natl. Acad. Sci. 99 8597–8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiocho, F.A., Spurlino, J.C., and Rodseth, L.E. 1997. Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport chemosensory receptor. Structure 5 997–1015. [DOI] [PubMed] [Google Scholar]
- Routzahn, K.M. and Waugh, D.S. 2002. Differential effects of supplementary affinity tags on the solubility of MBP fusion proteins. J. Struct. Funct. Genom. 2 83–92. [DOI] [PubMed] [Google Scholar]
- Rubin, S.M., Lee, S.Y., Ruiz, E.J., Pines, A., and Wemmer, D.E. 2002. Detection and characterization of xenon-binding sites in proteins by Xe-129 NMR spectroscopy. J. Mol. Biol. 322 425–440. [DOI] [PubMed] [Google Scholar]
- Sachdev, D. and Chirgwin, J.M. 1998. Solubility of proteins isolated from inclusion bodies is enhanced by fusion to maltose-binding protein or thioredoxin. Protein Expr. Purif. 12 122–132. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Fusions to maltose-binding protein: Control of folding and solubility in protein purification. Methods Enzymol. 326 312–321. [DOI] [PubMed] [Google Scholar]
- Saibil, H.R. 2000. Macromolecular structure determination by cryo-electron microscopy. Acta Crystallogr. D56 1215–1222. [DOI] [PubMed] [Google Scholar]
- Shih, Y.P., Kung, W.M., Chen, J.C., Yeh, C.H., Wang, A.H.J., and Wang, T.F. 2002. High-throughput screening of soluble recombinant proteins. Protein Sci. 11 1714–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerra, A. and Schmidt, T.G.M. 2000. Use of the Strep-tag and streptavidin for detection and purification of recombinant proteins. Methods Enzymol. 326 271–304. [DOI] [PubMed] [Google Scholar]
- Smith, D.B. 2000. Generating fusions to glutathione S-transferase for protein studies. Methods Enzymol. 326 254–270. [DOI] [PubMed] [Google Scholar]
- Spurlino, J.C., Lu, G.-Y., and Quiocho, F.A. 1991. The 2.3-Å resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J. Biol. Chem. 266 5202–5219. [DOI] [PubMed] [Google Scholar]
- Stanasila, L., Massotte, D., Kieffer, B.L., and Pattus, F. 1999. Expression of δ, κ and μ human opioid receptors in Escherichia coli and reconstitution of the high-affinity state for agonist with heterotrimeric G proteins. Eur. J. Biochem. 260 430–438. [DOI] [PubMed] [Google Scholar]
- Stevens, R.C. 2000. Design of high-throughput methods of protein production for structural biology. Struct. Fold. Des. 8 R177–R185. [DOI] [PubMed] [Google Scholar]
- Stoll, V.S., Manohar, A.V., Gillon, W., Macfarlane, E.L.A., Hynes, R.C., and Pai, E.F. 1998. A thioredoxin fusion protein of VanH, a D-lactate dehydrogenase from Enterococcus faecium: Cloning, expression, purification, kinetic analysis, and crystallization. Protein Sci. 7 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, H., Raymond, L., Rockey, D.D., Fischer, E., Hackstadt, T., and Caldwell, H.D. 1996. A recombinant Chlamydia trachomatis major outer membrane protein binds to heparan sulfate receptors on epithelial cells. Proc. Natl. Acad. Sci. 93 11143–11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate, C.G. 2001. Overexpression of mammalian integral membrane proteins for structural studies. FEBS Lett. 504 94–98. [DOI] [PubMed] [Google Scholar]
- Therien, A.G., Glibowicka, M., and Deber, C.M. 2002. Expression and purification of two hydrophobic double-spanning membrane proteins derived from the cystic fibrosis transmembrane conductance regulator. Protein Expr. Purif. 25 81–86. [DOI] [PubMed] [Google Scholar]
- Vitkup, D., Melamud, E., Moult, J., and Sander, C. 2001. Completeness in structural genomics. Nat. Struct. Biol. 8 559–566. [DOI] [PubMed] [Google Scholar]
- Weiss, H.M. and Grisshammer, R. 2002. Purification and characterization of the human adenosine A(2a) receptor functionally expressed in Escherichia coli. Eur. J. Biochem. 269 82–92. [DOI] [PubMed] [Google Scholar]
- Zhan, Y., Song, X., and Zhou, G.W. 2001. Structural analysis of regulatory protein domains using GST-fusion proteins. Gene 281 1–9. [DOI] [PubMed] [Google Scholar]
- Zhuang, J.P., Prive, G.G., Werner, G.E., Ringler, P., Kaback, H.R., and Engel, A. 1999. Two-dimensional crystallization of Escherichia coli lactose permease. J. Struct. Biol. 125 63–75. [DOI] [PubMed] [Google Scholar]