Abstract
The interaction between ligands and proteins usually induces changes in protein thermal stability with modifications in the midpoint denaturation temperature, enthalpy of unfolding, and heat capacity. These modifications are due to the coupling of unfolding with binding equilibrium. Furthermore, they can be attained by changes in protein structure and conformational flexibility induced by ligand interaction. To study these effects we have used bovine serum albumin (BSA) interacting with three different anilinonaphthalene sulfonate derivatives (ANS). These ligands have different effects on protein stability, conformation, and dynamics. Protein stability was studied by differential scanning calorimetry and fluorescence spectroscopy, whereas conformational changes were detected by circular dichroism and infrared spectroscopy including kinetics of hydrogen/deuterium exchange. The order of calorimetric midpoint of denaturation was: 1,8-ANS-BSA > 2,6-ANS-BSA > free BSA >> (nondetected) bis-ANS-BSA. Both 1,8-ANS and 2,6-ANS did not substantially modify the secondary structure of BSA, whereas bis-ANS induced a distorted α-helix conformation with an increase of disordered structure. Protein flexibility followed the order: 1,8-ANS-BSA < 2,6-ANS-BSA < free BSA << bis-ANS-BSA, indicating a clear correlation between stability and conformational flexibility. The structure induced by an excess of bis-ANS to BSA is compatible with a molten globule-like state. Within the context of the binding landscape model, we have distinguished five conformers (identified by subscript): BSA1,8-ANS, BSA2,6-ANS, BSAfree, BSAbis-ANS, and BSAunfolded among the large number of possible states of the conformational dynamic ensemble. The relative population of each distinguishable conformer depends on the type and concentration of ligand and the temperature of the system.
Keywords: Protein stability, ligand binding, dynamic landscape, calorimetry, H/D exchange
The interaction of proteins with small ligands often takes place with an increase in protein thermostability due to the coupling of binding with unfolding equilibrium (Fukada et al. 1983; Brandts and Lin 1990; Shrake and Ross 1990, 1992). Previous works from our laboratory have demonstrated that the tight binding of biotin to streptavidin or avidin increases the temperature of midpoint denaturation (Tm) from 75° and 85°C in the free form to 112° and 117°C, respectively, at full ligand saturation (González et al. 1997, 1999). The increase in the thermal stability of streptavidin goes together with an increase in the unfolding cooperativity and a substantial increase in the structural order as measured by infrared spectroscopy (González et al. 1997).
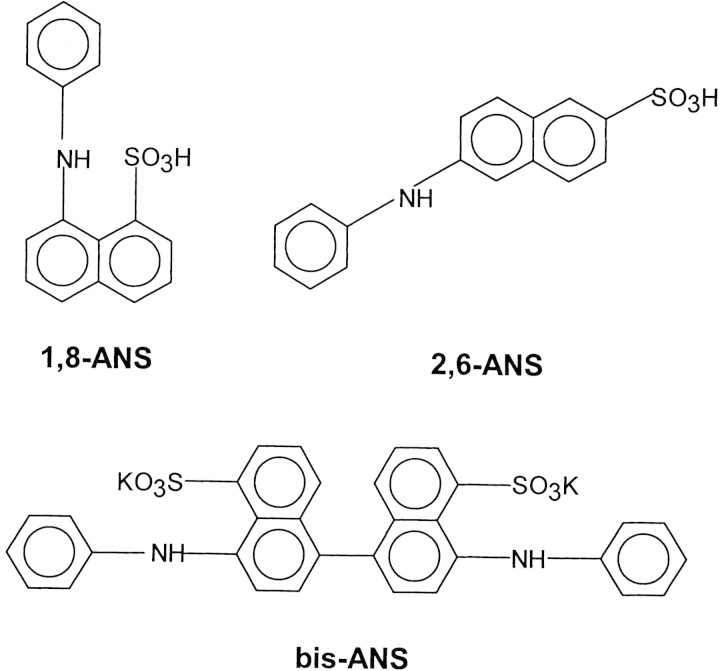
It has been observed that mutations that change the packing of the inner core modify protein stability (Munson et al. 1996; Richards 1997). In this connection, it could be expected that modifications in protein stability correlate with changes in the protein packing induced by ligand binding. Similar to the case of streptavidin and avidin, the binding of different ligands induces an increase in the thermal stability of serum albumin (Shrake and Ross 1988; Rosso et al. 1998). In the present study we used bovine serum albumin (BSA) and anilinonaphthalene sulphonate (ANS) derivatives as experimental system to study the correlation between changes in protein thermal stability and conformational dynamics induced by ligand binding. We have used three different ANS derivatives: 8-anilinonaphthalene-1-sulfonic acid (1,8-ANS), 6-anilinonaphthalene-2-sulfonic acid (2,6-ANS), and 4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid, dipotassium salt (bis-ANS). The chemical structure for each ligand is shown in Figure 1▶. These fluorescent probes have been frequently used to study hydrophobic sites, conformational changes, and to detect the presence of the molten globule state in proteins (Shi et al. 1994; Arighi et al. 1998). However, possible effects of these dyes on protein conformation and stability have been largely overlooked (Shi et al. 1994; Ali et al. 1999; Matulis et al. 1999). In the present work we have studied the effect of ANS derivatives binding on BSA thermostability by using differential scanning calorimetry (DSC) and fluorescence spectroscopy. The effect of binding on protein structure and conformational flexibility was studied by circular dichroism (CD) and Fourier transform infrared spectroscopy (FTIR) including kinetics of hydrogen/deuterium exchange. We have found a correlation between changes in protein thermostability and flexibility induced by ligand binding.
Figure 1.
Chemical structure of ANS dyes.
Results
Thermal-induced unfolding of BSA–ANS complexes
Figure 2▶ shows the thermograms of free BSA and BSA in the presence of different ANS derivatives. The thermodynamic parameters obtained from the calorimetric curves are summarized in Table 1▶.
Figure 2.
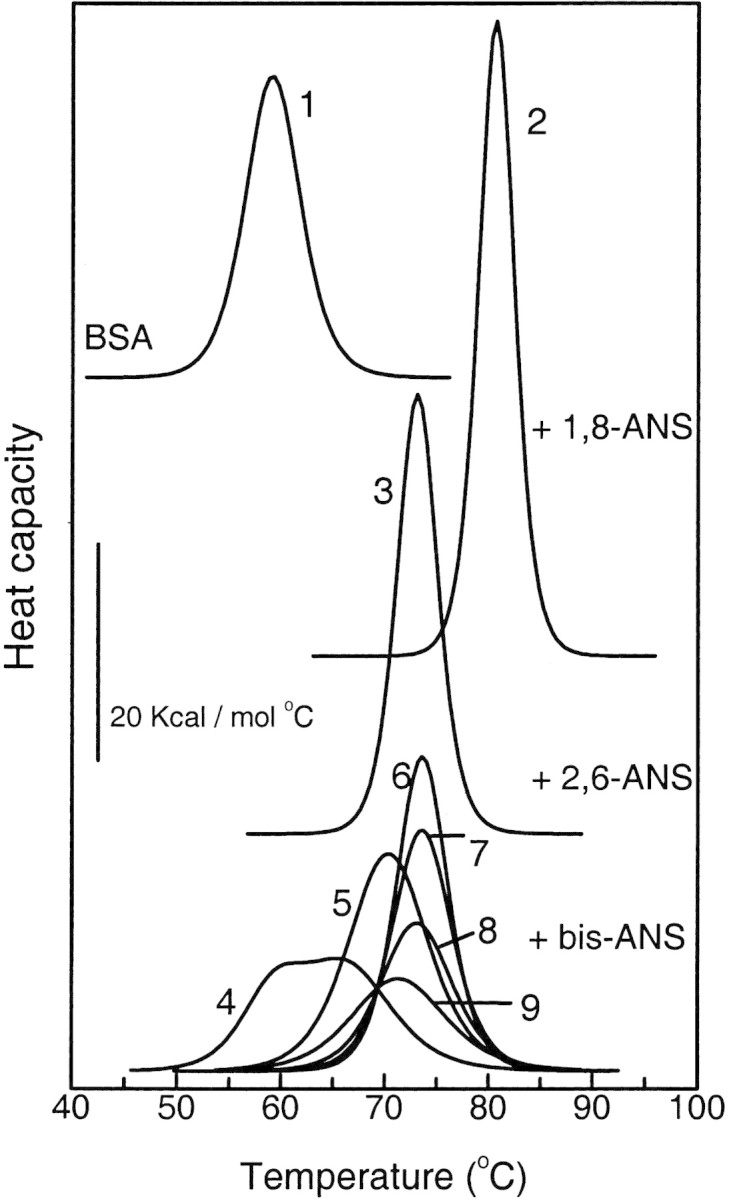
Thermal unfolding curves of BSA at different binding conditions. (1) Free BSA and its complexes with (2) 1,8-ANS, (3) 2,6-ANS, and (4–9) bis-ANS. Moles of ligand per mole of protein for each thermogram are: 0 (1), 50 (2), 50 (3), 1 (4), 2.5 (5), 5 (6), 10 (7), 15 (8) and 20 (9).
Table 1.
Summary of differential scanning calorimetry
| BSA | +1,8-ANS | +2,6-ANS | +bis-ANS | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| L:Pa | 50:1 | 50:1 | 1:1 | 2.5:1 | 5:1 | 10:1 | 15:1 | 20:1 | 50:1 | |
| Tmb (°C) | 59.0 | 79.8 | 73.2 | 60.2 | 70.6 | 73.6 | 73.5 | 72.9 | 71.4 | —e |
| 69.8 | ||||||||||
| ΔHcalc (kcal/mole) | 134 | 259 | 169 | 46 | 178 | 173 | 122 | 114 | 93 | —e |
| 48 | ||||||||||
| CUd | 0.9 | 1.3 | 0.9 | 0.4 | 2.0 | 1.3 | 0.9 | 1.1 | 1.2 | —e |
| 1.4 | ||||||||||
a Ligand:Protein mole ratio.
b Temperature of half-completion of thermal unfolding transition.
c Calorimetric enthalpy.
d Cooperative unit defined as the ratio ΔHcal/ΔHvH.
e Nondetected.
In the absence of ligands, BSA shows a typical two-state thermally induced unfolding with a midpoint of denaturation (Tm) centered at 59°C and an unfolding enthalpy (ΔHcal) of 134 kcal•mole−1 (Fig. 2▶). The effect of ligand binding on protein stability depends on the total ligand:protein mole ratio and the type of ANS derivative. The binding of either 1,8-ANS or 2,6-ANS increases the thermal stability of BSA. The thermostability is gradually augmented as the ligand:protein mole ratio is raised (not shown), reaching a maximal stability at saturating conditions (50:1 ligand: protein mole ratio; Table 1▶; Fig. 2▶). In this condition, 1,8-ANS and 2,6-ANS induce an increment in Tm and ΔHcal of 21°C and 125 kcal•mole−1, and 14°C and 35 kcal•mole−1, respectively, compared with unliganded BSA (Table 1▶; Fig. 2▶). The cooperative unit remains close to 1, indicating that unfolding is essentially a two-state process (Privalov 1979) also in the presence of 1,8-ANS and 2,6-ANS. The 1,8-ANS dye has the strongest effect on BSA thermostability (Table 1▶; Fig. 2▶).
In opposition to 1,8-ANS and 2,6-ANS, bis-ANS has a dual effect on Tm and ΔHcal depending on the ligand concentration. At low ligand:protein mole ratio (≤5:1) bis-ANS produces a similar effect than that observed for the other two ANS derivatives, but at higher proportions, a gradual decrease in protein stability is observed (Fig. 2▶; Table 1▶). At a 1:1 ligand:protein mole ratio, a biphasic endotherm is observed. This behavior at subsaturating ligand concentrations is characteristic of other systems (Brandts and Lin 1990; Shrake and Ross 1990, 1992). At a 5:1 ligand:protein mole ratio a single endothermic transition is observed, centered at 73.6°C, ΔHcal = 173 kcal•mole−1, and a cooperative unit close to 1. It must be noticed that in this condition, Tm and ΔHm have similar values to those obtained in the presence of saturating concentrations of 2,6-ANS. When the relative proportion of bis-ANS is increased above a 5:1 ligand:protein mole ratio, the stability is gradually reduced, as evidenced by the decrease in Tm and ΔHcal. At a 50:1 ligand:protein mole ratio no endothermic thermogram is observed. Under this condition and at room temperature, the infrared spectrum reveals that BSA retains a large amount of secondary structure and the H/D exchange experiment indicates a loosening of tertiary structure (see below). It must be concluded that the lack of heat absorption is due to the absence of cooperativity for the protein unfolding (Table 1▶; Fig. 2▶). Compensatory endothermic–exothermic processes (such as ligand–solvent, ligand–ligand, ligand–protein, and solvent–protein interactions) could cancel each other leading to the absence of heat absorption. However, this may not be the case because a complete absence of cooperative unfolding was observed when the process was monitored by fluorescence spectroscopy (see below).
Influence of ANS derivatives binding on BSA secondary structure
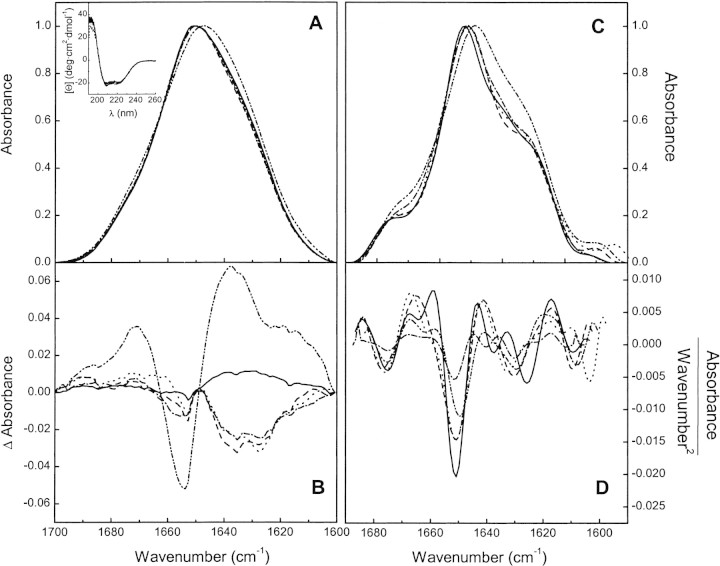
FTIR spectroscopy is suitable to detect conformational changes in proteins upon binding of different ligands (Trewhella et al. 1989; González et al. 1997). We have obtained the FTIR spectra of BSA free and in the presence of saturating amounts of the ANS derivatives in D2O solution. The amide I‘ absorption band, which is sensitive to changes in the protein secondary structure and the corresponding difference spectra obtained by subtracting the spectrum of free BSA from that of the liganded protein, are shown in Figure 3▶, A and B, respectively. In addition, Fourier-deconvolved and second derivative spectra are shown in Figure 3▶, C and D. Far-UV CD spectra are also shown in the inset of Figure 3A▶.
Figure 3.
Secondary structure of BSA at different binding conditions. (A) FTIR absorbance spectra, (B) difference spectra, (C) Fourier-deconvolved amide I‘ band, and (D) second derivative spectra of free BSA (solid line) and its complexes with 1,8-ANS (dashed line) and 2,6-ANS (dotted line) at 50:1 ligand:protein mole ratio and bis-ANS at 5:1 (dash-dot) and 50:1 (dash-dot-dot) ligand:protein mole ratio. (Inset) Far-UV CD spectra of free BSA (solid line) and its complexes with 1,8-ANS (dashed line) and 2,6-ANS (dotted line) at 50:1 and bis-ANS (dash-dot) at 5:1 ligand:protein mole ratio. The solid line in B represents the difference spectrum of two independent free BSA samples as control.
The amide I‘ band of free BSA is centered at 1652 cm−1, as expected for a protein with a high proportion of α-helix. The bands appearing at 1628 and 1676 cm−1 can be attributed to the low- and high-frequency components of the β-sheet, whereas the band centered 1665 and 1640 cm−1 (Fig. 3C▶) can be assigned to turns and unordered structure, respectively (Byler and Susi 1986; Arrondo et al. 1993; Surewicz et al. 1993).
Upon binding of 1,8-ANS, the main component centered at 1650 cm−1, assigned to α-helix, is still present. The component corresponding to unordered structure disappears, as evidenced by the negative band at 1640 cm−1 in the difference spectrum and the disappearance of the band at 1640 cm−1 in the second derivative spectrum (Fig. 3▶, B and D, respectively). This seems to be a relatively small change in the secondary structure, which is not evidenced by CD (Fig. 3A▶, inset).
For bis-ANS-BSA at a 5:1 ligand:protein mole ratio and 2,6-ANS–BSA complexes, the main band position at 1650 cm−1 is not modified and the IR spectra are similar to that obtained with 1,8-ANS (Fig. 3▶).
Conversely, the binding of bis-ANS at saturating condition substantially modifies the IR spectrum. The position of the main band shifts from 1652 to 1648 cm−1 and the intensity of the bands at 1640–1630 cm−1 is increased (Fig. 3A,C). These changes are evidenced in the difference spectrum as a large negative band around 1650 cm−1 and a positive region in the range 1640–1620 cm−1 (Fig. 3B▶). These modifications can be due to the acquirement of distorted α-helical structure as described by Trewhella et al. (1989). CD data acquisition at high bis-ANS concentration was not possible due to the strong absorption of the probe resulting in a very noisy blank.
Kinetics of hydrogen/deuterium exchange
The rate of exchange of a particular amide hydrogen with deuterium depends on the stability and exposure to the solvent of the hydrogen bond in which it is involved. This parameter has been used to obtain information about protein dynamics under different conditions (Goormaghtigh et al. 1994b; Heimburg et al. 1997; Gray and Tamm 1998). We have measured the H/D exchange rate of free BSA and BSA saturated with the different ANS ligands. A correlation was observed between the degree of packing and the thermostability induced by the ligand binding.
The replacement of amide hydrogen by deuterium produces a shift to lower wave numbers of both amide I and II bands. Amide II band shifts about 50 cm−1, from 1550 to 1450 cm−1, whereas amide I shifts only 5–10 cm−1 (Arrondo et al. 1993; Goormaghtigh et al. 1994a). Because of possible distortions in the amide II due to the overlapping with vibrational bands of HDO, we have used the changes in amide I to quantify the H/D exchange evidenced by difference spectra (Heimburg and Marsh 1993).
Figure 4A▶ shows representative difference spectra obtained after exposure to D2O at different times. The difference spectra were obtained by subtracting the spectrum of a completely deuterated sample from those measured at different times (see Materials and Methods). Positive areas of difference spectra (ΔA) were taken as a measurement of exchange degree. Figure 4B▶ shows the time course of H/D exchange obtained from the data such as those shown in Figure 4A▶. ΔA as a function of time was fitted by a multiexponential decay according to
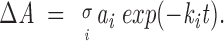 |
Figure 4.
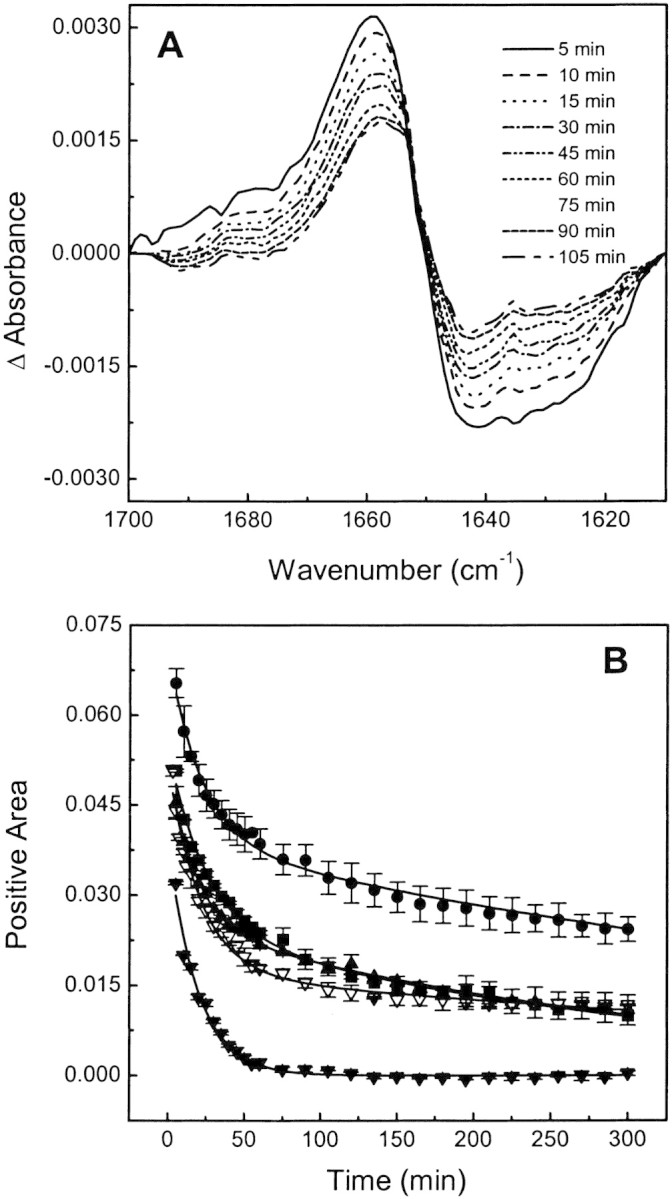
H/D exchange of BSA at different binding conditions. (A) Representative FTIR difference spectra showing the time-dependent changes in the amide I‘ region after sample dispersion in D2O buffer. (B) Kinetics of H/D exchange of free BSA (squares) and its complexes with 1,8-ANS (circles) and 2,6-ANS (solid triangles) at a 50:1 ligand:protein mole ratio and with bis-ANS at 5:1 (open triangles) and 50:1 (solid down triangles) ligand:protein mole ratio.
By this procedure we calculated an average kinetic exchange rate, as indicated in Materials and Methods. The best fit to the experimental data was obtained with a bi-exponential decay indicating two well-defined populations of exchangeable amide protons, defined as slow and fast components. The exchange rate values obtained for free or bound BSA are summarized in Table 2▶.
Table 2.
Analysis of the amide H/D exchange behavior of BSA and its complexes with ANS derivatives
| BSA |
+1,8-ANS |
+2,6-ANS |
+bis-ANS | |||
|---|---|---|---|---|---|---|
| L:Pa | 50:1 | 50:1 | 5:1 | 50:1 | ||
| Fastb | % | 59.3 | 43.3 | 53.3 | 67.0 | 100 |
| k (×103 min−1) | 40 | 49 | 49 | 47 | 96 | |
| Slowc | % | 40.7 | 56.6 | 46.7 | 33.0 | — |
| k (×103 min−1) | 2.7 | 1.5 | 2.3 | 1.5 | — | |
| Averaged | ≤k> (×103 min−1) | 6.2 | 2.6 | 4.6 | 4.3 | 96 |
a Ligand:Protein mole ratio.
b,c Fast and slow component of the bi-exponential decay.
d Average kinetic exchange rate.
Both 1,8-ANS and 2,6-ANS decrease the proportion of the fast component with a small increase in its exchange rate and increase the proportion of the slow component with a reduction in its exchange rate compared with unliganded BSA (Table 2▶). On the other hand, bis-ANS, at a low ligand:protein mole ratio, increases the proportion of the fast component with a small increase in its exchange rate and decreases the proportion of the slow component with a reduction in its exchange rate (Table 2▶). However, in all cases, the average exchange rate is decreased compared with free BSA. The decrease in the exchange rate can be interpreted as an average decrease in the conformational flexibility of BSA. The intermediate H/D exchange rate between free BSA and bound to 1,8-ANS found for bis-ANS at a low ligand:protein mole ratio and 2,6-ANS are in agreement with the lower increment in thermostability induced by these ANS derivatives compared with 1,8-ANS (Table 1▶; Fig. 2▶).
Binding of bis-ANS at a 50:1 ligand:protein mole ratio largely affects the H/D exchange rate (Fig. 4B▶). The exchange curve can be fitted with a single exponential decay. The average H/D exchange rate constant is substantially increased compared with free BSA or BSA bound to bis-ANS at a low proportion, 1,8-ANS or 2,6-ANS (Table 2▶; Fig. 4B▶). The increase in the exchange rate constant indicates that this ligand induces a more open and flexible structure with either a loosing or distortion of typical α-helix content, which is in agreement with a reduced protein thermostability.
Thermal unfolding monitored by fluorescence spectroscopy
We have studied the thermally induced unfolding of BSA by measuring the changes of either the intrinsic fluorescence of the tryptophan residues for free BSA or the fluorescence of ANS derivatives in the ANS–BSA complexes. Both the intensity and position of the fluorescence emission spectrum of Trp are sensitive to changes in the fluorophore environment and, consequently, to the protein structure. ANS derivatives have a very low quantum yield in aqueous solution but the fluorescence emission is dramatically increased in nonpolar environments like hydrophobic sites in proteins. This fluorescence increment is attended by a blue shift in the emission spectrum. In control experiments, we observed that the fluorescence intensity of Trp in buffer and ANS derivatives in organic solvent decreases monotonically when the temperature is raised without changes in the spectral position. We ruled out the possibility that 1,8-ANS and 2,6-ANS bind to the unfolded state because the fluorescent properties after denaturation are the same as those observed for the probes in buffer. Thus, we considered that the cooperative ANS fluorescence changes are due to the release of probe into the aqueous medium.
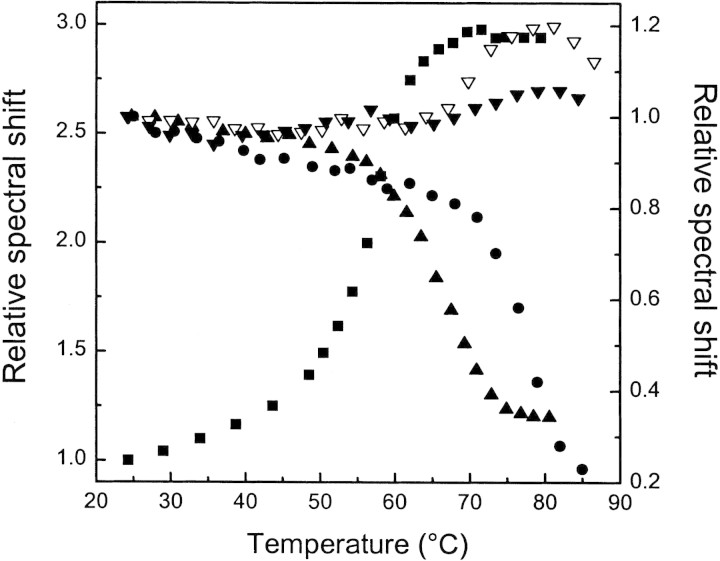
BSA has two Trp residues (Brown and Shockley 1982), with a fluorescence emission spectrum centered at 340 nm, indicating an inner localization in the native folded conformation. Unliganded BSA undergoes a cooperative blue shift in the fluorescence emission between 48°–63°C with a midpoint of 58.7°C (Fig. 5▶), which is the temperature of the global unfolding as determined by DSC (Table 1▶; Fig. 2▶).
Figure 5.
Fluorescence changes upon thermal unfolding of BSA at different binding conditions. (Left axis) Relative fluorescence emission spectral shift of free BSA (squares). (Right axis) Relative fluorescence emission spectral shift of BSA comlexed with 1,8-ANS (circles) and 2,6-ANS (solid triangles) at a 50:1 ligand:protein mole ratio and with bis-ANS at 5:1 (open down triangles) and 50:1 (solid down triangles) ligand:protein mole ratios.
At a 50:1 ligand:protein mole ratio, the release of 1,8-ANS to the aqueous solvent produces a decrease in the fluorescence intensity and the emission is red-shifted (Fig. 5▶). The midpoint temperature of fluorescence change for the 1,8-ANS-BSA complex is 77.5°C (Fig. 5▶). This value is close to the Tm = 79.8°C obtained by DSC (Table 1▶), indicating that the release of bound 1,8-ANS is concomitant with the cooperative thermal protein denaturation.
Similar to 1,8-ANS, the fluorescence of bound 2,6-ANS is red-shifted when the temperature is increased, but the midpoint of the fluorescence change is 67°C, 7°C lower than the Tm measured by DSC (compare Table 1▶ with Fig. 5▶). For this dye, the temperature-induced fluorescence change is already completed at the beginning of heat absorption (lift-off region of thermogram; see Fig. 2▶).
The fluorescence changes of bis-ANS–BSA complex with the temperature follow a different pattern than those obtained for the other two ANS derivatives, and they correlate with the results found by FTIR and DSC. At a low bis-ANS:BSA mole ratio, there is a blue shift in the spectra with a temperature midpoint of 75°C (Fig. 5▶), in agreement with that determined by DSC of 73.6°C (Table 1▶; Fig. 2▶). At a higher ligand proportion, the cooperative fluorescence changes vanish, in keeping with the complete absence of endothermic transition (Table 1▶; Fig. 5▶). The release of the dye from the protein into the aqueous medium should produce a red shift of the fluorescence spectrum. Instead, we have observed a blue shift, which could be attributed to the interactions between the dye and non-native states of the protein (Fig. 5▶). The possibility of ligand binding to native and non-native conformation may explain the initial increase and the subsequent decrease in Tm obtained by DSC when the ligand concentration is raised (Shrake and Ross 1992). Under saturating conditions the dye induces a considerable distortion of the BSA structure, abolishing the cooperative endothermic unfolding.
Discussion
The binding of small molecules to proteins and protein–protein interactions are key processes in cell biochemistry. The usual paradigm is that ligand binding induces a change in the conformation of the target protein that, in turn, produces a given response. In many cases an increase in protein thermostability as a consequence of the interaction has been reported. The simple model of BSA interacting with ANS derivatives that we have used in the present work has interesting features because it addressed evidences about the correlation between the observed thermostability and the ligand effects on protein flexibility.
In a previous work, we have found two binding sites for 1,8-ANS in human serum albumin using phase-modulation fluorescence spectroscopy (Bagatolli et al. 1996). The fluorescence properties of 1,8-ANS are different for each site. The high affinity site binds one ANS molecule with a binding constant of 8.7 × 105 M−1, whereas the low affinity site has a 2:1 ANS:albumin stoichiometry and a binding constant of 7.9 × 104 M−1. BSA shows the same trend. This strength of affinity is enough to account for the observed increase in thermostability (Table 1▶; Fig. 2▶) due to the coupling of binding and unfolding equilibriums (Shrake and Ross 1988, 1990, 1992). Thermodynamic considerations of thermograms of 2,6-ANS–BSA complexes at different ligand:protein mole ratios using the approach of Fukada et al. (1983) gives an overall number of sites of 2.5 (data not shown). Also, the increase in the Tm induced by 2,6-ANS is compatible with association constants similar to those determined for 1,8-ANS. The dual effect of bis-ANS on BSA thermostability (Table 1▶; Fig. 2▶) is compatible with the presence of high affinity sites in the folded conformation and multiple low affinity binding sites in the non-native conformations (Shrake and Ross 1992). The fluorescence data give evidence for bis-ANS binding to non-native conformation (Fig. 5▶).
FTIR spectroscopy can reveal changes both in the secondary and tertiary structure of a protein upon ligand binding. bis-ANS at low concentration, 1,8-ANS and 2,6-ANS, have similar effects on BSA secondary structure. They induce the disappearance of the band at 1640 cm−1 present in free BSA. Even when these changes are rather minor and not clearly observed by circular dichroism (Fig. 3A▶, inset), they indicate that dye binding results in a decrease of the unordered structure. The H/D exchange rate is also sensitive to ligand binding (Fig. 4▶). For free BSA and BSA bound to bis-ANS at a low ligand:protein mole ratio, 1,8-ANS or 2,6-ANS, the H/D exchange rate can be resolved in a bi-exponential decay with a slow and a fast component (Table 2▶; Fig. 4▶) from which we can calculate an average H/D exchange rate. Taking the average exchange rate constant as a measure of the protein fluctuations and global exposure to solvent, we conclude that the protein bound to bis-ANS at a low concentration, 1,8-ANS or 2,6-ANS, have less conformational flexibility than the native protein, because the effect of 1,8-ANS is stronger than the other binding conditions (Table 2▶).
On the other hand, bis-ANS, at a high ligand:protein mole ratio, substantially modifies the secondary structure of BSA, by judging the change observed in the main band position from 1652 cm−1 to 1648 cm−1 of the FTIR spectrum (Fig. 3▶). For a mainly α-helix protein, this change can be associated with the acquisition of a distorted helix conformation in which the carbonyl stretching frequencies are lowered due to stronger interaction with solvent molecules (Trewhella et al. 1989). Together with the large increase in H/D exchange rate in the presence of saturating amounts of bis-ANS (Table 2▶; Fig. 4▶), we conclude that the protein has a more flexible structure compared with free BSA. This is in agreement with the complete absence of cooperative temperature-induced unfolding (Table 1▶). The characteristics observed for BSA in the presence of an excess of bis-ANS correspond to a molten globule-like state according to the accepted general definition: presence of native-like secondary structure, a fluctuating tertiary structure, and noncooperative thermal unfolding (Kuwajima 1989; Ptitsyn 1992; Dobson 1994). Several reports have also indicated that bis-ANS per se induces protein structural changes in the direction of a more disordered conformation. Particularly, the interaction of bis-ANS with the DNAK, a 70-kD heat-shock protein of Escherichia coli, shifts the equilibrium from the native to an intermediate state, and a correlation is observed between the decrease of the unfolding temperature and dye concentration (Shi et al. 1994). In this connection, it has recently been reported that bis-ANS induces a more distorted structure of tubulin with a higher susceptibility for enzyme digestion (Gupta et al. 2003).
Serum albumins undergo reversible conformational changes at low pH. As early as in 1955, Tanford reported that BSA shows a reversible expansion in acidic medium (Tanford et al. 1955). More recently, it has been identified that this expanded and reversible state corresponds to a molten globule-like state for human albumin at low pH, clearly distinguishable from the GdnClH-denaturated state (Muzammil et al. 1999). The titration of carboxylic side chains of some amino acids in albumins seems to perturb the native conformation pulling the equilibrium to a molten globule-like conformer. Also, 1,8-ANS binds to albumin in the acidic expanded conformation but not in the completely unfolded state (Muzammil et al. 1999). This interaction induces shrinkage of the hydrodynamic volume of the protein (Matulis et al. 1999). The ability of 1,8-ANS to induce partial folding has been stated for both acid unfolded cytochrome c and pectate lyase C proteins (Ali et al. 1999; Kamen and Woody 2001).
On the other hand, there are evidences that bis-ANS induces disordered structures. It has been stated that bis-ANS is able to detect a more disordered conformation in the entry portal region of the ligand cavity site of the intestinal fatty acid binding protein (Arighi et al. 1998), and also, to destabilize the GroEl apical domain fragment inducing an intermediate conformation with increased hydrophobic surfaces (Smoot et al. 2001). In addition, it has been shown that this dye binds to glutaminyl-tRNA (Bhattacharyya et al. 2000), apo α-lactalbumin (Vanderheeren et al. 1998), and Salmonella typhimurium bacteriophage scaffolding proteins (Teschke et al. 1993), leading to a more disordered protein structure compatible with a molten globule state. According to our results, and in agreement with the data previously reviewed, an excess of bis-ANS induces in BSA a more open structure at neutral pH compatible with a molten globule state, whereas bis-ANS at low concentration, 1,8-ANS or 2,6-ANS, induce tighter conformers.
The correlation between conformational flexibility induced by ANS derivatives with thermostability clearly emerges when we compare the average H/D exchange rate with the calorimetric midpoint of denaturation (Tables 1▶,▶). The order of the average H/D exchange rates found is BSA-bis-ANS (high ligand concentration) >> free BSA > BSA-2,6-ANS ≈ BSA-bis-ANS (low ligand concentration) > BSA-1,8-ANS, whereas the order of Tms is 79.8°C (BSA-1,8-ANS) > 73.2°C (BSA-2,6-ANS) ≈ 73.6°C (BSA-bis-ANS low ligand concentration) > 59.0°C (free BSA) >> nondetected for BSA-bis-ANS (high ligand concentration). In other words, an increased thermal stability corresponds to a reduced conformational flexibility.
Several authors have observed a correlation between protein thermostability with protein flexibility detected by H/D exchange. The binding of Mg–ADP to the FoF1-type ATP synthase from the thermophilic Bacillus PS3 produces an increase in thermal stability and a decrease in protein flexibility, whereas Mg–ATP reduces the thermal stability with a concomitant increase in the flexibility (Villaverde et al. 1997). Munson et al. (1996), in a systematic redesign of the hydrophobic core of Rop protein from E. coli, found a correlation between the matching of the hydrophobic core with thermostability. The decrease in conformational flexibility is also stated as the main factor accounting for thermostability in extremophiles, organisms living in extreme conditions of temperature and pH. For example, adenylate kinase from thermoacidophilic archaeon Sulfolobus acidocaldarius was compared with that from porcine muscle. Both proteins have a similar global structure but the enzyme from the extremophile is characterized by a more compact core (Bonisch et al. 1996). Furthermore, a perusal comparative evaluation of key metabolic homologous enzymes from hyperthermophilic and mesophilic organisms arrived to the general conclusion that the additional increment in free energy of stabilization is not the result of a unique contribution, but it rather comes from additive weak interactions that lead to the optimization of the packing and the interactions with the solvent, solutes, and specific ligands (for review, see Landestein and Antranikian 1998; Jaenicke 2000).
Because biologic processes are carried out through binding events, changes in the environment and signaling pathways, the induced structural changes upon ligand binding have been a matter of extensive studies and modeling. The Koshland-Nemethy-Filmer sequential binding model was introduced to explain cooperativity under structural basis (Cantor and Schimmel 1980). In this connection, we have reported that the binding of one or two biotin to homotetrameric streptavidin induces structural cooperativity and conformational communication, which is evidenced by an increased thermostability of unliganded subunits. Thus, a particular and localized interaction in a subunit is transmitted to distal regions of the tetramer (González et al. 1997).
Miller and Dill (1997) have proposed that ligand binding can be better described in terms of ensembles and energy landscapes. Recently, Freire (1999) has demonstrated that the propagation of the stabilization effects to remote sites triggered by ligand binding is due to a redistribution of substates favoring those binding-competent states. Extending these concepts, a generalized funnel-shape energy landscape theory has recently been presented. It was successfully utilized in describing folding and binding as well as in explaining enzyme pathways, multimolecular associations, and allostery (Tsai et al. 1999a,b). The framework of this theory lies on the concept that in the bottom of the folding funnel around the native conformation, there are multiple protein conformational substates in equilibrium. A funnel-shape energy landscape can be dynamically shifted by the presence of ligands or changes in the surroundings, which in turn, modifies the population of the conformers. This model is a generalization that includes the induced fit model (Kumar et al. 2000). According to this theory, there is an ensemble of protein conformations with different affinities for the ligand. The conformer that binds a ligand is depleted from the solution, having the equilibrium shifted towards this conformation. Thus, it is not a conformational change induced by the ligand, but rather a change induced by shifting the equilibrium among conformers (Tsai et al. 1999b; Kumar et al. 2000).
On the basis of the above theory, it can be proposed that within the large dynamic ensemble of BSA states, at least three conformers besides the native and the unfolded states can be distinguished. They can be defined as (in order of increasing flexibility): BSA1,8-ANS, BSA2,6-ANS, BSAfree, BSAbis-ANS (high concentration of bis-ANS), and BSAunfolded, in which the subscript identifies each protein conformer. The relative population of each distinguishable substate depends on the type and ligand concentration and the temperature of the system. At saturating ligand condition the distribution shifts to a particular conformer that is differentially evidenced by thermal stability and FTIR spectroscopy.
It is well known that protein thermal stability is modified by ligand binding due to the coupling between two mutual processes under equilibrium: binding and unfolding (Fukada et al. 1983; Sturtevant 1987; Shrake and Ross 1990, 1992). The additional binding free energy is responsible for shifting the unfolding temperature, and it could include or not contributions from conformational changes. If the thermodynamics parameters of the ligand–protein interaction are known, the thermogram of the complex can be built from the binding data and the excess heat capacity trace of unliganded protein (Brandts and Lin 1990; Shrake and Ross 1990, 1992; Straume and Freire 1992). These concepts can be updated in the light of the dynamic energy landscape shifts: The presence of a given ligand shifts the conformer distribution, and then the thermogram must be accounted from the new distribution of substates.
For the 1,8-ANS–BSA complex, the thermal unfolding is concomitant with ligand release (Figs. 2,5▶▶). Instead, for 2,6-ANS, the ligand release precedes the unfolding by 7°C. This could be interpreted that the ligand modifies the energy landscape, and this new state distribution is retained even when the ligand is released at 67°C. However, to undoubtedly conclude for this "molecular imprinting" effect (i.e., the ligand induces a conformation that persists after dissociation) it is necessary for further experiments, and it is out of the focus of the present work. Alternative explanations could also be considered. For example, the ligand may persist in its bound state but in a more solvent-exposed mode.
Molecular chaperones participate in the cell to prevent protein misfolding (for recent reviews, see Ellgaard et al. 1999; Hartl and Hayer-Hartl 2002). It was recently emphasized that nonpeptide compounds such as glycerol, dimethylsulfoxide, or trimethylamine are able to reverse the intracellular retention of misfolded proteins, and have been called chemical chaperones (Morello et al. 2000). In this connection, Diamant et al. (2001) have reported that substances that normally accumulate in the cell (glycerol, trehalose, and proline) can also assist protein refolding, preventing aggregation during stress. Taking into account that chemical chaperones most probably stabilize folding intermediates, Morello et al. (2000) go a step forward, suggesting that ligands with more specificity for a particular target protein may improve functional folding, thus acting as pharmacologic chaperones with potential uses in conformational diseases. The interaction between bis-ANS and BSA could be a suitable model for pharmacologic chaperone molecules.
Finally, and in agreement with Matulis et al. (1999) and Ali et al. (1999), the results presented here indicate that special care must be taken in using ANS derivatives, because they should not be seen as an "inert" probe for sensing protein conformation.
Materials and methods
Materials
Fatty acid- and globulin-free bovine serum albumin (BSA) and D2O were obtained from Sigma Chemical Co. 1,8-ANS, 2,6-ANS, and bis-ANS were obtained from Molecular Probes Inc. They were used without further purification. The buffers were prepared with analytical grade reagents.
Differential scanning calorimetry
Calorimetric curves were obtained in a MicroCal MC-2D scanning calorimeter. BSA and the fluorescent dyes were dissolved in 100 mM phosphate buffer pH = 7.2. The protein concentration was 50 μM. Samples were exhaustively degassed before the injection into the calorimeter cell to prevent the formation of air bubbles. The reference cell was filled with buffer. Nitrogen pressure of 2 atm was applied to both cells. The calorimetric data were processed with the software provided by the manufacturer. The data were analyzed assuming a reversible non-two-state denaturation model, which provides the calorimetric (ΔHcal) and the van’t Hoff (ΔHvH) enthalpies. ΔHcal is the actual heat absorption of protein unfolding, whereas ΔHvH is the theoretical heat of the transition assuming a two-state model. The total molar heat capacity for n independent non-two-state transitions after baseline subtraction is
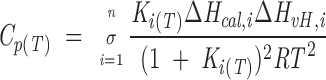 |
where the unfolding equilibrium constants are calculated as
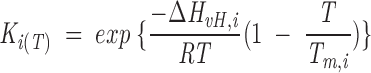 |
The cooperative unit is defined by the ratio ΔHcal/ΔHvH (Privalov 1979).
Infrared spectroscopy
Fourier transform infrared spectra were obtained in a Nicolet Nexus equipped with a DTGS KBr detector. The sample was placed in a cell for liquid samples with CaF2 windows and 100-μm Teflon spacer. The instrument was purged with dry N2 to decrease atmospheric H2O vapor. When necessary, residual H2O vapor was subtracted prior to deconvolution or spectral subtraction. All spectra were obtained at room temperature with a nominal resolution of 2 cm−1. Typically, 0.2 mg of BSA was dissolved in 30 μL of D2O buffer (Tris 100 mM, pD 7.2) or buffer containing the ANS derivative.
Steady-state experiments
To allow complete exchange of hydrogen by deuterium, pure BSA was incubated for 48 h in D2O buffer previous to the addition of ligand. Fourier transform self-deconvolution of the unsmoothed spectra was performed with Omnic 5.0 software using a triangular apodization function, a bandwidth HWHH = 17 cm−1, and line narrowing factor K = 2. For each measurement, 100 interferograms were acquired and averaged.
Kinetics of hydrogen/deuterium exchange
The protein lyophylized from aqueous solution was resuspended in pure D2O buffer or D2O buffer containing the ANS derivative and immediately placed in the cell. For each time point, 10 interferograms were acquired and averaged. After buffer subtraction, spectra were normalized between 1700 cm−1 and 1600 cm−1 to an area of 1 × cm−1. The normalized spectrum of the completely deuterated sample was subtracted from the normalized spectra taken at different times. The time course of the amide H/D exchange was evaluated from the area of the positive region of difference spectra (ΔA). Three independent experiments were averaged. The data were analyzed using a multiexponential decay function representing the different groups i:
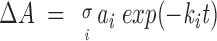 |
where ai is proportional to the relative fractions of each type of amide hydrogen that exchange with a deuteration rate constant ki. The fitted parameters were used to calculate the average H/D exchange rate constant ≤k>, defined as (Heimburg and Marsh 1993):
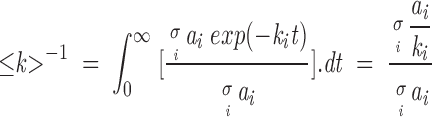 |
Circular dichroism spectroscopy
Circular dichroism spectra were measured on a Jasco 810 spectropolarimeter under constant N2 flush. BSA was dissolved in 10 mM phosphate buffer, pH = 7.2. Protein concentration was 0.2 mg/mL for all measurements. CD data were expressed as the mean residue ellipticity ([θ]) in deg•cm2•dmole−1, from the equation ([θ]) = M0ξ/(10l × C). M0 is the mean residue weight; ξ, ellipticity in degrees; l, cell path length in cm, and C, protein concentration in g•cm−3.
Fluorescence spectroscopy
Steady-state emission spectra were recorded in an SLM-Aminco 4800C spectrofluorometer, equipped with a Xenon arc lamp and thermostated cell holder. All slits were set at 4 nm. The temperature was controlled by a circulating bath and measured with a thermocouple placed into the cell. The sample was allowed to equilibrate for 5 min at the desired temperature before acquisition of the spectra. The global heating rate was similar to that used in DSC experiments. The protein concentration was around 2 μM. The fluorescence emissions of L-Trp and ANS derivatives were obtained in phosphate buffer and n-buthanol, respectively. Trp, 1,8-ANS, 2,6-ANS, and bis-ANS were selectively excited at 295, 380, 331, and 400 nm, respectively.
The spectral shifts were taken as a measure of protein unfolding. They were measured by taking the ratio of fluorescence intensity at the blue edge and the intensity at the red edge. This ratio was used as a more sensitive parameter to detect changes in the band position than the wavelength of maximum emission (Jiang and London 1990; Wharton et al. 1988). An increase in this ratio reflects a blue shift of the spectrum. The used ratios were FI310/FI375 for free BSA and FI450/FI525, FI400/FI475, and FI460/FI540 for its complexes with 1,8-ANS, 2,6-ANS, and bis-ANS, respectively. For comparative purpose, each value was normalized to that obtained at room temperature.
Acknowledgments
This work was supported by grants from CONICET, FONCYT, SECYT-UNC, and Agencia Córdoba Ciencia. M.S.C. is a fellow from CONICET. G.D.F. and G.G.M. are members of the Career of Investigator from CONICET. We would like to thank J.M. Ruysschaert for critical suggestions about FTIR experiments.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0240003.
References
- Ali, V., Prakash, K., Kulkarni, S., Ahmad, A., Madhusudan, K.P., and Bhakuni, V. 1999. 8-Anilino-1-naphthalene sulfonic acid (ANS) induces folding of acid unfolded cytochrome c to molten globule state as a result of electrostatic interactions. Biochemistry 38 13635–13642. [DOI] [PubMed] [Google Scholar]
- Arighi, C., Rossi, J., and Delfino, J. 1998. Temperature-induced conformational transition of intestinal fatty acid binding protein enhancing ligand binding: A functional, spectroscopy and molecular modeling study. Biochemistry 37 16802–16814. [DOI] [PubMed] [Google Scholar]
- Arrondo, J.L., Muga, A., Castresana, J., and Goñi, F. 1993. Quantitative studies of the structure of proteins in solution by Fourier transform infrared spectroscopy. Prog. Biophys. Mol. Biol. 59 23–56. [DOI] [PubMed] [Google Scholar]
- Bagatolli, L., Kivatinitz, S.C., Aguilar, F., Soto, M.A., Sotomayor, P., and Fidelio, G.D. 1996. Two distinguishable fluorescent modes of 1-anilino-8-napthalenesulfonate bound to human albumin. J. Floresc. 6 33–40. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya, A., Mandal, A.K., Banerjee, R., and Roy, S. 2000. Dynamics of compact denatured states of glutaminyl-tRNA synthetase probed by bis-ANS binding kinetics. Biophys. Chem. 87 201–212. [DOI] [PubMed] [Google Scholar]
- Bonisch, H., Backmann, J., Kath, T., Naumann, D., and Schafer, G. 1996. Adenylate kinase from Sulfolobus acidocaldarius: Expression in Escherichia coli and characterization by Fourier transform infrared spectroscopy. Arch. Biochem. Biophys. 333 75–84. [DOI] [PubMed] [Google Scholar]
- Brandts, J.F. and Lin, L.N. 1990. Study of strong to ultratight protein interaction using differential scanning calorimetry. Biochemistry 29 6927–6940. [DOI] [PubMed] [Google Scholar]
- Brown, J.R. and Shockley, P. 1982. Serum albumins: Structure and characterization of its ligand sites. In Lipid—protein interactions (eds. P.C. Jost and O.H. Griffith), vol. 1, pp. 25–68. John Wiley & Sons, New York.
- Byler, M. and Susi, H. 1986. Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers 25 469–487. [DOI] [PubMed] [Google Scholar]
- Cantor, C.R. and Schimmel, P.R. 1980. Biophysical chemistry, part III: The behavior of biological macromolecules, 1st ed. W.H. Freeman and Co., San Francisco.
- Diamant, S., Eliahu, N., Rosenthal, D., and Goloubinoff, P. 2001. Chemical chaperones regulate molecular chaperones in vitro and in cells under combined salt and heat stresses. J. Biol. Chem. 276 39586–39591. [DOI] [PubMed] [Google Scholar]
- Dobson, C.M. 1994. Protein folding. Solid evidence for molten globules. Curr. Biol. 4 636–640. [DOI] [PubMed] [Google Scholar]
- Ellgaard, L., Molinari, M., and Helenius, A. 1999. Setting the standards: Quality control in the secretory pathway. Science 286 1882–1888. [DOI] [PubMed] [Google Scholar]
- Freire, E. 1999. The propagation of binding interactions to remote sites in proteins: Analysis of the binding of the monoclonal antibody D1.3 to lysozyme. Proc. Natl. Acad. Sci. 96 10118–10122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada, H., Sturtevant, J., and Quiocho, F. 1983. Thermodynamics of the binding of L-arabinose and of D-galactose to the L-arabinose-banding protein of Escherichia coli. J. Biol. Chem. 258 13193–13198. [PubMed] [Google Scholar]
- González, M., Bagatolli, L., Echabe, I., Arrondo, J., Argaraña, C., Cantor, C., and Fidelio, G. 1997. Interaction of biotin with streptavidin. J. Biol. Chem. 272 11288–11294. [DOI] [PubMed] [Google Scholar]
- González, M., Argaraña, C., and Fidelio, G. 1999. Extremely high thermal stability of streptavidin and avidin upon biotin binding. Biomol. Eng. 16 67–72. [DOI] [PubMed] [Google Scholar]
- Goormaghtigh, E., Cabiaux, V., and Ruysschaert, J.M. 1994a. Determination of soluble and membrane protein structure by Fourier transform infrared spectroscopy. Subcell. Biochem. 23 363–404. [DOI] [PubMed] [Google Scholar]
- Goormaghtigh, E., Vigneron, L., Scarborough, G., and Ruysschaert, J.M. 1994b. Tertiary conformational changes of the Neurospora crassa plasma membrane H+-ATPase monitored by hydrogen/deuterium exchange kinetics. J. Biol. Chem. 269 27409–27413. [PubMed] [Google Scholar]
- Gray, C. and Tamm, L.K. 1998. pH-induced conformational changes of membrane-bound influenza hemagglutinin and its effect on target lipid bilayers. Protein Sci. 7 2359–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S., Chakraborty, S., Poddar, A., Sarkar, N., Das, K.P., and Bhattacharyya, B. 2003. bis-ANS binding to tubulin: Isothermal titration calorimetry and the site-specific proteolysis reveal the GTP-induced structural stability of tubulin. Proteins 2 283–289. [DOI] [PubMed] [Google Scholar]
- Hartl, F.U. and Hayer-Hartl, M. 2002. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 295 1852–1858. [DOI] [PubMed] [Google Scholar]
- Heimburg, T. and Marsh, D. 1993. Investigation of secondary and tertiary structural changes of cytrochrome c in complexes with anionic lipids using amide hydrogen exchange measurement: An FTIR study. Biophys. J. 65 2408–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimburg, T., Esmann, M., and Marsh, D. 1997. Characterization of the secondary structure and assembly of the transmembrane domains of trypsinized Na,K-ATPase by Fourier transform infrared spectroscopy. J. Biol. Chem. 65 2408–2417. [DOI] [PubMed] [Google Scholar]
- Jaenicke, R. 2000. Stability and stabilization of globular proteins in solution. J. Biotechnol. 79 193–203. [DOI] [PubMed] [Google Scholar]
- Jiang, J.X. and London, E.J. 1990. Involvement of denaturation-like changes in Pseudomonas exotoxin hydrophobicity and membrane penetration determined by characterization of pH and thermal transitions. Roles of two distinct conformationally altered state. J. Biol. Chem. 265 8636–8641. [PubMed] [Google Scholar]
- Kamen, D.E. and Woody, R.W. 2001. A partially folded intermediate conformation is induced in pectate lyase C by the addition of 8-anilino-1-naphthalenesulfonate (ANS). Protein Sci. 10 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Buyong, M.A., Tsai, C.J., Sinha, N., and Nussinov, R. 2000. Folding and binding cascades: Dynamic landscape and population shifts. Protein Sci. 9 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwajima, K. 1989. The molten globule state as a clue for understanding the folding and cooperativity of globular–protein structure. Proteins 6 87–103. [DOI] [PubMed] [Google Scholar]
- Landestein, R. and Antranikian, G. 1998. Proteins from hyperthermophiles: Stability and enzimatic catalysis close to the boiling point of water. Adv. Biochem. Eng. 61 37–85. [DOI] [PubMed] [Google Scholar]
- Matulis, D., Baumann, C.G., Bloomfield, V.A., and Lovrien, R.E. 1999. 1-Anilino-8-naphthalene sulfonate as a protein conformational tightening agent. Biopolymers 49 451–458. [DOI] [PubMed] [Google Scholar]
- Miller, D.W. and Dill, K.A. 1997. Ligand binding to proteins: The binding landscape model. Protein Sci. 6 2166–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello, J.P., Petaja-Repo, U.E., Bichet, D.G., and Bouvier, M. 2000. Pharmacological chaperones: A new twist on receptor folding. Trends Pharmacol. Sci. 21 466–469. [DOI] [PubMed] [Google Scholar]
- Munson, M., Balasubramanian, S., Fleming, K., Nagi, A., O’Brien, R., Sturtevant, J., and Regan, L. 1996. What makes a protein a protein? Hydrophobic core designs that specify stability and structural properties. Protein Sci. 5 1584–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzammil, S., Kumar, Y., and Tayyab, S. 1999. Molten globule-like state of human serum albumin at low pH. Eur. J. Biochem 266 26–32. [DOI] [PubMed] [Google Scholar]
- Privalov, P. 1979. Stability of proteins. Small globular proteins. Adv. Protein Chem. 33 167–241. [DOI] [PubMed] [Google Scholar]
- Ptitsyn, O.B. 1992. The molten globule state. In Protein folding, 1st ed. (ed. T.E. Creighton), pp. 243–300. W.H. Freeman and Co., New York.
- Richards, F. 1997. Protein stability: Still an unsolved problem. Cell. Mol. Life Sci. 53 790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, S., González, M., Bagatolli, L., Duffard, R., and Fidelio, G. 1998. Evidence of a strong interaction of 2,4-dichlorophenoxyacetic acid herbicide with human serum albumin. Life Sci. 63 2343–2351. [DOI] [PubMed] [Google Scholar]
- Shrake, A. and Ross, P.D. 1988. Biphasic denaturation of human albumin due to ligand redistribution during unfolding. J. Biol. Chem. 263 15392–15399. [PubMed] [Google Scholar]
- ———. 1990. Ligand-induced biphasic protein denaturation. J. Biol. Chem. 265 5055–5059. [PubMed] [Google Scholar]
- ———. 1992. Origin and consequences of ligand-induced multiphasic thermal protein denaturation. Biopolymers 32 925–940. [DOI] [PubMed] [Google Scholar]
- Shi, L., Palleros, D.R., and Fink, A.L. 1994. Protein conformational changes induced by 1,1′-Bis(4-anilino-5-napthalenesulfonic acid): Preferential binding to the molten globule of DnaK. Biochemistry 33 7536–7546. [DOI] [PubMed] [Google Scholar]
- Smoot, A.L., Panda, M., Brazil, B.T., Buckle, A.M., Fersht, A.R., and Horowitz, P.M. 2001. The binding of bis-ANS to the isolated GroEL apical domain fragment induces the formation of a folding intermediate with increased hydrophobic surface not observed in tetradecameric GroEL. Biochemistry 40 4484–4492. [DOI] [PubMed] [Google Scholar]
- Straume, M. and Freire, E. 1992. Two-dimentional differential scanning calorimetry: Simultaneous resolution of intrinsic protein structural energetics and ligand binding interaction by global linkage analysis. Anal. Biochem. 203 259–268. [DOI] [PubMed] [Google Scholar]
- Sturtevant, J.M. 1987. Biochemical application of differential scanning calorimetry. Annu. Rev. Phys. Chem. 38 463–488. [Google Scholar]
- Surewicz, W., Mantsch, H., and Chapman, D. 1993. Determination of protein secondary structure by Fourier transform infrared spectroscopy. Biochemistry 32 289–394. [DOI] [PubMed] [Google Scholar]
- Tanford, C., Buzzel, J.G., Rands, D.G., and Swanson, S.A. 1955. The reversible expansion of bovine serum albumin in acid solutions. J. Am. Chem. Sci. 77 6421–6428. [Google Scholar]
- Teschke, C.M., King, J., and Prevelige Jr., P.E. 1993. Inhibition of viral capsid assembly by 1,1′-bi(4-anilinonaphthalene-5-sulfonic acid). Biochemistry 32 10658–10665. [DOI] [PubMed] [Google Scholar]
- Trewhella, J., Liddle, W.K., Heidorn, D.B., and Strynadka, N. 1989. Calmodulin and Troponin C structures studied by Fourier transform infrared spectroscopy: Effects of Ca2+ and Mg2+ binding. Biochemistry 28 1294–1301. [DOI] [PubMed] [Google Scholar]
- Tsai, C.J., Kumar, S., Buyong, M.A., and Nussinov, R. 1999a. Folding funnels, binding funnels, and protein function. Protein Sci. 8 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, C.J., Ma, B., and Nussinov, R. 1999b. Folding and binding cascade: Shift in energy landscape. Proc. Natl. Acad. Sci. 96 9970–9972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderheeren, G., Hanssens, I., Noyelle, K., Van Dael, H., and Joniau, M. 1998. The perturbations of the native state of goat α-lactalbumin induced by 1,1′-bis(4-anilino-5-naphthalenesulfonate) are Ca2+-dependent. Biophys. J. 75 2195–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaverde, J., Cladera, J., Padros, E., Rigaud, J.L., and Dunach, M. 1997. Effect of nucleotides on the thermal stability and on the deuteration kinetics of the thermophilic F0F1 ATP synthase. Eur. J. Biochem. 244 441–448. [DOI] [PubMed] [Google Scholar]
- Wharton, S.A., Martin, S.R., Ruigrok, R.W.H., Skehel, J.J., and Wiley, D.C. 1988. Membrane fusion by peptide analogues of influenza virus haemagglutinin. J. Gen. Virol. 69 1847–1857. [DOI] [PubMed] [Google Scholar]