Abstract
Acid-sensing ion channels (ASICs) are thought to be important ion channels, particularly for the perception of pain. Some of them may also contribute to synaptic plasticity, learning, and memory. Psalmotoxin 1 (PcTx1), the first potent and specific blocker of the ASIC1a proton-sensing channel, has been successfully expressed in the Drosophila melanogaster S2 cell recombinant expression system used here for the first time to produce a spider toxin. The recombinant toxin was identical in all respects to the native peptide, and its three-dimensional structure in solution was determined by means of 1H 2D NMR spectroscopy. Surface characteristics of PcTx1 provide insights on key structural elements involved in the binding of PcTx1 to ASIC1a channels. They appear to be localized in the β-sheet and the β-turn linking the strands, as indicated by electrostatic anisotropy calculations, surface charge distribution, and the presence of residues known to be implicated in channel recognition by other inhibitor cystine knot (ICK) toxins.
Keywords: Spider toxin, NMR structure, ASIC, ICK motif
Venoms of snakes, frogs, scorpions, spiders, cone snails, sea anemones, and some insects contain highly complex mixtures of bioactive peptides, many of which act primarily against ion channels. Peptide toxins isolated from these venoms have been invaluable tools for structural and physiological investigations of voltage-dependent Na+, Ca2+, and K+ ion channels as well as for mechanosensitive channels (Olivera et al. 1991; Escoubas et al. 2000b; Lewis 2000).
Numerous three-dimensional structures of peptide toxins acting on ion channels have been solved (Craik et al. 2001). In invertebrates, these peptides comprise ∼15 to 70 amino acids and are usually reticulated by disulfide bridges. A large number of the peptides studied to date belong to two major structural motifs. The first one is the cystine stabilized α/β scaffold (CSαβ) comprising a short α-helix and a double- or triple-stranded antiparallel β-sheet stabilized by three or four disulfide bridges. This structural organization is essentially found in scorpion venom toxins acting on voltage-sensitive K+ and Na+ channels (Possani et al. 2000). The second type is the inhibitor cystine knot (ICK) motif which comprises several loops that emerge from a double- or triple-stranded antiparallel β-sheet structure reticulated by at least three disulfide bridges (Pallaghy et al. 1994; Norton and Pallaghy 1998). Two of the disulfide bridges together with the amino-acid backbone form a ring, which is penetrated by a third disulfide bridge. The ICK motif is widespread in animal, plant, and fungal proteins, and has mainly been associated with inhibitory activities (Craik et al. 2001). Animal toxins structurally organized around this motif have been characterized from cone snails and spider venoms (Narasimhan et al. 1994; Norton and Pallaghy 1998; Escoubas et al. 2000b). In these peptides, the ICK fold is associated with a wide pharmacological profile, as these toxins can block voltage-dependent Na+ (Adams et al. 1989; Hill et al. 1997), K+ (Swartz and MacKinnon 1995; Savarin et al. 1998; Bernard et al. 2000), and Ca2+ (Adams et al. 1990; Olivera et al. 1991) channels as well as ryanodine-sensitive calcium channels (Mosbah et al. 2000).
Proton-sensitive inward currents activated by a drop in external pH were identified in the 1980’s (Krishtal and Pidoplichko 1981), and the underlying channels [acid-sensing ion channels (ASIC)] were cloned (Waldmann et al. 1997b). The predicted membrane topology of ASICs suggests a large extracellular loop connecting two transmembrane domains with the N and C termini inside the cell. This class of proton-gated cation channels appears to be involved in brain function and nociception (Reeh and Steen 1996; Waldmann and Lazdunski 1998; Kress and Zeilhofer 1999; McCleskey and Gold 1999; Chen et al. 2002). Several ASIC subunits and splice variants have now been described: ASIC1a (Waldmann et al. 1997b), ASIC1b (Chen et al. 1998), ASIC2a (Waldmann et al. 1997b; Champigny et al. 1998), ASIC2b (Lingueglia et al. 1997), ASIC3 (Waldmann et al. 1997a), and ASIC4 (Grunder et al. 2000). The different subtypes produce channels with different kinetics, external pH sensitivities, and tissue distribution. These different subunits can form homo- and heteromultimers in both the central nervous system and in nociceptor sensory neurons (Waldmann and Lazdunski 1998).
Pharmacological studies will be essential to understand the exact role of these channels in brain function and nociception. Amiloride blocks the inactivating phase of all of these channels, but it does so at relatively high concentrations at which the drug also blocks other transport systems.
The first-identified potent and specific peptide blocker of ASIC1 channels is psalmotoxin 1 (PcTx1). It was isolated from the venom of the South American tarantula Psalmopoeus cambridgei (Escoubas et al. 2000a) and very specifically blocks homomultimers of ASIC1a. It has been essential in characterizing the stoichiometry and role of these homomeric assemblies of ASIC1a in different neuron types (Escoubas et al. 2000a). The primary sequence of PcTx1 was found to be related to that of other spider venom ICK peptide toxins identified as inhibitors of voltage-dependent calcium or potassium channels. This paper reports the recombinant production of PcTx1 in Drosophila melanogaster S2 cells, and the determination of its solution structure by means of 1H 2D NMR spectroscopy.
Results and Discussion
Recombinant expression of PcTx1
Various recombinant expression systems have been used for small peptide toxins reticulated by disulfide bridges. They are produced either as linear peptides that are then reoxidized in vitro or directly in their native, refolded form. Methods leading to the production of reduced toxins often have the advantage of reduced costs and higher initial yields. The most widely used methods have been chemical synthesis or recombinant production in E. coli. However, in vitro reoxidative formation of disulfide bridges can be challenging, in particular for peptide toxins conforming to the ICK fold. This probably relates to the higher flexibility of the peptide chain compared to the αβ hairpin fold of scorpion toxins, which readily refold in vitro. Thus, direct production of refolded peptides is most desirable when attainable.
Examples of recombinant production of unfolded peptide toxins in E. coli followed by in vitro disulfide formation include potassium channel scorpion toxins (Park et al. 1991), huwentoxin I (Li et al. 2000), and the J-atracotoxins (Maggio and King 2002). Although production yields can be up to 10–20 mg/L (Li et al. 2000), the refolding step is usually the limiting factor in obtaining sufficient amounts of active toxin, as the refolding yield can be less than 10% of the original product. Other strategies based on fusion proteins have permitted the production of soluble, folded forms of the toxins in E. coli using a thioredoxin reductase-deficient strain (Johnson et al. 2000) or two protein A IgG-binding domains (ZZ) leading to periplasmic expression (Bouhaouala-Zahar et al. 1996; Korolkova et al. 2001). A yeast expression system has also been used successfully with yields in the 8–10 mg/L range (Wu et al. 2002). Several drawbacks of the methodology include the need for proteolytic or chemical cleavage of the fusion proteins, lowering final yields and sometimes leaving undesired residues in the toxin sequence, as well as the difficulty in obtaining posttranslational modifications, often necessary for toxin activity.
In addition, our experience with oxidative refolding of synthetic PcTx1 and other ICK spider peptide toxins has shown that obtaining the correct disulfide bridge pairing under classical redox conditions is difficult (P. Escoubas and G. Lambeau, unpubl.), yielding amounts of toxin insufficient for structural and physiological studies.
The Drosophila S2 cell production system permits direct recombinant production of correctly folded toxins that can be easily purified from the culture medium. This system also permits the production of glycosylated proteins and thus offers significant advantages over chemical synthesis for ICK peptides, in particular for mutagenesis studies. It was therefore chosen for the production of recombinant PcTx1.
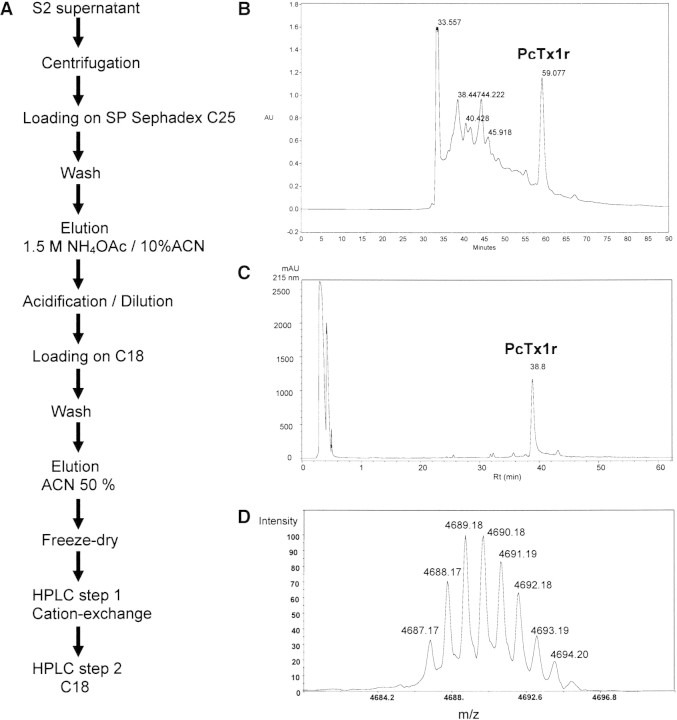
Pilot purification of PcTx1 from 2L of S2 cell culture supernatant demonstrated the presence of active folded recombinant PcTx1 (PcTx1r). An optimized purification scheme (Fig. 1A▶) was developed after extensive pilot studies in which either ion-exchange HPLC or RP-HPLC was used as the primary separation method. The complexity of the S2 cell supernatant and the very low relative amounts of the recombinant peptide did not permit resolution by direct HPLC fractionation. Two batch separations involving elution steps were found to be necessary to obtain a fraction containing PcTx1, which was manageable by RP-HPLC for final purification (Fig. 1B▶). The final ion-exchange HPLC step was done for final polishing to >99% purity. Treatment of a total of 12L of cell culture supernatant permitted the purification of 5.5 mg of PcTx1r (final yield 0.48 mg/L). Throughout the purification procedure, MALDI-TOF MS analysis of the chromatographic fractions allowed easy monitoring of the presence of PcTx1r as indicated by observation of a peptide ion at m/z 4690 in linear mode (Fig. 1B,C▶). MALDI-TOF MS thus proved to be a rapid, sensitive, and reliable method for the assessment of recombinant toxin production.
Figure 1.
Purification of recombinant PcTx1. (A) Purification scheme. (B) Cation-exchange HPLC of PcTx1r after two steps of batch purification (HPLC step 1). Linear gradient of ammonium acetate, on a Tosoh SP5PW cation-exchange semipreparative column (70 × 4.6 mm). (C) Reversed-phase HPLC final purifcation of PcTx1r (HPLC step 2). Linear gradient of acetonitrile on a semipreparative C18 column. (D) Reflector MALDI-TOF spectrum of purified PcTx1r, internal calibration. Calculated monoisotopic m/z 4687.1824, measured 4687.1722 (accuracy 2.1 ppm).
The purified recombinant toxin co-eluted with the native toxin in two separate, orthogonal chromatographic systems. Its full sequence and measured molecular mass (4686.19 Da, monoisotopic) corresponded to the sequence and calculated molecular mass (4686.18 Da, 2.1 ppm accuracy) of native PcTx1 (Fig. 1D▶). The inhibitory activity of PcTx1r on heterologously expressed ASIC1a was the same as that of native and synthetic PcTx1 (Escoubas et al. 2000a), and a dose-response curve yielded an IC50 value of 1.2 nM (data not shown).
A properly folded, active form of the PcTx1 toxin can thus be produced in the Drosophila S2 cell expression system, in amounts compatible with the requirements of 2-D NMR analysis and physiological experiments. Further improvement of the purification process (fraction recycling) could improve the final production yield. It should be noted that the toxin is produced folded, without the need for a fusion protein. This strategy avoids the supplementary steps associated with removal of the fusion protein (Bernard et al. 2000; Li et al. 2000) and thus limits losses of material. Direct production of a correctly folded form of the toxin also avoids the in vitro disulfide reoxidation step necessary for synthetic peptides and responsible for the very low final yields that have been obtained for other ICK toxins (Pennington et al. 1992; Lew et al. 1997; Sasaki et al. 1999).
The recombinant production strategy which has been used here for the first time for an ICK spider toxin holds promise for further production of short arthropod toxins.
NMR resonance assignment
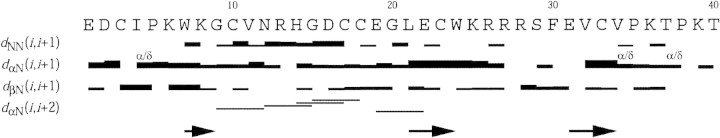
Sequential assignment was obtained by the now standard method first described by Wüthrich (Wüthrich 1986) and successfully applied to various arthropod toxins such as HpTx2, Ptu1, and Maurocalcine, respectively from the spider Heteropoda venatoria, the assassin bug Peirates turpis, and the scorpion Scorpio maurus (Bernard et al. 2000, 2001; Mosbah et al. 2000). The spin systems were identified on the basis of both COSY and TOCSY spectra recorded at 300 and 283K. The use of two temperatures for recording allowed us to resolve overlapping signals in the fingerprint region, and thus intraresidue HN-Hα cross-peaks were unambiguously assigned. At the end of the sequential assignment procedure, almost all protons were identified and their resonance frequency determined (BMRB ID 5495). The repartition of the Hα/HN and HN/HN correlations showed that the toxin is essentially organized in loops, beside three extended regions characterized by strong Hα/HN correlations (Fig. 2▶).
Figure 2.
Sequence of PcTx1 and sequential assignment. Collected sequential NOEs are classified into strong and weak NOEs and are indicated by thick and thin bars, respectively. The secondary elements (β-strands) are indicated by arrows.
Structure calculation
The structure of PcTx1 was determined by using 374 NOE-based distance restraints, including 195 intraresidue restraints, 111 sequential restraints, 24 medium-range restraints, and 44 long-range restraints. The repartition of these NOE along the sequence is shown in Figure 3▶. In addition, 12 hydrogen bond restraints and 21 dihedral angle constraints derived respectively from proton exchange and coupling constants have been included, as well as nine distance restraints derived from the three disulfide bridges which had been deduced from visual analysis of the previously obtained solutions and which are identical to that of other toxins folded into the same ICK motif (Escoubas et al. 2000b). Altogether, the final experimental set corresponded to 10.4 constraints per residue on average. The structures were calculated with DIANA using a distance geometry protocol, and energy minimized by CNS. The best-fit superimposition of backbone atoms for the 20 best structures gives RMSD values of 2.51 +/−0.51 Å for backbone atoms and 3.58 +/−0.40 Å if all nonhydrogen atoms are included. These rather high values are explained by the existence of structurally undefined N and C termini and two flexible regions encompassing residues 10 to 17 and 26 to 30. A summary of the structural statistics and the values of RMSD calculated on the well-defined regions are given in Table 1▶. All of the calculated structures have good nonbonded contacts and good covalent geometry, as shown by the low values of CNS energy terms and low RMSD values for bond lengths, valence angles, and improper dihedral angles. Correlation with the experimental data shows no NOE-derived distance violation greater than 0.2 Å. The analysis of the Ramachandran plot for the ensemble of the 20 calculated structures (in PROCHECK software nomenclature) shows 92.5% of the residues in the most favored and additional regions, 7.5% in the generously allowed regions, and none in the disallowed regions (data not shown).
Figure 3.
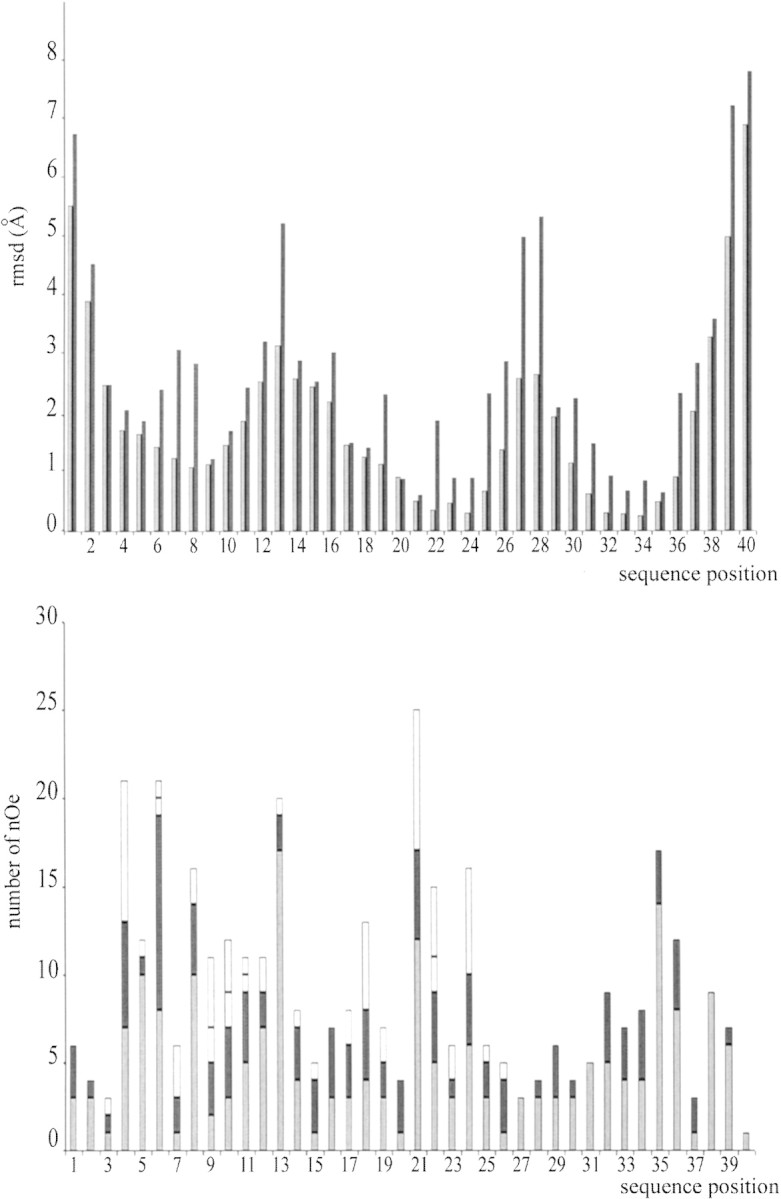
RMSD (top) and NOE (bottom) distribution vs. sequence. The RMSD values for backbone atoms and all heavy atoms are in gray and black, respectively. Intraresidue NOEs are in dark gray, sequential NOEs are in black, medium NOEs are in light gray, and long-range NOEs are in white.
Table 1.
Structural statistics of the PcTx1 20 best structures obtained by distance geometry and minimization
| Coordinate precisiona | |
| r.m.s. deviation (Å) | |
| Region 1–40 | |
| Backbone | 2.51 ± 0.51 |
| All heavy atoms | 3.58 ± 0.48 |
| Region 6–35 | |
| Backbone | 1.68 ± 0.36 |
| All heavy atoms | 2.92 ± 0.48 |
| Region 6–9, 18–25, 31–35 | |
| Backbone | 0.90 ± 0.22 |
| All heavy atoms | 1.95 ± 0.36 |
| Statistics for structure determination | |
| r.m.s. deviation from experimental restraints | |
| NOE distances (Å) | 0.029 ± 0.001 |
| Dihedral angles (degrees) | 0.14 ± 0.01 |
| r.m.s. deviation over secondary structure | |
| Bonds (Å) | 0.0033 ± 0.0001 |
| Angles (degrees) | 0.42 ± 0.02 |
| Impropers (degrees) | 0.26 ± 0.02 |
| Procheck analysis | |
| Most favored and additional allowed (%) | 92.5 |
| Generously allowed (%) | 7.5 |
| Disallowed region (%) | 0 |
a Determined from the 20 lowest energy structures obtained after energy minimization by XPLOR.
The minimized mean structure was obtained by averaging the coordinates of the individual DG structures best fitted to each other.
Structure description
The structure was calculated using 10.4 constraints per residue on the average. The repartition of these constraints along the sequence is highly heterogeneous as shown in Figure 3▶, and this reveals a differential flexibility with regard to the secondary structure. N and C termini as well as loops are less resolved than the central core of the toxin. This does come from a lack of NOEs in these regions, resulting from the internal dynamics of the protein. This feature has already been mentioned in most of the solution structures of ICK motifs solved to date.
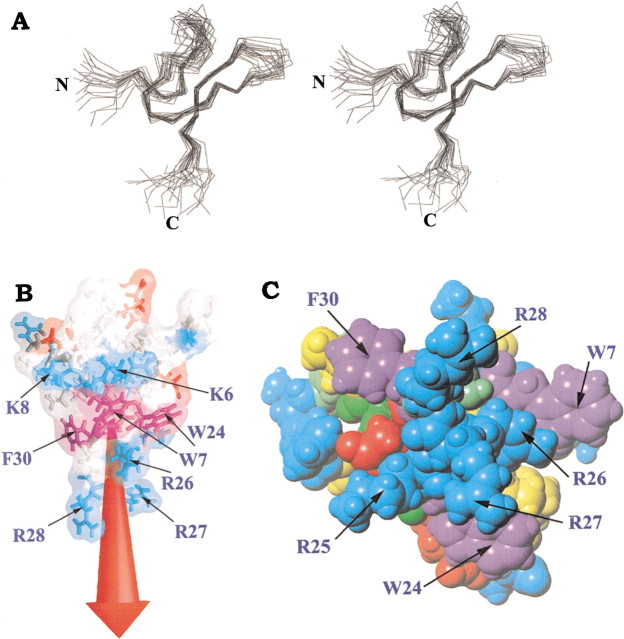
The three-dimensional structure (PDB accession code 1LMM) of PcTx1 consists of a compact disulfide-bonded core from which three loops and the N and C termini emerge. Figure 4A▶ shows a stereopair representation of the best-fit superimposition of the Cα traces of the 20 best structures. The main element of secondary structure is a three-stranded antiparallel β-sheet comprising residues 21–24 and 31–34. Two of the three strands are stabilized by NH/CO hydrogen bonds involving amide protons from residues 22, 24, 32, and 34 and are well detected by the PROCHECK-NMR software. The third peripherally extended strand composed of residues 7 to 9 is very poorly defined and is not detected by the PROCHECK-NMR software. Nevertheless, the existence of HN8-HN33, Hα9-Hα32 correlations, and the fact that the amide proton of residue 33 is in slow exchange (engaged in an hydrogen bond with the carbonyl group of residue 8) allowed us to describe this region as the third strand of the β-sheet. Overall, the structure shows a strong geometric anisotropy and can be related to a truncated cone.
Figure 4.
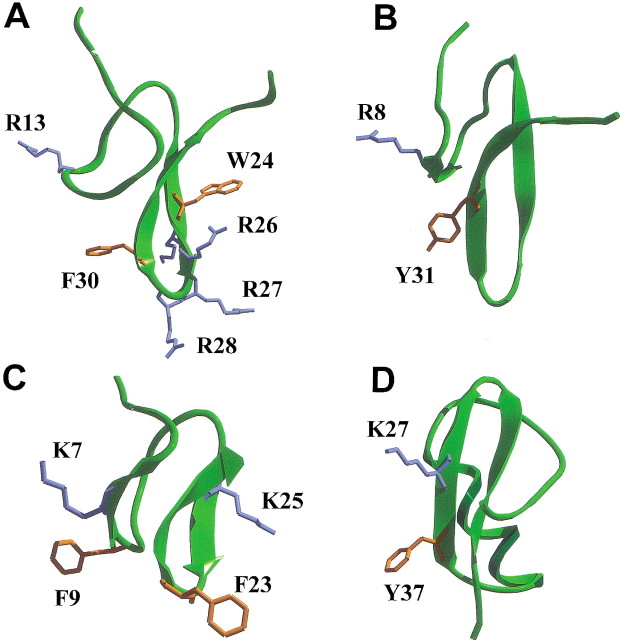
(A) Stereoview of the best fit of 20 solution structures of PcTx1. Cα are shown. The N and C termini are labeled "N" and "C". (B) Molecular surface colored according to the electrostatic charge (red for acidic and blue for basic) and the resulting dipolar moment (red arrow). The aromatic residues are indicated and colored in purple. (C) CPK representation of the proposed functional surface of PcTx1. The residues suspected to be important for the interaction of the toxin with the channel are labeled. Residues are colored as follows: green for polar uncharged residues, blue for basic residues, red for acidic residues, purple for aromatic residues, and yellow for aliphatic residues.
Analysis of the overall charge distribution of PcTx1 reveals a marked electrostatic anisotropy. This can be represented by a dipole moment, which is collinear to the geometric main axis of the molecule (the structure is described as a truncated cone, and the main geometric axis is the symmetry axis of the cone). It emerges from the turn between the second and third β-sheet strands, essentially composed of basic residues (Lys25, Arg26, Arg27, and Arg28; Fig. 4B▶). This positive patch is surrounded on its top by three aromatic residues (Trp7, Trp24, and Phe30). Another ring of four basic residues lies on top of the aromatic residues (Fig. 4C▶).
Comparison with related toxins
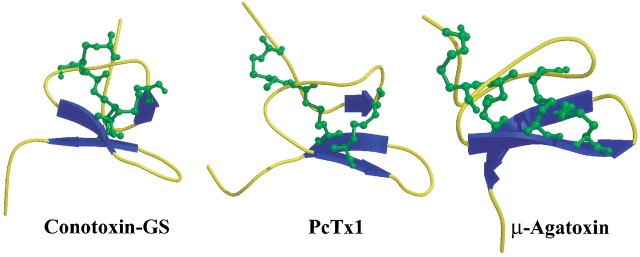
The PcTx1 fold can be classified as an inhibitor cystine knot (ICK) fold already described for numerous toxic and inhibitory animal venom peptides (Norton and Pallaghy 1998). It belongs to the class of small disulfide-bridged peptides classified as knottins in the SCOP database (http://scop.mrc-lmb.cam.ac.uk/scop/data/scop.b.h.d.html). Many ion channel effectors from marine snail, spider, and scorpion venoms share the same ICK fold, although they possess very different pharmacological profiles (Fig. 5▶; Olivera et al. 1991; Norton and Pallaghy 1998; Escoubas et al. 2000b; Craik et al. 2001). Among ICK spider toxins, PcTx1 is the only peptide known to act on ASIC1a channels, whereas μ-agatoxin I (Omecinsky et al. 1996), the δ-palutoxins (Corzo et al. 2000), and the δ-atracotoxins (Fletcher et al. 1997) are active on Na+ channels. In contrast, hanatoxins, HpTx2, and stromatoxins, respectively from the spiders Grammostola spatulata (Takahashi et al. 2000), Heteropoda venatoria (Bernard et al. 2000), and Stromatopelma calceata (Escoubas et al. 2002) act against voltage-dependent K+ channels. Other spider ICK toxins have been reported to act on Ca2+ channels, such as the ω-agatoxins IVA and IVB (Adams et al. 1993; Mintz and Bean 1993), huwentoxin I (Peng et al. 2001), SNX482 (Newcomb et al. 1998), ω-GsTx SIA (Lampe et al. 1993), and the ω-atracotoxins (Wang et al. 1999), respectively from the venoms of the spiders Agelenopsis aperta, Selenocosmia huwena, Hysterocrates gigas, Grammostola spatulata, and Hadronyche spp. More recently, the ICK fold has also been found in the spider peptide toxins GsMTx-4 and GsMTx-2, which block mechanosensitive cationic ion channels in heart cells (Oswald et al. 2002).
Figure 5.
Comparison of three-dimensional structures of structurally related toxins blocking Na+ channels with PcTx1, in Molscript representation.
In addition, the ICK fold has also been associated with other inhibitory functions such as phenoloxidase inhibition in houseflies (Daquinag et al. 1999) and antimicrobial activity of peptides from horseshoe crab hemocytes (Osaki et al. 1999) and spider venom (Yan and Adams 1998).
PcTx1 has a unique pharmacology, which distinguishes it from all other ICK toxins. Although its overall structure fits into the canonical ICK fold of related Ca2+, Na+, and K+ channel toxins, its selectivity and mode of action are totally different and place it in a unique pharmacological class. Previous results (Escoubas et al. 2000a) demonstrated that PcTx1 has a very high selectivity for the homomultimeric ASIC1a channel subtype and does not affect other ion channels, including other subtypes or heteromultimeric assemblies of ASIC subunits. PcTx1 perfectly illustrates the often mentioned capability of the ICK scaffold reticulated by three or four disulfides to bear a variety of pharmacological activities through selective mutagenesis of the solvent-exposed loops. The evolutionary mechanism leading to the extraordinary pharmacological diversity of venom peptides has been extensively studied in cone snails (Espiritu et al. 2001), and a similar model would be suitable to describe the molecular diversity of spider venom peptides.
Functional surface of PcTx1
To analyze the putative functional surface of peptide toxins, we used a prediction method based on the orientation of the dipole moment resulting from the electrostatic anisotropy of the toxin. The dipole is used as a guideline to predict a putative functional surface (Cui et al. 2001, 2002; Fu et al. 2002). The highly undefined N and C termini (residues 1, 2, and 38 to 40) were not included in the calculation of the dipole of PcTx1, but it was clearly the same as the dipole calculated for the averaged complete molecule. Assuming that the dipole may work as an orientation force in the electric field of the receptor (Fremont et al. 1997; Bernard et al. 2000; Mosbah et al. 2000), a proposed functional surface for PcTx1 is presented in Figure 4C▶. Two possible sites of action for peptide toxin blockade of ion channels have been proposed. Current inhibition may result from pore occlusion, as observed for the scorpion and cone snail toxins blocking voltage-dependent potassium channels, or via steric hindrance of ion conduction by binding at the mouth of the pore. The latter has been proposed as the binding mechanism of voltage-gating modifiers such as the hanatoxins, which might also impede the movement of ion channel transmembrane segments in response to a depolarizing voltage and therefore affect ion conduction. Recent work also supports the "hot spot" model of protein-protein interaction (Maggio and King 2002) for toxin binding to ion channels. The hot spot on the toxin surface has been proposed to involve a functional dyad comprising a critical lysine residue involved in pore blocking and a neighboring aromatic residue such as tryptophan or phenylalanine (Dauplais et al. 1997).
This toxin-channel interaction model has been validated by mutagenesis studies on scorpion (Dauplais et al. 1997) and cone snail toxins (Jacobsen et al. 2000), and the sea anemone toxins BgK (Alessandri-Haber et al. 1999) and ShK (Pennington et al. 1996a,b), and more recently on the insecticidal J-atracotoxins from spider venom (Maggio and King 2002). This structural motif appears to be found in various channel inhibitors such as snake dendrotoxins, charybdotoxin, agitoxin, noxiustoxin, and related scorpion peptides (Dauplais et al. 1997), the newly discovered κ-hefutoxins (Srinivasan et al. 2002), κ-conotoxin (Savarin et al. 1998), and BgK and ShK from sea anemones (Pennington et al. 1996a,b; Alessandri-Haber et al. 1999). All these toxins block ion channels through pore occlusion, and the association of a hydrophobic residue with a patch of positive residues or a single basic amino acid such as Lys or Arg has been demonstrated to be crucial for channel recognition and blockade (Pennington et al. 1996a). As novel peptides are described and characterized, evidence is mounting to support the crucial role of the functional dyad in many toxin-channel interactions.
Another interaction model has been proposed for gating-modifier toxins such as the hanatoxins (Takahashi et al. 2000). In this case, the interaction hot spot would be represented by a patch of hydrophobic residues forming a contact surface surrounded by charged residues anchoring the toxin to its target surface through formation of salt bridges. In the hot spot model, residues surrounding the toxin hot spot are proposed to serve as "gaskets," preventing interaction of water molecules with the target residues on the channel surface. Although the hydrophobic patch model appears to be relevant in other potassium channel blockers such as the stromatoxins (Escoubas et al. 2002), in which a dyad Lys-Phe can also be found, no mutagenesis data are available to validate its relevance.
Examination of the structure of PcTx1 reveals that it does not conform to the hydrophobic patch model but bears a considerable number of positively charged residues in the β-turn linking the two β-strands (loop 4). The stretch K25R26R27R28 in fact forms a contiguous positive surface protruding from the rest of the molecule (Fig. 4C▶). Additionally, three aromatic residues (Trp7, Trp24, and Phe30) are found in the vicinity of the basic residues and may possibly be involved in the formation of functional dyads equivalent to that of other ion channel toxins. Judging from interatomic distances, only the dyad Arg26/Trp24 (6.8 Å) would fit the proposed pattern in which distances between basic and aromatic residues vary between 5.8 and 7.6 Å (Srinivasan et al. 2002). Arg28 and Phe30 are located further apart (14 Å) in the PcTx1 structure. Figure 6▶ shows the structural similarity of basic and aromatic residue side chains in several of these toxins. Loop 4 in PcTx1 thus appears to be a potentially important structural feature, in accordance with the electrostatic anisotropy data presented above. We propose that the positively charged patch formed mostly by residues K25–R28, accompanied by aromatic side chains and other surrounding hydrophobic or negatively charged residues could form the channel recognition surface of PcTx1. It should be noted that negatively charged residues are located primarily in the N-terminal part of the peptide, and on the opposite side of the K25–R28 loop, forming a "negative crown" above the positively charged patch.
Figure 6.
Comparison of putative functional dyads of PcTx1 (A) and related ICK toxins J-atracotoxin Hv1c (PDB 1DL0; B), κ-conotoxin PVIIA (PDB 1KCP; C), and agitoxin 2 (PDB 1AGT; D). Basic residues side chains are colored in blue; aromatic residues side chains, in orange.
In conclusion, we have successfully expressed the ASIC1a channel blocker PcTx1 in a recombinant expression system used for the first time for a spider toxin, and determined its structure in solution. Although the mode of action of PcTx1 cannot be inferred solely from examination of its surface characteristics, key structural elements for PcTx1 binding to its ASIC1a target appear to be localized in the two β-strands and the β-turn linking them, as indicated by anisotropy calculations, surface charge distribution, and the presence of residues known to be implicated in channel recognition of ICK toxins. These results will allow the development of further research on the molecular determinants involved in recognition of the ASIC channel binding site, and will guide selective mutagenesis studies. As the toxin is the only known high-affinity ligand for the ASIC class of channels that probably play an important role in nociception, PcTx1 can be an important tool for designing new classes of drugs acting directly against pain at the nociceptor level. In addition, the ASIC1a channel is widely expressed in brain, and is thought to be involved in numerous functions involving synaptic plasticity, learning, and memory. Therefore, PcTx1 may also prove to be an invaluable tool in examining these central processes.
Materials and methods
Preparation of recombinant PcTx1
A synthetic gene coding for the mature PcTx1 protein was designed based on the sequence of purified PcTx1 (Escoubas et al. 2000b). Because the strategy was to produce PcTx1 in Drosophila S2 cells, the codon usage for Drosophila highly expressed genes found in the GCG package database was used to design the PcTx1 gene. The synthetic gene was fused to the signal peptide (MKFLVNVALVFMVVYISYIYA) of mellitin, a highly expressed bee venom peptide, to allow for the efficient secretion of PcTx1 into cell medium. To prepare the PcTx1 synthetic gene, two long, partially overlapping oligonucleotides of 91 and 88 bases (2 μg each) were annealed and elongated using a high-fidelity Pwo polymerase (Roche Molecular Biochemicals) in a reaction volume of 50 μL. Annealing, elongation, and denaturation steps were performed at 50°C, 72°C, and 95°C for 30 sec each for a total of 10 cycles. Forward and reverse PcTx1 primers were: 5′-tcgtgtacatttcttacatctatgcgGAGGACTGCATCCCCAAGTGGAAGGGCTGCG TGAACCGCCACGGCGACTGCTGCGAGGGCCTGGA-3′, 5′-aaagtcgactattaGGTCTTGGGGGTCTTGGGCACGCACACCTC GAAGGAGCGGCGGCGCTTCCAGCACTCCAGGCCCTCGC AGCAGT-3′. Bases in lowercase letters of the forward primer code for the 3′-end of the mellitin signal peptide and contain a BsrGI restriction site. Those in the reverse primer code for a stop codon followed by a SalI restriction site. Bases in uppercase letters in the two primers code for the entire PcTx1 protein sequence. After analysis on a 3% agarose gel and purification with the wizard PCR prep kit (Promega), the PcTx1 DNA fragment was ligated into a pGEM-T easy vector (Promega) in which the mellitin signal peptide containing the BsrGI and SalI restriction sites had been previously inserted. After sequencing, the DNA construct was subcloned into the Drosophila expression vector pMT/V5-His (InvitroGen) to allow for the recombinant expression of PcTx1 in S2 cells according to the InvitroGen Drosophila expression system. The sequence of the complete PcTx1 synthetic gene is available from the authors upon request. S2 cells were grown at 24°–27°C in Drosophila-SFM medium (InvitroGen #10797-025) supplemented with 1% fetal bovine serum and antibiotics (InvitroGen #15240-021). After sequencing, the expression plasmid PcTx1-pMT/V5-His and a plasmid carrying a G418-resistance selection cassette were co-transfected into Drosophila S2 cells using Ca2+/PO4. The Ca2+/PO4-DNA precipitate was incubated with cells for 4 h, after which the cells were washed by centrifugation and resuspended in fresh medium. Two d after transfection, G418 sulfate (2mg/mL) was added to the cell culture medium to select for stably transfected cells. Fresh selection medium was added every 4–5 d. Five d after transfection and 4 wks after selection, PcTx1 expression was examined by evaluating the presence of PcTx1 in the cell medium. For these assays, cells were plated at 3 × 106 cells/mL in 24-well plates and induced 1 d later with 500 μM CuSO4 for 3 to 7 d. Fifty microliters of cell medium were loaded on a C18 ZipTip (Millipore) to remove salts, and the presence of PcTx1 was determined by MALDI-TOF mass spectrometry. For large-scale production of PcTx1, stably transfected cells were seeded into 500 mL of complete medium at a density of 2–3 × 106 cells/mL in 1-L spinner flasks (Integra Biosciences). Upon reaching a density of 5–6 × 106 cells/mL, cells were induced with 500 μM CuSO4. Three to four d after induction, the cells were spun down and the medium was collected for further purification of PcTx1.
Recombinant toxin purification and characterization
Drosophila S2 cell culture medium was centrifuged at 5000 rpm for 30 min. One liter of the supernatant was then diluted twofold with equilibration buffer (H2O/10% acetonitrile/1% acetic acid) and loaded overnight in batch mode on 300 mL of SP Sephadex C-25 cation-exchange resin (Amersham Pharmacia Biotech) prewashed and equilibrated in H2O/10% acetonitrile/1% acetic acid. Two washing steps were performed with (1) equilibration buffer and (2) equilibration buffer containing 0.4M ammonium acetate/10% acetonitrile. Recombinant PcTx1 was then eluted with 1.5M ammonium acetate/10% acetonitrile. After a twofold dilution with water and acidification with TFA to pH 3.0, the PcTx1 fraction was loaded in batch mode on 100 mL of C18 reversed-phase silica (20–43 μm; Merck) pre-equilibrated in 0.1% TFA. Three consecutive washing steps (500 mL each) were performed with (1) water/0.1% TFA; (2) 5% aqueous acetonitrile/0.1% TFA; (3) 10% aqueous acetonitrile/0.1% TFA. PcTx1 was eluted with 500 mL of 50% aqueous acetonitrile in 0.1% TFA, and the elution volume was diluted threefold with water and freeze-dried.
Throughout the batch purification scheme, the presence of recombinant PcTx1 was monitored in all fractions by analysis of a small aliquot by MALDI-TOF MS in linear mode. For diluted fractions, a solid-phase extraction with C18 ZipTip microconcentrating devices (Millipore) was performed prior to mass spectrometry analysis.
Further purification was performed by cation-exchange HPLC on a preparative SP5PW column (150 × 21 mm, Tosoh-Haas) equilibrated with 10% aqueous acetonitrile/1% acetic acid, using a linear gradient of 10% to 90% of 2M ammonium acetate/10% acetonitrile (1%/min), at 6 mL/min. PcTx1 fractions from several HPLC runs were pooled and then freeze-dried, and a final purification step was done by reversed-phase HPLC on a C18 semipreparative column (250 × 10 mm, 5μm, 100 Å, Nacalai Tesque), using a linear gradient of acetonitrile/0.1% TFA in water/0.1%TFA (5% to 35% acetonitrile in 60 min, 2 mL/min). Final quantification of the toxin was done by optical density measurement at 280 nm, using a calculated ɛ280 of 11740.
Full characterization of recombinant PcTx1 was achieved by automated N-terminal Edman sequencing on an Applied Biosystems Procise gas-phase sequencer, monoisotopic molecular weight analysis by MALDI-TOF MS, and HPLC co-elution experiments with the native toxin. Co-elution of recombinant and native PcTx1 was performed by cation-exchange on an analytical SP5PW column (75 × 7.5 mm, Tosoh-Haas) using a linear gradient of 20mM to 1M ammonium acetate in 50 min (0.5 mL/min) and by reversed-phase on a Merck Purospher STAR 55–4 column (55 × 4 mm, 3 μm), using a linear gradient of acetonitrile in water (constant 0.1% TFA) at 1 mL/min and 0% to 35% acetonitrile/min over 35 min.
MALDI-TOF mass spectrometry
Peptides or HPLC fractions mixed with α-cyano-4-hydroxycinnamic acid (α-CHCA, Aldrich) matrix (10 mg/mL) were analyzed on an Applied Biosystems Voyager DE-Pro system in positive reflector (pure peptides) or linear (fractions) mode. Mass spectra (200–300 scans) were calibrated with external or internal standards and analyzed with Data Explorer software.
Electrophysiological characterization
Activity of recombinant PcTx1 was tested against cloned ASIC1a channels heterologously expressed in COS cells as described (Escoubas et al. 2000b). Recombinant PcTx1 was applied in a range of concentrations to establish its dose-response curve (N=5 per dose).
Sample preparation for NMR
Recombinant PcTx1 (4 mg) was solubilized in 450 μL of an H2O/D2O mixture (9:1 v/v), to give a protein concentration of 2.9 mM at pH 3.0. This rather low pH is used to reduce as much as possible the amide proton exchange rate. The exchange rate was determined after lyophilization of this sample and dissolution in 100% D2O to check that the conformation (on the sight of NH resonance frequencies) was the same at pH 3 than the one at pH 5, which is used to test the pharmacological activity of the toxin.
NMR experiments
All 1H NMR spectra were recorded on a BRUKER DRX500 spectrometer equipped with an HCN probe, and self-shielded triple-axis gradients were used. The experiments were performed at two different temperatures in order to solve assignment ambiguities (283 K and 300 K). Two-dimensional spectra were acquired using the states-TPPI method to achieve F1 quadrature detection. Water suppression was achieved using presaturation during the relaxation delay (1.5 sec), and during the mixing time in the case of NOESY experiments (Kumar et al. 1980), or using a watergate 3–9–19 pulse train using a gradient at the magic angle obtained by applying simultaneous x-, y-, and z-gradients prior to detection. NOESY spectra were acquired using mixing times of 100 msec and 120 msec. TOCSY was performed with a spin locking field strength of 8 kHz and spin lock time of 80 msec. The amide proton exchange experiments were recorded immediately after dissolution of the peptides in D2O. Four series of NOESY spectra with a mixing time of 80 msec were recorded at 283K, the first series for 1 h, the subsequent three for 4 h each.
The identification of amino acid spin systems and the sequential assignment were done using the standard strategy described by Wüthrich (1986) and regularly used by our group (Bernard et al. 2000, 2001; Mosbah et al. 2000), with the graphic software XEASY (Bartels et al. 1995). The comparative analysis of TOCSY spectra recorded in water gave the spin system signatures of the protein. The spin systems were then sequentially connected using the NOESY spectra.
The integration of NOE data was done by measuring the peak volumes using a specific routine of the XEASY package. These volumes were then translated into upper limit distances by the CALIBA routine of the DIANA software (Güntert and Wüthrich 1991). The lower limit was systematically set at 0.18 nm. The φ torsion angle constraints resulted from the 3JHN-Hα coupling constant measurements. They were estimated with the INFIT program (Szyperski et al. 1992). For a given residue, separated NOESY cross-peaks with the backbone amide proton in the ω2 dimension were used. Several cross-sections through these cross-peaks that exhibited a good signal-to-noise ratio were selected and then summed, and only those data points of the peak region that were above the noise level were retained. The left and right ends of the peak region were then brought to zero intensity by linear baseline correction. After extending the baseline-corrected peak region with zeros on both sides, which is equivalent to oversampling in the time domain, an inverse Fourier transformation was performed. The value of the 3JHN-Hα coupling constant was obtained from the first local minimum. 3JHN-Hα coupling constants were translated into angle restraints using HABAS from the DIANA package.
We checked that increasing the pH to 5, closer to the actual activation pH of the channel, does not alter the conformation of the protein. This was checked by comparing NH resonance frequencies at both pHs. A NOESY recorded at pH 5 does not show any significant modification in NOEs collected for the structure determination.
Structure calculation
Distance geometry calculations were performed with the variable target function program DIANA 2.8. A preliminary set of 1000 structures was initiated including only intraresidual and sequential upper limit distances. From these, the 500 best as shown by the value of the target function were kept for a second round, including medium-range distances. The resulting 250 best solutions were kept for a third round, using the whole set of upper restraints. The 100 best solutions were then refined by adding the restraints coming out from the disulfide pairing. This pairing was assessed from the visual analysis of the structures obtained solely from the NOE constraints, and found to be identical to that of other ICK-folded animal toxins. Starting from the 100 best structures, one REDAC cycle (Güntert and Wüthrich 1991) was used in a last step in order to include the dihedral constraints together with the additional distance restraints coming from hydrogen bonds.
To remove residual bad Van der Waals contacts, the 30 best structures were refined by restrained molecular dynamics annealing, slow cooling, and energy minimization (parameter file: protein-allhdg in CNS). Visual analysis was done using TURBO software (Roussel and Cambillau 1989), and the geometric quality of the obtained structures was assessed by PROCHECK 3.3 (Laskowski et al. 1993) and PROCHECK-NMR software (Laskowski et al. 1996).
Acknowledgments
We thank Dr. A. Baron for electrophysiological measurements, M. Jodar, C. Le Calvez, and S. Scarzello for technical assistance, Y. Benhamou and V. Briet for editorial assistance, and Dr. L.D. Rash for English editing and critical reading of the manuscript. This work was partially supported by a grant from the Association Française contre les Myopathies (AFM).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
PcTx1, psalmotoxin 1
NOESY, nuclear Overhauser effect spectroscopy
TOCSY, total correlation spectroscopy
COSY, correlation spectroscopy
CNS, crystallography and NMR system
ICK, inhibitor cystine knot
The PDB coordinate files have been deposited in the Brookhaven Data Bank (PDB code 1LMM).
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0307003.
References
- Adams, M.E., Herold, E.E., and Venema, V.J. 1989. Two classes of channel-specific toxins from funnel web spider venom. J. Comp. Physiol. 164 333–342. [DOI] [PubMed] [Google Scholar]
- Adams, M.E., Bindokas, V.P., Hasegawa, L., and Venema, V.J. 1990. ω-agatoxins: Novel calcium channel antagonists of two subtypes from funnel web spider (Agelenopsis aperta) venom. J. Biol. Chem. 265 861–867. [PubMed] [Google Scholar]
- Adams, M.E., Mintz, I.M., Reily, M.D., Thanabal, V., and Bean, B.P. 1993. Structure and properties of ω-agatoxin IVB, a new antagonist of P-type calcium channels. Mol. Pharmacol. 44 681–688. [PubMed] [Google Scholar]
- Alessandri-Haber, N., Lecoq, A., Gasparini, S., Grangier-Macmath, G., Jacquet, G., Harvey, A.L., de Medeiros, C., Rowan, E.G., Gola, M., Ménez, A., et al. 1999. Mapping the functional anatomy of BgK on Kv1.1, Kv1.2, and Kv1.3. Clues to design analogs with enhanced selectivity. J. Biol. Chem. 274 35653–35661. [DOI] [PubMed] [Google Scholar]
- Bartels, C., Xia, T.-H., Billeter, M., Güntert, P., and Wüthrich, K. 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 5 1–10. [DOI] [PubMed] [Google Scholar]
- Bernard, C., Legros, C., Ferrat, G., Bischoff, U., Marquardt, A., Pongs, O., and Darbon, H. 2000. Solution structure of HpTX2, a toxin from Heteropoda venatoria spider that blocks Kv4.2 potassium channel. Protein Sci. 9 2059–2067. [PMC free article] [PubMed] [Google Scholar]
- Bernard, C., Corzo, G., Mosbah, A., Nakajima, T., and Darbon, H. 2001. Solution structure of Ptu1, a toxin from the assassin bug Peirates turpis that blocks the voltage-sensitive calcium channel N-type. Biochemistry 40 12795–12800. [DOI] [PubMed] [Google Scholar]
- Bouhaouala-Zahar, B., Ducancel, F., Zenouaki, I., Ben Khalifa, R., Borchani, L., Pelhate, M., Boulain, J.C., El Ayeb, M., Ménez, A., and Karoui, H. 1996. A recombinant insect-specific α-toxin of Buthus occitanus tunetanus scorpion confers protection against homologous mammal toxins. Eur. J. Biochem. 238 653–660. [DOI] [PubMed] [Google Scholar]
- Champigny, G., Voilley, N., Waldmann, R., and Lazdunski, M. 1998. Mutations causing neurodegeneration in Caenorhabditis elegans drastically alter the pH sensitivity and inactivation of the mammalian H+-gated Na+ channel MDEG1. J. Biol. Chem. 273 15418–15422. [DOI] [PubMed] [Google Scholar]
- Chen, C.C., England, S., Akopian, A.N., and Wood, J.N. 1998. A sensory neuron-specific, proton-gated ion channel. Proc. Natl. Acad. Sci. 95 10240–10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.C., Zimmer, A., Sun, W.H., Hall, J., and Brownstein, M.J. 2002. A role for ASIC3 in the modulation of high-intensity pain stimuli. Proc. Natl. Acad. Sci. 99 8992–8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corzo, G., Escoubas, P., Stankiewicz, M., Pelhate, M., Kristensen, C.P., and Nakajima, T. 2000. Isolation, synthesis and pharmacological characterization of δ-palutoxins IT, novel insecticidal toxins from the spider Paracoelotes luctuosus (Amaurobiidae). Eur. J. Biochem. 267 5783–5795. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Daly, N.L., and Waine, C. 2001. The cystine knot motif in toxins and implications for drug design. Toxicon 39 43–60. [DOI] [PubMed] [Google Scholar]
- Cui, M., Shen, J., Briggs, J.M., Luo, X., Tan, X., Jiang, H., Chen, K., and Ji, R. 2001. Brownian dynamics simulations of interaction between scorpion toxin Lq2 and potassium ion channel. Biophys. J. 80 1659–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, M., Shen, J., Briggs, J.M., Fu, W., Wu, J., Zhang, Y., Luo, X., Chi, Z., Ji, R., Jiang, H., et al. 2002. Brownian dynamics simulations of the recognition of the scorpion toxin P05 with the small-conductance calcium-activated potassium channels. J. Mol. Biol. 318 417–428. [DOI] [PubMed] [Google Scholar]
- Daquinag, A.C., Sato, T., Koda, H., Takao, T., Fukuda, M., Shimonishi, Y., and Tsukamoto, T. 1999. A novel endogenous inhibitor of phenoloxidase from Musca domestica has a cystine motif commonly found in snail and spider toxins. Biochemistry 38 2179–2188. [DOI] [PubMed] [Google Scholar]
- Dauplais, M., Lecoq, A., Song, J., Cotton, J., Jamin, N., Gilquin, B., Roumestand, C., Vita, C., de Medeiros, C.L., Rowan, E.G., et al. 1997. On the convergent evolution of animal toxins. Conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures. J. Biol. Chem. 272 4302–4309. [DOI] [PubMed] [Google Scholar]
- Escoubas, P., De Weille, J.R., Lecoq, A., Diochot, S., Waldmann, R., Champigny, G., Moinier, D., Ménez, A., and Lazdunski, M. 2000a. Isolation of a tarantula toxin specific for a class of proton-gated Na+ channels. J. Biol. Chem. 275 25116–25121. [DOI] [PubMed] [Google Scholar]
- Escoubas, P., Diochot, S., and Corzo, G. 2000b. Structure and pharmacology of spider venom neurotoxins. Biochimie 82 893–907. [DOI] [PubMed] [Google Scholar]
- Escoubas, P., Diochot, S., Célérier, M.L., Nakajima, T., and Lazdunski, M. 2002. Novel tarantula toxins for subtypes of voltage-dependent potassium channels in the Kv2 and Kv4 subfamilies. Mol. Pharmacol. 62 48–57. [DOI] [PubMed] [Google Scholar]
- Espiritu, D.J., Watkins, M., Dia-Monje, V., Cartier, G.E., Cruz, L.J., and Olivera, B.M. 2001. Venomous cone snails: Molecular phylogeny and the generation of toxin diversity. Toxicon 39 1899–1916. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.I., Chapman, B.E., Mackay, J.P., Howden, M.E., and King, G.F. 1997. The structure of versutoxin (δ-atracotoxin-Hv1) provides insights into the binding of site 3 neurotoxins to the voltage-gated sodium channel. Structure 5 1525–1535. [DOI] [PubMed] [Google Scholar]
- Fremont, V., Blanc, E., Crest, M., Eauclaire, M.F., Gola, M., Darbon, H., and Van Rietscoten, J. 1997. Dipole moments of scorpion toxins direct the interaction towards small- or large-conductance Ca++-activated K+ channels. Lett. Peptide Sci. 4 305–312. [Google Scholar]
- Fu, W., Cui, M., Briggs, J.M., Huang, X., Xiong, B., Zhang, Y., Luo, X., Shen, J., Ji, R., Jiang, H., et al. 2002. Brownian dynamics simulations of the recognition of the scorpion toxin maurotoxin with the voltage-gated potassium ion channels. Biophys. J. 83 2370–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güntert, P. and Wüthrich, K. 1991. Improved efficiency of protein structure calculations from NMR data using the program DIANA with redundant dihedral angle constraints. J. Biomol. NMR 1 447–456. [DOI] [PubMed] [Google Scholar]
- Grunder, S., Geissler, H.S., Bassler, E.L., and Ruppersberg, J.P. 2000. A new member of acid-sensing ion channels from pituitary gland. Neuroreport 11 1607–1611. [DOI] [PubMed] [Google Scholar]
- Hill, J.M., Alewood, P.F., and Craik, D.J. 1997. Solution structure of the sodium channel antagonist conotoxin GS: A new molecular caliper for probing sodium channel geometry. Structure 5 571–583. [DOI] [PubMed] [Google Scholar]
- Jacobsen, R.B., Koch, E.D., Lange-Malecki, B., Stocker, M., Verhey, J., Van Wagoner, R.M., Vyazovkina, A., Olivera, B.M., and Terlau, H. 2000. Single amino acid substitutions in κ-conotoxin PVIIA disrupt interaction with the shaker K+ channel. J. Biol. Chem. 275 24639–24644. [DOI] [PubMed] [Google Scholar]
- Johnson, T.M., Quick, M.W., Sakai, T.T., and Krishna, N.R. 2000. Expression of functional recombinant scorpion β-neurotoxin Css II in E. coli. Peptides 21 767–772. [DOI] [PubMed] [Google Scholar]
- Korolkova, Y.V., Kozlov, S.A., Lipkin, A.V., Pluzhnikov, K.A., Hadley, J.K., Filippov, A.K., Brown, D.A., Angelo, K., Strobaek, D., Jespersen, T., et al. 2001. An ERG channel inhibitor from the scorpion Buthus eupeus. J. Biol. Chem. 276 9868–9876. [DOI] [PubMed] [Google Scholar]
- Kress, M. and Zeilhofer, H.U. 1999. Capsaicin, protons and heat: New excitement about nociceptors. Trends Pharmacol. Sci. 20 112–118. [DOI] [PubMed] [Google Scholar]
- Krishtal, O.A. and Pidoplichko, V.I. 1981. Receptor for protons in the membrane of sensory neurons. Brain Res. 214 150–154. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Ernst, R.R., and Wuthrich, K. 1980. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton–proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 95 1–6. [DOI] [PubMed] [Google Scholar]
- Lampe, R.A., Defeo, P.A., Davison, M.D., Young, J., Herman, J.L., Spreen, R.C., Horn, M.B., Mangano, T.J., and Keith, R.A. 1993. Isolation and pharmacological characterization of ω-grammotoxin SIA, a novel peptide inhibitor of neuronal voltage-sensitive calcium channel responses. Mol. Pharmacol. 44 451–460. [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.M., and Thornton, J.M. 1993. A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26 283–291. [Google Scholar]
- Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8 477–486. [DOI] [PubMed] [Google Scholar]
- Lew, M.J., Flinn, J.P., Pallaghy, P.K., Murphy, R., Whorlow, S.L., Wright, C.E., Norton, R.S., and Angus, J.A. 1997. Structure-function relationships of ω-conotoxin GVIA. Synthesis, structure, calcium channel binding, and functional assay of alanine-substituted analogues. J. Biol. Chem. 272 12014–12023. [DOI] [PubMed] [Google Scholar]
- Lewis, R.J. 2000. Ion channel toxins and therapeutics: From cone snail venoms to ciguatera. Ther. Drug. Monit. 22 61–64. [DOI] [PubMed] [Google Scholar]
- Li, M., Li, L.Y., Wu, X., and Liang, S.P. 2000. Cloning and functional expression of a synthetic gene encoding huwentoxin-I, a neurotoxin from the Chinese bird spider (Selenocosmia huwena). Toxicon 38 153–162. [DOI] [PubMed] [Google Scholar]
- Lingueglia, E., de Weille, J.R., Bassilana, F., Heurteaux, C., Sakai, H., Waldmann, R., and Lazdunski, M. 1997. A modulatory subunit of acid sensing ion channels in brain and dorsal root ganglion cells. J. Biol. Chem. 272 29778–29783. [DOI] [PubMed] [Google Scholar]
- Maggio, F. and King, G.F. 2002. Scanning mutagenesis of a Janus-faced atracotoxin reveals a bipartite surface patch that is essential for neurotoxic function. J. Biol. Chem. 277 22806–22813. [DOI] [PubMed] [Google Scholar]
- McCleskey, E.W. and Gold, M.S. 1999. Ion channels of nociception. Annu. Rev. Physiol. 61 835–856. [DOI] [PubMed] [Google Scholar]
- Mintz, I.M. and Bean, B.P. 1993. Block of calcium channels in rat neurons by synthetic ω-Aga-IVA. Neuropharmacology 32 1161–1169. [DOI] [PubMed] [Google Scholar]
- Mosbah, A., Kharrat, R., Fajloun, Z., Renisio, J.G., Blanc, E., Sabatier, J.M., El Ayeb, M., and Darbon, H. 2000. A new fold in the scorpion toxin family, associated with an activity on a ryanodine-sensitive calcium channel. Proteins 40 436–442. [DOI] [PubMed] [Google Scholar]
- Narasimhan, L., Singh, J., Humblet, C., Guruprasad, K., and Blundell, T. 1994. Snail and spider toxins share a similar tertiary structure and "cystine motif." Nat. Struct. Biol. 1 850–852. [DOI] [PubMed] [Google Scholar]
- Newcomb, R., Szoke, B., Palma, A., Wang, G., Chen, X., Hopkins, W., Cong, R., Miller, J., Urge, L., Tarczy-Hornoch, K., et al. 1998. Selective peptide antagonist of the class E calcium channel from the venom of the tarantula Hysterocrates gigas. Biochemistry 37 15353–15362. [DOI] [PubMed] [Google Scholar]
- Norton, R.S. and Pallaghy, P.K. 1998. The cystine knot structure of ion channel toxins and related polypeptides. Toxicon 36 1573–1583. [DOI] [PubMed] [Google Scholar]
- Olivera, B.M., Rivier, J., Scott, J.K., Hillyard, D.R., and Cruz, L.J. 1991. Conotoxins. J. Biol. Chem. 266 22067–22070. [PubMed] [Google Scholar]
- Omecinsky, D.O., Holub, K.E., Adams, M.E., and Reily, M.D. 1996. Three-dimensional structure analysis of μ-agatoxins: Further evidence for common motifs among neurotoxins with diverse ion channel specificities. Biochemistry 35 2836–2844. [DOI] [PubMed] [Google Scholar]
- Osaki, T., Omotezako, M., Nagayama, R., Hirata, M., Iwanaga, S., Kasahara, J., Hattori, J., Ito, I., Sugiyama, H., and Kawabata, S. 1999. Horseshoe crab hemocyte-derived antimicrobial polypeptides, tachystatins, with sequence similarity to spider neurotoxins. J. Biol. Chem. 274 26172–26178. [DOI] [PubMed] [Google Scholar]
- Oswald, R.E., Suchyna, T.M., McFeeters, R., Gottlieb, P., and Sachs, F. 2002. Solution structure of peptide toxins that block mechanosensitive ion channels. J. Biol. Chem. 277 34443–34450. [DOI] [PubMed] [Google Scholar]
- Pallaghy, P.K., Nielsen, K.J., Craik, D.J., and Norton, R.S. 1994. A common structural motif incorporating a cystine knot and a triple-stranded β-sheet in toxic and inhibitory polypeptides. Protein Sci. 3 1833–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.S., Hausdorff, S.F., and Miller, C. 1991. Design, synthesis, and functional expression of a gene for charybdotoxin, a peptide blocker of K+ channels. Proc. Natl. Acad. Sci. 88 2046–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, K., Chen, X.D., and Liang, S.P. 2001. The effect of Huwentoxin-I on Ca(2+) channels in differentiated NG108–15 cells, a patch-clamp study. Toxicon 39 491–498. [DOI] [PubMed] [Google Scholar]
- Pennington, M.W., Festin, S.M., Maccecchini, M.L., and Kem, W.R. 1992. Synthesis and characterization of a disulfide bond isomer of ω-conotoxin GVIA. Toxicon 30 755–764. [DOI] [PubMed] [Google Scholar]
- Pennington, M.W., Mahnir, V.M., Khaytin, I., Zaydenberg, I., Byrnes, M.E., and Kem, W.R. 1996a. An essential binding surface for ShK toxin interaction with rat brain potassium channels. Biochemistry 35 16407–16411. [DOI] [PubMed] [Google Scholar]
- Pennington, M.W., Mahnir, V.M., Krafte, D.S., Zaydenberg, I., Byrnes, M.E., Khaytin, I., Crowley, K., and Kem, W.R. 1996b. Identification of three separate binding sites on SHK toxin, a potent inhibitor of voltage-dependent potassium channels in human T-lymphocytes and rat brain. Biochem. Biophys. Res. Commun. 219 696–701. [DOI] [PubMed] [Google Scholar]
- Possani, L.D., Merino, E., Corona, M., Bolivar, F., and Becerril, B. 2000. Peptides and genes coding for scorpion toxins that affect ion-channels. Biochimie 82 861–868. [DOI] [PubMed] [Google Scholar]
- Reeh, P.W. and Steen, K.H. 1996. Tissue acidosis in nociception and pain. Prog. Brain Res. 113 143–151. [DOI] [PubMed] [Google Scholar]
- Roussel, A. and Cambillau, C. 1989. Silicon graphics geometry partner directory, pp. 77–78. Silicon Graphics, Mountain View, CA.
- Sasaki, T., Feng, Z.P., Scott, R., Grigoriev, N., Syed, N.I., Fainzilber, M., and Sato, K. 1999. Synthesis, bioactivity, and cloning of the L-type calcium channel blocker ω-conotoxin TxVII. Biochemistry 38 12876–12884. [DOI] [PubMed] [Google Scholar]
- Savarin, P., Guenneugues, M., Gilquin, B., Lamthanh, H., Gasparini, S., Zinn-Justin, S., and Ménez, A. 1998. Three-dimensional structure of κ-conotoxin PVIIA, a novel potassium channel-blocking toxin from cone snails. Biochemistry 37 5407–5416. [DOI] [PubMed] [Google Scholar]
- Srinivasan, K.N., Sivaraja, V., Huys, I., Sasaki, T., Cheng, B., Kumar, T.K., Sato, K., Tytgat, J., Yu, C., Brian Chia, C.S., et al. 2002. κ-Hefutoxin1, a novel toxin from the scorpion heterometrus fulvipes with unique structure and function: Importance of the functional diad in potassium channel selectivity. J. Biol. Chem. 277 30040–30047. [DOI] [PubMed] [Google Scholar]
- Swartz, K.J. and MacKinnon, R. 1995. An inhibitor of the Kv2.1 potassium channel isolated from the venom of a Chilean tarantula. Neuron 15 941–949. [DOI] [PubMed] [Google Scholar]
- Szyperski, T., Güntert, P., Otting, G., and Wüthrich, K. 1992. Determination of scalar coupling constants by inverse Fourier transformation of in-phase multiplets. J. Magn. Reson. 99 552–560. [Google Scholar]
- Takahashi, H., Kim, J.I., Min, H.J., Sato, K., Swartz, K.J., and Shimada, I. 2000. Solution structure of hanatoxin1, a gating modifier of voltage-dependent K+ channels: Common surface features of gating modifier toxins. J. Mol. Biol. 297 771–780. [DOI] [PubMed] [Google Scholar]
- Waldmann, R. and Lazdunski, M. 1998. H+-gated cation channels: Neuronal acid sensors in the NaC/DEG family of ion channels. Curr. Opin. Neurobiol. 8 418–424. [DOI] [PubMed] [Google Scholar]
- Waldmann, R., Bassilana, F., de Weille, J., Champigny, G., Heurteaux, C., and Lazdunski, M. 1997a. Molecular cloning of a noninactivating proton-gated Na+ channel specific for sensory neurons. J. Biol. Chem. 272 20975–20978. [DOI] [PubMed] [Google Scholar]
- Waldmann, R., Champigny, G., Bassilana, F., Heurteaux, C., and Lazdunski, M. 1997b. A proton-gated cation channel involved in acid-sensing. Nature 386 173–177. [DOI] [PubMed] [Google Scholar]
- Wang, X., Smith, R., Fletcher, J.I., Wilson, H., Wood, C.J., Howden, M.E., and King, G.F. 1999. Structure-function studies of ω-atracotoxin, a potent antagonist of insect voltage-gated calcium channels. Eur. J. Biochem. 264 488–494. [DOI] [PubMed] [Google Scholar]
- Wu, J.J., He, L.L., Zhou, Z., and Chi, C.W. 2002. Gene expression, mutation, and structure-function relationship of scorpion toxin BmP05 active on SK(Ca) channels. Biochemistry 41 2844–2849. [DOI] [PubMed] [Google Scholar]
- Wüthrich, K. 1986. NMR of proteins and nucleic acids. Wiley, New York.
- Yan, L. and Adams, M.E. 1998. Lycotoxins, antimicrobial peptides from venom of the wolf spider Lycosa carolinensis.J. Biol. Chem. 273 2059–2066. [DOI] [PubMed] [Google Scholar]