Abstract
The effects of deimination (conversion of arginyl to citrullinyl residues) of myelin basic protein (MBP) on its binding to calmodulin (CaM) have been examined. Four species of MBP were investigated: unmodified recombinant murine MBP (rmMBP-Cit0), an engineered protein with six quasi-citrullinyl (i.e., glutaminyl) residues per molecule (rmMBP-qCit6), human component C1 (hMBP-Cit0), and human component C8 (hMBP-Cit6), both obtained from a patient with multiple sclerosis (MS). Both rmMBP-Cit0 and hMBP-Cit0 bound CaM in a Ca2+-dependent manner and primarily in a 1:1 stoichiometry, which was verified by dynamic light scattering. Circular dichroic spectroscopy was unable to detect any changes in secondary structure in MBP upon CaM-binding. Inherent Trp fluorescence spectroscopy and a single-site binding model were used to determine the dissociation constants: Kd = 144 ± 76 nM for rmMBP-Cit0, and Kd = 42 ± 15 nM for hMBP-Cit0. For rmMBP-qCit6 and hMBP-Cit6, the changes in fluorescence were suggestive of a two-site interaction, although the dissociation constants could not be accurately determined. These results can be explained by a local conformational change induced in MBP by deimination, exposing a second binding site with a weaker association with CaM, or by the existence of several conformers of deiminated MBP. Titration with the collisional quencher acrylamide, and steady-state and lifetime measurements of the fluorescence at 340 nm, showed both dynamic and static components to the quenching, and differences between the unmodified and deiminated proteins that were also consistent with a local conformational change due to deimination.
Keywords: Myelin basic protein, calmodulin, multiple sclerosis, deimination, citrulline, intrinsic Trp fluorescence, fluorescence lifetime, dynamic light scattering, circular dichroism
Myelin basic protein (MBP) is one of the most abundant proteins of the myelin sheath of the central nervous system (CNS), and its primary role is generally considered to be maintenance of the stability of the sheath by holding together the apposing cytoplasmic leaflets of the oligodendrocyte membrane (Smith 1992; Moscarello 1997). However, the MBP family comprises numerous developmentally regulated isoforms, of which the 18.5-kD species is the most abundant in adult human myelin, and has been the most studied (Givogri et al. 2000, 2001). This isoform itself undergoes a complex series of posttranslational modifications, giving rise to charge isomers designated as components C1 to C8 (Moscarello 1997; Wood and Moscarello 1997; Zand et al. 1998). In addition to its associations with lipids, MBP has been shown to interact with calmodulin (CaM) and cytoskeletal proteins such as actin and tubulin (Chan et al. 1990; Boggs and Rangaraj 2000). Thus, functional roles for MBP in phosphoinositide-mediated signal transduction and other processes have been postulated (Staugaitis et al. 1996; Dyer et al. 1997; Harauz et al. 2000; Lintner and Dyer 2000; Arvanitis et al. 2002).
One important posttranslational modification of MBP that correlates with the severity of the autoimmune disease multiple sclerosis (MS) is deimination, the enzymatic conversion of arginine to citrulline by peptidylarginine deiminase (EC 3.5.3.15; Finch et al. 1971; Moscarello et al. 1994; Whitaker and Mitchell 1996; Wood et al. 1996). Deimination reduces the net positive charge of the protein, yielding the C8 component, and limits its ability to maintain a compact myelin sheath by disrupting both its tertiary structure and its interactions with lipids (Lamensa and Moscarello 1993; Boggs et al. 1999; Cao et al. 1999; Beniac et al. 2000; Pritzker et al. 2000a,b; Ishiyama et al. 2001). This posttranslational modification is potentially involved in antigen recognition and autoimmunity (Doyle and Mamula 2001, 2002). Deimination of MBP elicits another biological response in that the citrulline-containing C8 charge isomer does not stimulate phospholipase Cα activity like the least-modified C1 component does (Tompkins and Moscarello 1991, 1993).
We have begun to investigate, by fluorescence microscopy and spectroscopy and by gel shift assays, the interactions of MBP with Ca2+-CaM (Libich and Harauz 2002a,b). We have reported the apparent dissociation constants of the putative 1:1 interaction to be 2.1 ± 0.1 μM and 2.0 ± 0.2 μM for the natural bovine C1 charge isomer (bMBP/C1, equivalently bMBP-Cit0) and a recombinant murine product (rmMBP, equivalently rmMBP-Cit0), respectively. We also showed that the C-terminal domain of MBP interacted with Ca2+-CaM, consistent with a theoretical prediction (Rhoads and Friedberg 1997; Harauz et al. 2000; Yap et al. 2000). For these interactions, the apparent dissociation constants were 1.78 ± 0.17 μM and 2.81 ± 0.91 μM for the bMBP-Cit0 and rmMBP-Cit0 C-terminal fragments, respectively. Although the physiological significance of this interaction is unknown, investigating 18.5-kD MBP:CaM interactions is the logical place to begin to elucidate the potential roles of MBP in signal transduction pathways (Dyer et al. 1997; Boggs and Rangaraj 2000; Lintner and Dyer 2000). Moreover, because MBP is an "intrinsically unstructured" or "natively unfolded" protein (Hill et al. 2002), its three-dimensional structure might only be determined in its heterocomplex with another protein. For this reason also, the MBP:CaM interaction requires precise characterization.
In the present work, we have extended our previous studies primarily to test the hypothesis that deimination affects the interaction of MBP with Ca2+-CaM, to define more rigorously the stoichiometry and strength of the interaction, and to assess if there are any structural changes induced in MBP by CaM-binding. We used four different MBP preparations: rmMBP-Cit0 (the unmodified Leu-Glu-His6-tagged recombinant protein with no citrullines; Bates et al. 2000), rmMBP-qCit6 (a mutant protein with six specific Arg/Lys→Gln conversions to mimic the effects of deimination; Bates et al. 2002), hMBP-Cit0 (the least-modified C1 charge isomer of human MBP obtained from autopsy material of an MS patient), and hMBP-Cit6 (the deiminated C8 charge isomer obtained from the same patient, with an average of six citrullinyl residues per molecule of MBP; Moscarello 1997; Wood and Moscarello 1997). For control and dynamic light scattering (DLS) experiments, we have also used the bovine protein, bMBP/C1 (or bMBP-Cit0). For reference, the aligned and gapped sequences of these proteins are shown in Figure 1▶.
Figure 1.
Aligned sequences of the 18.5-kD isoforms of human MBP (hMBP, 170 residues), bovine MBP (bMBP, 169 residues), murine MBP (mMBP, 168 residues), unmodified recombinant mMBP (rmMBP, 176 residues), and the recombinant mutant (rmMBP-qCit6, 176 residues). The LEH6 tag that distinguishes the recombinant murine protein is given in lowercase letters. The number at the beginning or end of each row refers to the first or last residue, respectively, for that particular sequence. Gaps are labeled by the symbol ". .". All arginyl residues are in bold type; six of these are labeled with an asterisk in hMBP (human sequence numbering: Arg25, Arg33, Arg122, Arg130, Arg159, and Arg170). These particular arginyl residues are most commonly deiminated, giving rise to the C8 isoform (hMBP-Cit6) that predominates in patients with chronic multiple sclerosis. The Arg122 in hMBP is replaced conservatively by Lys121 and Lys119 in the bovine and murine proteins, respectively. The arginyl residues labeled with an "m" (human Arg107, bovine Arg106, murine Arg104) are either unmethylated, monomethylated, or symmetrically dimethylated in the natural form. The segment rmMBP(132–167) was predicted to be a Ca2+-CaM–binding site. The C-terminal domain of MBP also is comparable to the KR-rich (lysine- and arginine-rich) phosphoinositide-binding regions of proteins such as MARCKS, vinculin, and ActA (Ishiyama et al. 2001).
Results and Discussion
Interactions of deiminated and quasi-deiminated MBP with Ca2+-CaM via Trp fluorescence emission spectroscopy
We have measured the changes in the intrinsic fluorescence emission spectra of the single Trp residue in each MBP species (Fig. 1▶), on titrating with CaM in the presence of 1 mM Ca2+. (There is no Trp in CaM.) For all four MBP samples investigated (hMBP-Cit0, hMBP-Cit6, rmMBP-Cit0, and rmMBP-qCit6), increasing concentrations of CaM resulted in spectra exhibiting a blue shift and an increase in maximum fluorescence intensity (Fig. 2▶), consistent with our previous results for bMBP-Cit0 and rmMBP-Cit0 (Libich and Harauz 2002a). These results are also fully consistent with those obtained for other CaM-binding proteins and peptides (Zhan et al. 1991; Yuan et al. 1995; Gomes et al. 2000; Murase and Iio 2002; Noonan et al. 2002). There was no significant difference in the steady-state Trp fluorescence spectra of hMBP-Cit0 versus hMBP-Cit6 or of rmMBP-Cit0 versus rmMBP-qCit6 (data not shown). Previously, the steady-state spectrum of 18.5-kD MBP has been observed to change as ionic strength or glycerol or urea concentrations were increased (Jones and Rumsby 1975; Vadas et al. 1981; Nowak and Berman 1991). Here, deimination did not have such a noticeable effect.
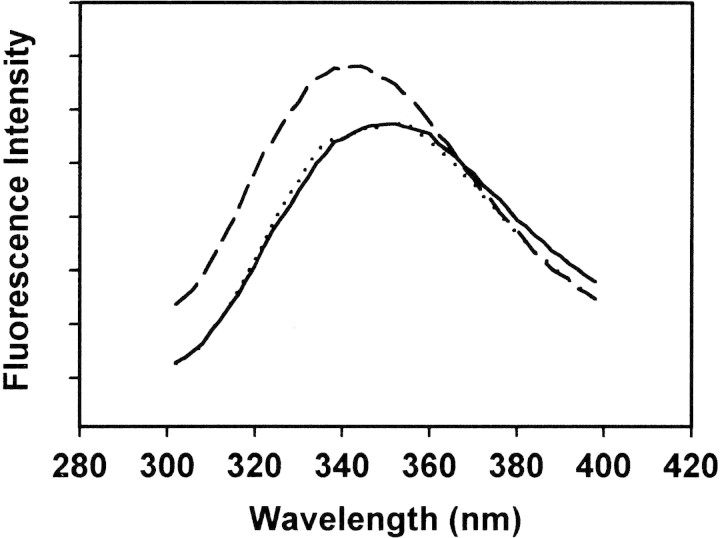
Figure 2.
Representative intrinsic Trp fluorescence emission spectra for hMBP-Cit6 at 2 μM (40 μg/mL) concentration. All spectra have been averaged (three replicates) and corrected for dilution only. The buffer comprised 50 mM TRIS-HCl (pH 7.4), 250 mM NaCl, and 1 mM CaCl2 (or 8 mM EDTA and 2 mM EGTA instead of 1 mM CaCl2). The dotted curve is the emission spectrum of hMBP-Cit6 alone in either the presence or absence of Ca2+. The solid curve is the emission spectrum of hMBP-Cit6 in the presence of an equimolar amount of CaM (which contains no Trp) but in the absence of Ca2+. The dashed curve is the emission spectrum of hMBP-Cit6 in the presence of both Ca2+ and an equimolar amount of CaM; the spectrum is shifted toward smaller wavelengths and the maximum fluorescence intensity increases.
Control experiments were performed in the absence of Ca2+ (i.e., in the presence of 8 mM EDTA and 2 mM EGTA). Upon titration with CaM, there was no change in the fluorescence spectra of any of the four MBP species examined, confirming the calcium-dependence of their interaction with CaM (Fig. 2▶). Moreover, the salt conditions of our experiments were designed to minimize electrostatic attraction between MBP and acidic CaM (net charges at neutral pH: 19 for hMBP-Cit0, 13 for hMBP-Cit6, 19 for rmMBP-Cit0, 13 for rmMBP-qCit6, and −16 for [Ca2+]4-CaM). Clearly, nonspecific (purely electrostatic) MBP:CaM binding was not observed here. Although apocalmodulin does bind target peptides in certain cases (Hill et al. 2000), the Ca2+-dependence observed here lends credence to the specificity and hence potential physiological significance of the interaction. In the presence of Ca2+, there was no increase in the absorbance at 450 nm of any MBP solution upon titration with CaM, indicating that the protein was probably not merely aggregating. We address this issue again below with DLS.
Determination of dissociation constants
After correction for dilution, etc., using Equation 1 (see Materials and Methods), the changes in fluorescence intensity at 340 nm, for minimally modified hMBP-Cit0 and unmodified rmMBP-Cit0 (Fig. 3A,B▶, respectively), were regressed against Equation 4 to yield dissociation constants of Kd = 42 ± 15 nM for hMBP-Cit0 (r2 = 0.999), and Kd = 144 ± 76 nM for rmMBP-Cit0 (r2 = 0.995). These values are lower than those previously determined (at 330 nm) for bMBP-Cit0 and rmMBP-Cit0, respectively (Libich and Harauz 2002a). The main difference is that we have now corrected for fractions of unbound proteins (Heyduk and Lee 1990; Winzor and Sawyer 1995; Ninfa and Ballou 1998). Thus, the interaction between MBP and CaM is stronger than indicated by the previously reported apparent dissociation constants, and is consistent with other qualitative observations that CaM binds MBP even in 8-M urea (Chan et al. 1990) or dissociates MBP from actin and phospholipid vesicles (Boggs and Rangaraj 2000). The affinity of the interaction suggests that the MBP:CaM reaction might indeed be biologically significant (cf., Gell et al. 2002), and that future structural studies of the heterocomplex by solution NMR spectroscopy are feasible, for which an upper Kd limit of 10 μM has been suggested (Zuiderweg 2002).
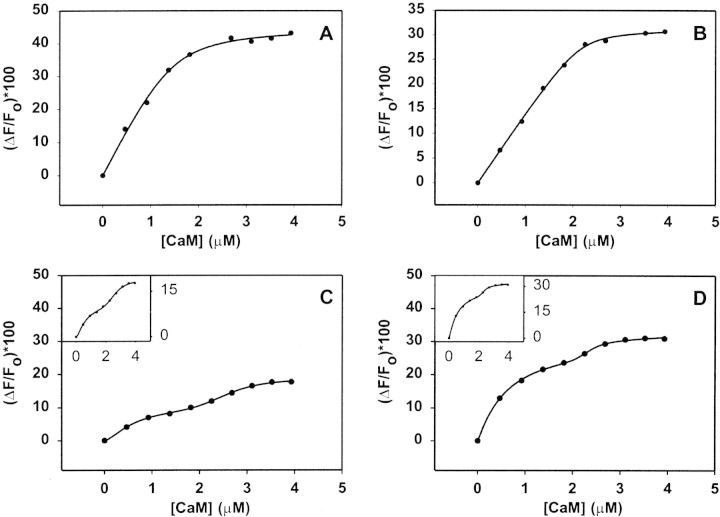
Figure 3.
Intrinsic Trp fluorescence emission (at 340 nm) spectroscopy of hMBP-Cit0 (A), rmMBP-Cit0 (B), hMBP-Cit6 (C), and rmMBP-qCit6 (D). Each sample was titrated with increasing molar ratios of CaM (which contains no Trp) in buffer comprising 50 mM TRIS-HCl (pH 7.4), 250 mM NaCl, and 1 mM CaCl2. The MBP concentration was 2 μM (40 μg/mL). All panels show the percentage change in fluorescence intensity at 340 nm as a function of CaM concentration (μM). All data have been corrected for dilution and the inner filter effect. The insets in panels (C) and (D) present the curves expanded vertically to accentuate their biphasic nature. The fitted curves in panels (A) and (B) represent Equation 4 in the text. The fitted curves in panels (C) and (D) represent a weighted sum of two Hill equations (Weiss 1997; Döppenschmitt et al. 1999). All measurements represent an average of three experiments.
The titration data for deiminated hMBP-Cit6 and quasi-deiminated rmMBP-qCit6 (Fig. 3C,D▶, respectively) could not be adequately described by Equations 3 and 4. The curves in Figure 3▶, C and D, were clearly biphasic; for illustrative purposes, the lines connecting the points represent a fit of a weighted sum of two Hill equations, which have been used by others to model the binding of multiple ligands to a protein (Weiss 1997; Döppenschmitt et al. 1999; Romsicki and Sharom 1999). Although the summed Hill equations are not the best way to describe the interaction between two proteins of comparable size (like MBP and CaM), the phenomenon that could be modeled here is the potential formation of a heterotrimeric complex of MBP1:CaM2 in addition to the heterodimer MBP1:CaM1. This situation would then require the determination of two dissociation constants via modeling and analysis extended from Equations 2 to 4. Unfortunately, it was not possible to develop a closed-form analytical solution because of the inability to separate variables. An attempt to treat the two portions of the curves separately also did not yield good fits. We discuss the differences in CaM-binding between unmodified and deiminated forms of MBP in more detail below.
Stoichiometry of interaction of MBP with Ca2+-CaM by DLS
The analysis of the fluorescence data presupposed a 1:1 interaction of MBP:CaM, which has been reported in previous work (Chan et al. 1990; Libich and Harauz 2002a,b). Here, the titration curves for hMBP-Cit0 and rmMBP-Cit0 (Fig. 3A,B▶, respectively) both started to level off at (CaM)T/(MBP)T ratios of 1.04 ± 0.04 (using corrected [MBP]T values obtained from the regression analysis above), confirming the supposition of primary 1:1 binding stoichiometry (Schleiff et al. 1996).
There are direct methods of determining the precise stoichiometry of binding of a (usually small) ligand to a protein: fluorescence spectroscopy and "stoichiometric titration", as well as equilibrium dialysis (Winzor and Sawyer 1995; Ninfa and Ballou 1998). However, these approaches are inapplicable here, because of the inability to work over large (orders of magnitude) ranges of protein concentration, and because MBP and CaM have similar molecular masses, ∼18.5 kD and 16.8 kD, respectively. Recently, DLS has been used to show changes in CaM shape upon binding of defined target peptides (Papish et al. 2002). Here, we have used DLS to address the question of aggregation and to define, under physiological solution conditions, the populations of complexes present when MBP and CaM interacted.
Initial DLS experiments were performed by using the bovine 18.5-kD isoform bMBP-Cit0 in the same buffer used for the fluorescence studies. The intensity-weighted fits showed the presence of some aggregates (Fig. 4A▶), which were shown via a volume-weighted fit to constitute a vanishing proportion of the particle population (Fig. 4B▶). Thus, there was minimal self-aggregation of MBP under our experimental conditions, and changes in fluorescence intensity could not be due to this phenomenon. Moreover, these data demonstrated a unimodal population of MBP of the expected size, consistent with our previous DLS data used during crystallization experiments (Hill et al. 2002). However, an inspection of the correlation function obtained under these conditions showed that the signal-to-noise ratio was poor (Fig. 4C▶). Thus, it would have been exceedingly difficult to use DLS to discern small changes in particle diameter under these conditions.
Figure 4.
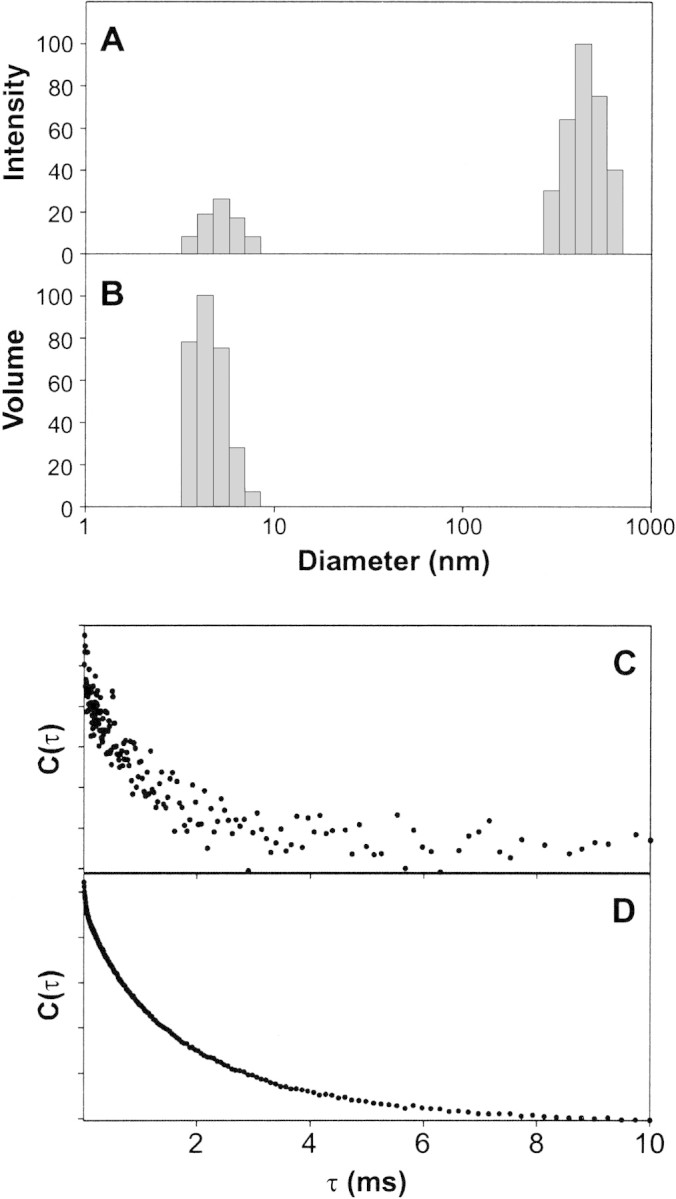
Dynamic light scattering of MBP at different pH values. Intensity-weighted (A) and volume-weighted (B) size distributions of 0.1 mg/mL bMBP-Cit0 alone in 20 mM HEPES-NaOH (pH 7.4), 200 mM NaCl, and 1 mM CaCl2. (C, D) Correlation functions for bMBP-Cit0 at pH 7.4 in the HEPES buffer (C) and at pH 6.0 in 20 mM MES-NaOH, 200 mM NaCl, and 1 mM CaCl2 (D).
Because previous studies (Liebes et al. 1975; Moskaitis and Campagnoni 1986) have indicated that MBP self-associates at pH >7.0 primarily by its histidyl residues, subsequent DLS experiments were performed at pH 6.0. A dramatic improvement in signal-to-noise ratio was obtained (Fig. 4D▶). Several key fluorescence experiments were repeated for bMBP-Cit0 at pH values of 5.4, 6.3, 7.0, and 8.3, and essentially identical results were obtained as in Figures 2 and 3▶▶, A and B, at pH 7.4 (data not shown). The degree of self-association at pH 7.4 is still quite small, based on present (Fig. 4B▶) and previous studies (Liebes et al. 1975; Moskaitis and Campagnoni 1986; Libich and Harauz 2002b), and is thus considered to have a negligible effect on the fluorescence data. The results presented here at pH 7.4 are also considered to be more physiologically relevant.
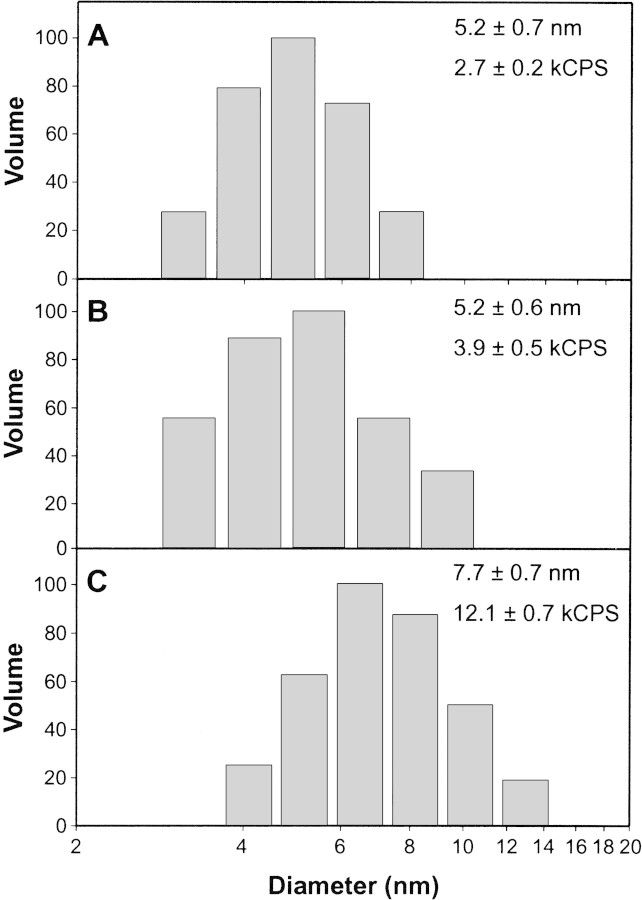
The results of subsequent DLS experiments performed at pH 6.0 are shown in Figure 5▶. Both bMBP-Cit0 and CaM alone showed unimodal particle size distributions (Fig. 5A,B▶, respectively). When the two proteins were mixed in equimolar ratios, a unimodal distribution was again obtained, showing 1:1 heterocomplex formation (Fig. 5C▶). The mean diameter of the heterodimer (7.7 ± 0.7 nm) was greater than either that of bMBP-Cit0 alone (5.2 ± 0.7 nm) or CaM alone (5.2 ± 0.6 nm), with statistical significance ascertained by using Student t tests. Thus, the DLS data support the conclusion of formation of a 1:1 MBP:CaM heterodimer and support the conclusions obtained below in quenching experiments.
Figure 5.
Dynamic light scattering of MBP:Ca2+-CaM complexes. Representative volume-weighted size distributions of 0.1 mg/mL bMBP-Cit0 alone (A), 0.1 mg/mL Ca2+-CaM alone (B), and bMBP-Cit0 and Ca2+-CaM mixed in equimolar ratios (C). Buffer conditions were 20 mM MES-NaOH (pH 6.0), 200 mM NaCl, and 1 mM CaCl2. Mean particle diameters (taken to be the center of each distribution) and count rates (kCPS indicates thousands of counts per second) are indicated in each panel.
Circular dichroic spectroscopy
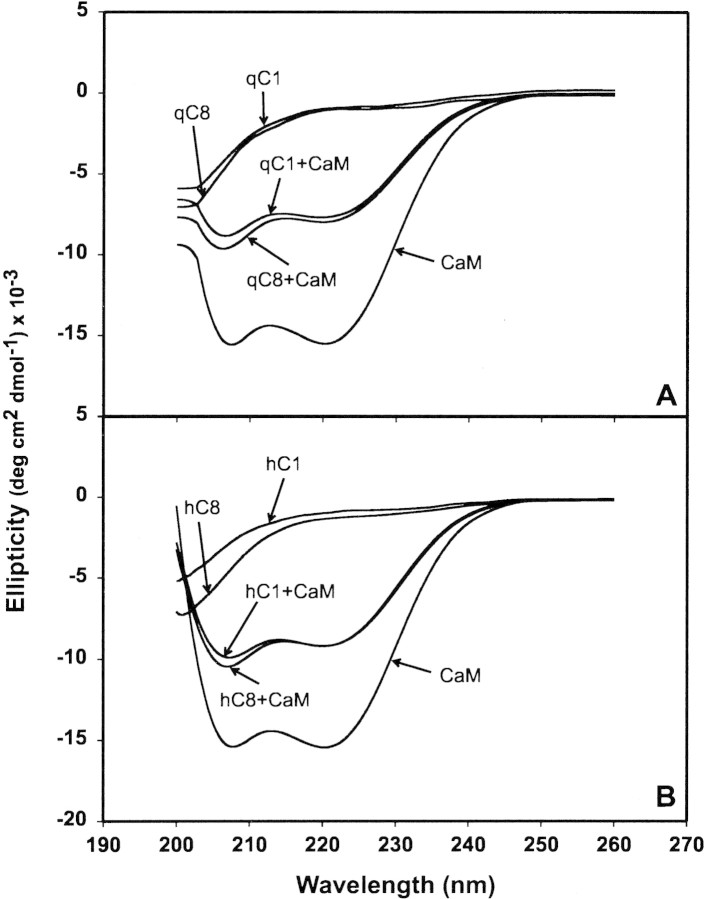
To ascertain if there were major changes in secondary structure composition of MBP upon CaM-binding, we used circular dichroic (CD) spectroscopy. This approach has previously been used in studies of the interactions of other proteins and peptides with CaM (Gomes et al. 2000; Yuan et al. 2001). Our results (Fig. 6▶) showed that all four species of MBP were primarily disordered in aqueous solution, and that CaM had a large α-helical component, all as expected. The deiminated species had a greater proportion of random component than did the unmodified proteins, as evidenced by the increased magnitude of negative ellipticity at low wavelengths and as reported previously (Bates et al. 2002). The far-UV CD spectrum of the complex between peptides and CaM has been used in other instances to estimate the effect of an increase in the α-helical content of the target peptide (Yuan et al. 2001). Because Ca2+-CaM does not gain or lose a significant amount of secondary structure when it binds to a target region, any changes in the CD spectrum can be attributed to the peptide alone (Ikura et al. 1992). Although MBP comprises mainly random coil in aqueous solution, it adopts a larger proportion of α-helix in the presence of detergent, lipid, or organic solvents (Anthony and Moscarello 1971a,b; Keniry and Smith 1979; Polverini et al. 1999; Bates et al. 2000), showing that segments of the protein have the propensity for formation of this secondary structure motif.
Figure 6.
CD spectroscopy of MBP:Ca2+-CaM complexes. (A) rmMBP-Cit0 alone (labelled qC1, for brevity), rmMBP-qCit6 alone (denoted qC8). (B) hMBP-Cit0 alone (denoted hC1) and hMBP-Cit6 alone (denoted hC8). (A, B) CaM alone, and all four MBP:Ca2+-CaM complexes.
Here, the MBP:Ca2+-CaM complexes appeared to have a high proportion of α-helix as indicated by the shape of their CD spectrum; however, the effect appeared to be simply additive. When the individual rmMBP-Cit0 and rmMBP-qCit6 spectra were added to the CaM spectrum and corrected for the change in concentration, the resulting spectrum was nearly identical to those of the rmMBP-Cit0:Ca2+-CaM and rmMBP-qCit6:Ca2+-CaM complexes (data not shown). This result indicated that there was no great change in secondary structure in MBP upon CaM binding. (Similarly, it has been suggested that MARCKS remains disordered upon binding CaM [Porumb et al. 1997].) However, this observation did not preclude large tertiary structure changes such as domain movement that might be involved in cooperativity of association, nor did it preclude local conformational changes.
Accessibility of Trp residue via acrylamide quenching: Steady-state measurements
Deimination of MBP has been shown by numerous means to result in a more open structure (Lamensa and Moscarello 1993; Cao et al. 1999; Beniac et al. 2000; Pritzker et al. 2000a,b). Here, titration of all MBP preparations with the collisional quencher acrylamide was used to probe the environment of the Trp residue in the control (minimally modified hMBP-Cit0 and unmodified rmMBP-Cit0) and deiminated (hMBP-Cit6 and rmMBP-qCit6) proteins. The results indicated both dynamic and static components to the quenching mechanism, because of the deviation from linearity (Fig. 7A▶; Nowak and Berman 1991; Lakowicz 1999). Regression analyses yielded values of the coefficients for Equation 5 that are summarized in Table 1▶, and that compare favorably with our previous data on NATA, bMBP-Cit0, and rmMBP-Cit0 (Libich and Harauz 2002a). The data indicated that this residue was fairly exposed to the aqueous milieu, but the differences in steady-state quenching constants among the various MBP forms were not appreciable.
Figure 7.
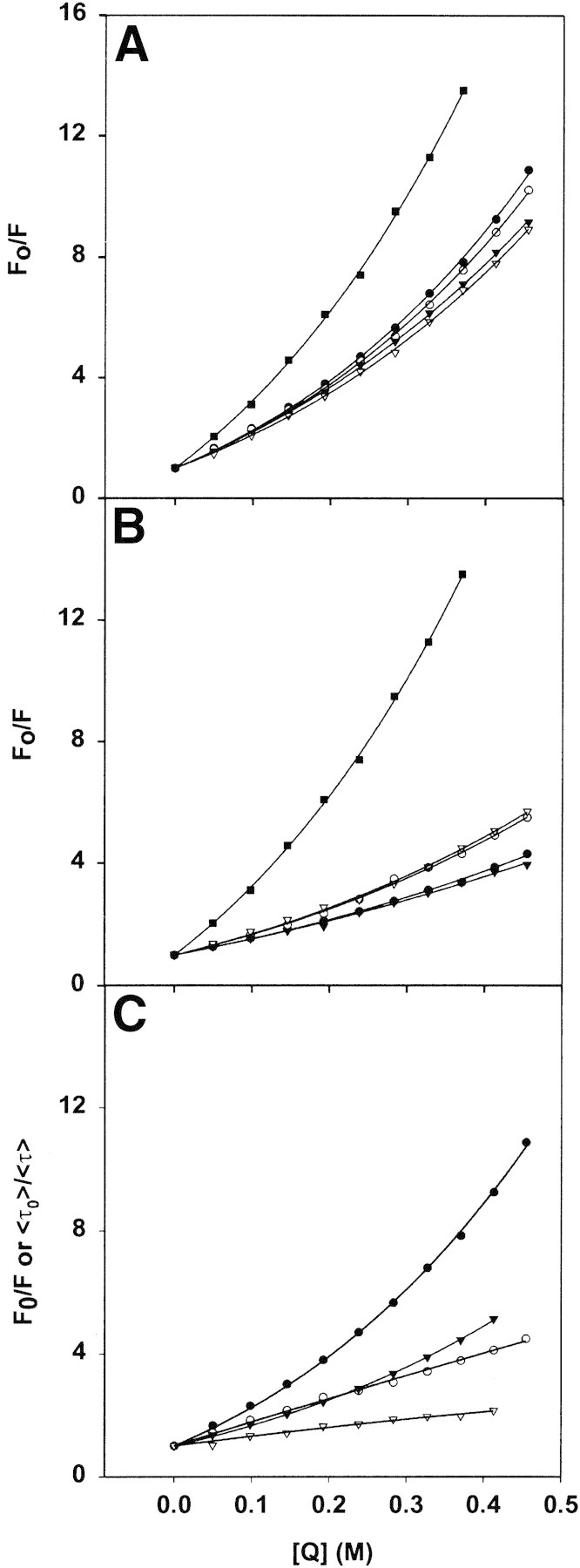
(A, B) Steady-state measurements of acrylamide quenching by MBP and MBP:Ca2+-CaM complexes. (A) Stern-Volmer plots for quenching of the intrinsic Trp fluorescence emission at 340 nm of MBP (2 μM, 40 μg/mL), in buffer comprising 50 mM TRIS-HCl (pH 7.4), 250 mM NaCl, and 1 mM CaCl2. The soluble Trp analog NATA was used as a reference. All measurements represent an average of three experiments. (B) The same experiment as in A obtained with CaM also present at an equimolar concentration. The symbols are the same in both panels A and B: NATA (filled squares), rmMBP-Cit0 (filled circles), rmMBP-qCit6 (open circles), hMBP-Cit0 (filled triangles), and hMBP-Cit6 (open triangles). The values of the Stern-Volmer coefficients obtained by regression analysis (Equation 5 in text) are given in Table 1▶. (C) Comparison of steady-state (filled symbols) and lifetime (open symbols) data for rmMBP-Cit0 in the absence (circles) and presence (triangles) of CaM at an equimolar concentration. The values of all Stern-Volmer coefficients obtained by regression analysis (Equation 7 in text) of fluorescence lifetime measurements are given in Table 3▶. In all panels, the concentration of acrylamide quencher is given in molar values.
Table 1.
Stern-Volmer constants obtained for dynamic and static modes of quenching by acrylamide via steady-state measurements
| Without CaM |
With CaM |
|||
|---|---|---|---|---|
| Sample | KSV (M−1) | V (M−1) | KSV (M−1) | V (M−1) |
| NATA | 17.9 ± 0.6 | 1.5 ± 0.1 | — | — |
| hMBP-Cit0 | 9.60 ± 0.20 | 1.19 ± 0.04 | 4.73 ± 0.21 | 0.53 ± 0.08 |
| hMBP-Cit6 | 8.44 ± 0.27 | 1.35 ± 0.06 | 4.81 ± 0.39 | 1.28 ± 0.14 |
| rmMBP-Cit0 | 9.31 ± 0.33 | 1.58 ± 0.07 | 4.14 ± 0.12 | 0.88 ± 0.05 |
| rmMBP-qCit6 | 9.11 ± 0.26 | 1.49 ± 0.06 | 4.54 ± 0.18 | 1.30 ± 0.06 |
All experiments were performed in 50 mM TRIS-HCl (pH 7.4) 250 mM NaCl, and 1 mM CaCl2. The MBP and CaM concentrations were 2 μM. The data represent averages of three experiments. The regression coefficients for fits to Equation 5 in the text were all r2 > 0.99.
When CaM was present in equimolar amounts to each MBP species, the quenching data again were nonlinear, but this time, the deiminated proteins behaved differently from the Cit0 ones (Fig. 7B▶). The values of KSV (representing dynamic quenching) for all four MBP preparations were comparable and were roughly half of those obtained in the absence of Ca2+-CaM (Table 1▶). This decrease indicated that the Trp residue was less exposed in the MBP:Ca2+-CaM complex than in MBP alone, an interpretation consistent with other acrylamide quenching experiments involving single-Trp–containing CaM-binding peptides (Gomes et al. 2000). The values of V (essentially representing a correction factor for static quenching) decreased only for the Ca2+-CaM complexes of hMBP-Cit0 and rmMBP-Cit0, but not of hMBP-Cit6 and rmMBP-qCit6. Thus, there was an additional quenching process in the complexes with Ca2+-CaM of the modified proteins, consistent with either an altered MBP conformation or a different mode of binding Ca2+-CaM.
Fluorescence lifetime measurements of MBP and of MBP:Ca2+-CaM complexes
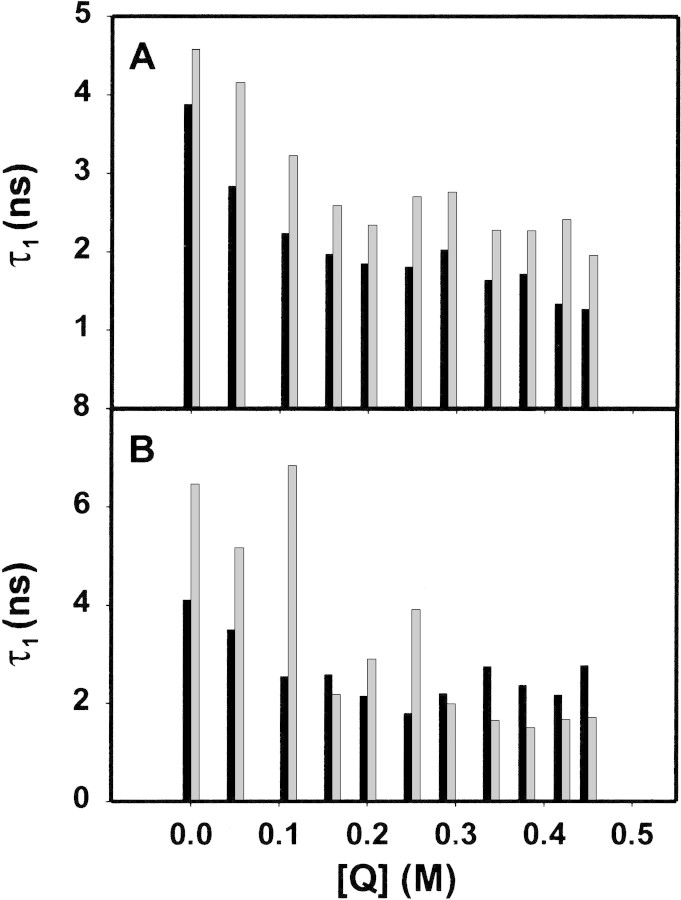
Further insight into the accessibility of the sole Trp residue, and its altered environment due to deimination, was obtained via fluorescence lifetime measurements (Calhoun et al. 1986). The lifetime measurements revealed a heterogeneous behavior of all four proteins, and decays were fit by double- or triple-exponential functions (Table 2▶, Fig. 8▶). For hMBP-Cit6, fits with single- or double-exponential functions yielded larger residuals (Fig. 8D,E▶) than did fits with triple-exponential functions (Fig. 8C▶). The nature of the multiexponential behavior is not obvious, but complex decays are rather common for Trp fluorescence in proteins, even for those with a single Trp residue (Beechem and Brand 1985; Chen and Barkley 1998; Clayton and Sawyer 1999). Previously, the multiple decays of MBP have been interpreted as representing different conformers (Cavatorta et al. 1994). Here, individual decay times were generally greater for the deiminated proteins than for the unmodified ones (Table 2▶, Fig. 9▶). This phenomenon can be due to deimination changing the conformation of MBP, especially that of the microenvironment of the Trp with a concomitant increase in structural microheterogeneity (cf., Kim et al. 1993; Kroes et al. 1998).
Table 2.
Weighted-average lifetimes (given in nanoseconds) and preexponential values of the four species of MBP in the presence of increasing concentrations of the bimolecular quencher (Q) acrylamide (given in molar values), but in the absence of CaM.
| rmMBP-Cit0 |
rmMBP-qCit6 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| [Q] | α1 | τ1 | α2 | τ2 | ≤τ> | α1 | τ1 | α2 | τ2 | ≤τ> |
| 0.000 | 0.429 | 3.878 | 0.571 | 1.360 | 2.440 | 0.239 | 4.587 | 0.239 | 1.564 | 3.075 |
| 0.050 | 0.364 | 2.840 | 0.636 | 0.960 | 1.645 | 0.125 | 4.153 | 0.393 | 1.568 | 2.192 |
| 0.110 | 0.348 | 2.242 | 0.652 | 0.748 | 1.274 | 0.141 | 3.224 | 0.448 | 0.902 | 1.456 |
| 0.160 | 0.307 | 1.971 | 0.693 | 0.610 | 1.028 | 0.143 | 2.586 | 0.504 | 0.591 | 1.031 |
| 0.200 | 0.223 | 1.847 | 0.777 | 0.618 | 0.892 | 0.126 | 2.339 | 0.579 | 0.454 | 0.791 |
| 0.250 | 0.180 | 1.813 | 0.820 | 0.625 | 0.838 | 0.070 | 2.698 | 0.524 | 0.626 | 0.870 |
| 0.290 | 0.098 | 2.026 | 0.902 | 0.670 | 0.802 | 0.067 | 2.757 | 0.506 | 0.580 | 0.835 |
| 0.340 | 0.132 | 1.641 | 0.868 | 0.534 | 0.681 | 0.079 | 2.271 | 0.658 | 0.351 | 0.557 |
| 0.380 | 0.094 | 1.718 | 0.906 | 0.503 | 0.617 | 0.063 | 2.266 | 0.489 | 0.493 | 0.694 |
| 0.420 | 0.143 | 1.340 | 0.857 | 0.351 | 0.493 | 0.047 | 2.416 | 0.445 | 0.576 | 0.752 |
| 0.450 | 0.134 | 1.273 | 0.866 | 0.335 | 0.461 | 0.056 | 1.956 | 0.567 | 0.370 | 0.513 |
| hMBP-Cit0 |
hMBP-Cit6 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| α1 | τ1 | α2 | τ2 | ≤τ> | α1 | τ1 | α2 | τ2 | α3 | τ3 | ≤τ> |
| The data were analyzed using a discrete one- to four-exponential fitting program. A double-exponential model was used for rmMBP-Cit0, rmMBP-qCit6, and hMBP-Cit0. A triple-exponential model was used for hMBP-Cit6. Average lifetimes ≤τ> were calculated by using Equation 6 in the text. All lifetimes are given in nanoseconds and, at [Q] = 0, are comparable to previous data obtained with unfractionated MBP (heterogeneous mixtures of all charge isomers) under different conditions (Nowak and Berman 1991; Cavatorta et al. 1994). | |||||||||||
| 0.246 | 4.102 | 0.389 | 1.383 | 2.437 | 0.078 | 6.460 | 0.402 | 2.724 | 0.519 | 0.427 | 1.824 |
| 0.139 | 3.504 | 0.430 | 1.212 | 1.771 | 0.040 | 5.157 | 0.444 | 2.358 | 0.516 | 0.525 | 1.526 |
| 0.149 | 2.553 | 0.418 | 0.880 | 1.319 | 0.008 | 6.839 | 0.362 | 2.151 | 0.630 | 0.541 | 1.174 |
| 0.090 | 2.588 | 0.405 | 0.858 | 1.172 | 0.062 | 2.178 | 0.140 | 0.775 | 0.834 | 0.040 | 0.249 |
| 0.095 | 2.146 | 0.385 | 0.685 | 0.975 | 0.045 | 2.898 | 0.315 | 1.256 | 0.640 | 0.305 | 0.722 |
| 0.097 | 1.801 | 0.389 | 0.536 | 0.788 | 0.012 | 3.911 | 0.255 | 1.514 | 0.733 | 0.391 | 0.720 |
| 0.064 | 2.197 | 0.519 | 0.610 | 0.785 | 0.080 | 1.978 | 0.238 | 0.950 | 0.683 | 0.311 | 0.595 |
| 0.029 | 2.744 | 0.531 | 0.596 | 0.709 | 0.091 | 1.654 | 0.539 | 0.381 | 0.370 | 0.015 | 0.361 |
| 0.048 | 2.369 | 0.601 | 0.599 | 0.730 | 0.148 | 1.501 | 0.502 | 0.315 | 0.350 | 0.308 | 0.488 |
| 0.056 | 2.167 | 0.703 | 0.446 | 0.573 | 0.077 | 1.668 | 0.189 | 0.667 | 0.733 | 0.246 | 0.435 |
| 0.023 | 2.769 | 0.567 | 0.557 | 0.643 | 0.060 | 1.706 | 0.028 | 0.693 | 0.912 | 0.245 | 0.345 |
Figure 8.
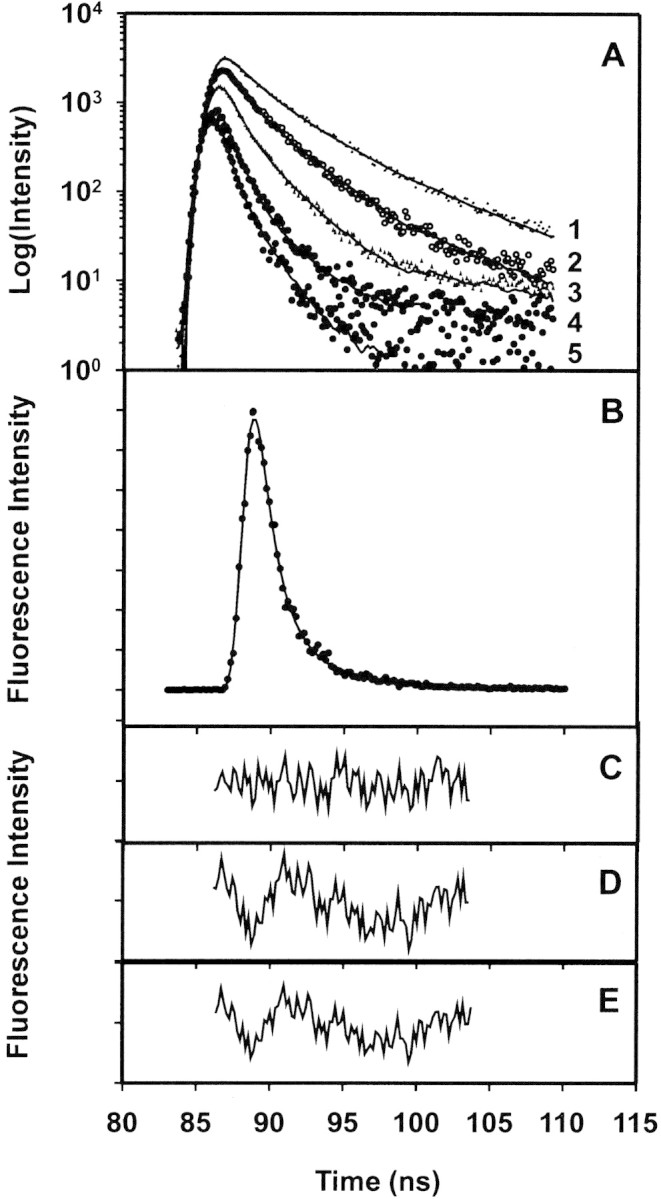
Fluorescence lifetime measurements of MBP. (A) Representative decay curve for hMBP-Cit6 in the presence of increasing amounts of acrylamide. Curve 1 indicates 0 M; curve 2, 0.05 M; curve 3, 0.15 M; curve 4, 0.30 M; and curve 5, 0.50 M acrylamide. The vertical scale is logarithmic. (B) Decay curve for hMBP-Cit6 in 0 M acrylamide, on a linear vertical scale, fitted with a triple-exponential function. (C) Residuals for fit with a triple-exponential function (χ2 = 0.89). (D) Residuals for fit with a double-exponential function (χ2 = 1.90). (E) Residuals for fit with a single-exponential function (χ2 = 2.00).
Figure 9.
Fluorescence lifetime measurements of MBP. The first lifetime components of each species of MBP as a function of the concentration of the bimolecular quencher (Q) acrylamide (Table 2▶). (A) Recombinant rmMBP-Cit0 (solid bars) and rmMBP-qCit6 (shaded bars). (B) Human hMBP-Cit0 (solid bars) and hMBP-qCit6 (shaded bars).
The weighted-average lifetime value (Equation 6) was used as a practical observable parameter to monitor the effect of acrylamide. The Stern-Volmer plots obtained from the weighted-average lifetime, at different acrylamide concentrations for all four MBP and MBP:Ca2+-CaM species, were all linear. Representative results for rmMBP-Cit0 only are presented in Figure 7C▶ (averages of three experiments) and are also compared with the steady-state results. The KSV values calculated from the slopes of the plots of ≤τo>/≤τ> versus [Q] are presented for all four MBP species in Table 3▶. These values are also very close to KSV values obtained from the steady-state experiment (Table 1▶). This similarity supports the assumption that the KSV values in the steady-state experiment indeed represent the dynamic quenching of Trp residues. Binding of Ca2+-CaM results in the reduction of the quenching constants to about half of those observed in its absence. These data are consistent with the notion that the Trp residue is less exposed in the MBP:Ca2+-CaM complex. Changes in Trp accessibility as a consequence of conformational change have been reported for other proteins in the literature (Wells et al. 1994).
Table 3.
Stern-Volmer constants obtained from fluorescence lifetime measurements of MBP alone, and of MBP:Ca2+-CaM complexes, respectively, using weighted-average values
| Sample | Without CaM (KSV [M−1]) | With CaM (KSV [M−1]) |
|---|---|---|
| hMBP-Cit0 | 5.99 ± 0.18 | 2.87 ± 0.11 |
| hMBP-Cit6 | 8.56 ± 0.15 | 3.15 ± 0.25 |
| rmMBP-Cit0 | 8.14 ± 0.07 | 3.05 ± 0.08 |
| rmMBP-qCit6 | 5.28 ± 0.18 | 4.25 ± 0.21 |
The data represent averages of three experiments. The regression coefficients for fits to Equation 7 in the text were all r2 > 0.91.
The steady-state and lifetime data for rmMBP-Cit0 are presented together in Figure 7C▶ for comparison. As stated above, the nonlinearity of the Stern-Volmer plots in steady-state experiments (Fig. 7A,B▶) indicates that both dynamic and static quenching occur. The proportional decreases in the fluorescence lifetimes in time-domain experiments show that part of the quenching observed is due to the diffusion controlled processes, and part is due to static interactions. The static quenching can be due to a number of factors, one of which can be conformational interactions. For example, ring-stacking has been observed to result in static quenching between purine and pyrimidine nucleotides and a number of fluorophores (Seidel et al. 1996).
Fluorescence lifetime measurements have previously been performed on unfractionated MBP, that is, heterogeneous mixtures of all charge isomers comprising all possible posttranslational modifications and under different buffer conditions (Nowak and Berman 1991; Cavatorta et al. 1994). Nowak and Berman (1991) studied the self-association of MBP at high-salt and high-protein concentrations; they modeled their fluorescence decays by using a double-exponential equation. Upon titration with acrylamide up to 0.1 M concentration, the short-lifetime component underwent dynamic quenching, whereas the long-lifetime component underwent static quenching. Overall, however, the dynamic component of quenching predominated. At high-salt concentrations at which the protein aggregated, the lifetimes of both decay components increased. Cavatorta et al. (1994) studied the Zn2+-induced aggregation of MBP in a 1 mM phosphate buffer (pH 7.5); their time-resolved fluorescence measurements were modeled by a triple-exponential fit. Upon aggregation, the relative proportions of each decay component did not change appreciably, but the decay times increased. Our measurements (Fig. 8▶, Tables 2, 3▶▶) are the first to be obtained on specific charge isomers of MBP but are qualitatively and quantitatively consistent with previous data inasmuch as they can be compared. Both aggregation (Cavatorta et al. 1994) and CaM-binding (here) appear to have resulted in a more hydrophobic environment for the fluorophore, as evidenced by the increased fluorescence decays.
The effect of deimination of MBP on its structure and interactions with CaM
An α-helix constructed from a C-terminal segment of MBP (hMBP[150–165]) is amphipathic (Ishiyama et al. 2001), and this segment represents a putative Ca2+-CaM–binding motif by virtue of having a cluster of basic residues on one face (Yap et al. 2000). We have recently examined the interactions between bMBP-Cit0, rmMBP-Cit0, and CaM in vitro, and results were consistent with the C terminus being the primary binding site (Libich and Harauz 2002a,b). In this work, rmMBP-qCit6 and hMBP-Cit6 were predicted to have an altered affinity for Ca2+-CaM compared with unmodified rmMBP-Cit0 and hMBP-Cit0, respectively. Previously, in vitro deimination of Arg residues in the CNS phosphatase calcineurin lowered the affinity of this protein for CaM only by a factor of 10 (Imparl et al. 1995). Here, the overall net charge reduction by deimination, and the two replaced arginyl residues in the latter part of the predicted Ca2+-CaM–binding domain (Fig. 1▶), did not appear to weaken the strength of this interaction significantly because the fluorescence intensity at 340 nm changed in the micromolar range of concentration of added CaM.
Nevertheless, the results presented here indicate that the interaction of posttranslationally modified MBP with Ca2+-CaM is more complex than anticipated from either the prediction of a single binding site or a single conformational state, or fluorescence spectroscopy with minimally modified or unmodified forms of the protein (Libich and Harauz 2002a). The interaction of deiminated hMBP-Cit6 or quasi-deiminated rmMBP-qCit6 with Ca2+-CaM was different in nature than for hMBP-Cit0 or rmMBP-Cit0. Generally, CaM binds with high affinity to relatively small, positively charged, amphipathic α-helical regions of proteins. However, there are numerous examples of proteins that do not follow this model (Schleiff et al. 1996; Porumb et al. 1997), and new structural classes of recognition targets are still being discovered (Osawa et al. 1999; Hoeflich and Ikura 2002). Recently, CaM has been shown to bind also to more complicated motifs formed from disparate regions of target membrane proteins (Han et al. 2000; Schumacher et al. 2001; Hoeflich and Ikura 2002). It is entirely possible that CaM might bind separate segments of MBP simultaneously, but we focus here on the question of predicted α-helical targets.
It is known that deimination disrupts the structure of MBP (Lamensa and Moscarello 1993; Cao et al. 1999; Pritzker et al. 2000a,b), as well as of other proteins such as trichohyalin (Tarcsa et al. 1996). The quasi-deiminated rmMBP-qCit6 also displayed a greater random coil component than did unmodified rmMBP-Cit0 (Bates et al. 2002). Thus, one interpretation of the present results is that deimination changed the conformation or physicochemical properties of MBP sufficiently to expose or create a second, lower-affinity Ca2+-CaM–binding site. Assuming that the first site is the C-terminal segment with the signature sequence of a known CaM-binding motif (Rhoads and Friedberg 1997; Yap et al. 2000), the second site could be another amphipathic α-helical segment that competes with the first one. Several segments of MBP with the potential to form amphipathic α-helices have previously been identified (Mendz et al. 1990, 1995; Polverini et al. 1999). Here, the interaction of deiminated MBP with CaM could possibly induce one of the other potentially amphipathic α-helical segments. However, if such a change occurred here, it was too subtle to be detected by CD.
Another potential explanation for the present results is that deiminated MBP has a number of interchangeable conformational states. This suggestion is plausible based on the fluorescence lifetime measurements (Figs. 8, 9▶▶). Thus, one subpopulation of MBP would have its C-terminal CaM-target site in a conformation with a slightly lower affinity for CaM. Fluorescence spectroscopic measurements of MBP titrated with CaM would thus yield data as in Figure 3▶, C and D, appearing as if there is a second site. Further work involving a more sensitive and specific approach like NMR spectroscopy is required to elucidate further the details of the MBP:Ca2+-CaM interaction (Kranz et al. 2002a,b; Zuiderweg 2002).
Biological significance of MBP:Ca2+-CaM interactions
The biological significance of CaM-binding to MBP is not known. By analogy with MARCKS (Arbuzova et al. 2002), it can be speculated that membrane-bound MBP might modulate local levels of free CaM in noncompact myelin. Boggs and Rangaraj (2000) have shown that MBP can bind actin filaments while bound to a membrane, and that CaM can disrupt the MBP:actin association. Thus, CaM can potentially regulate the ability of MBP to anchor microfilaments to the membrane and/or participate in signaling pathways. Whatever the functional roles of MBP beyond maintenance of myelin structure, posttranslational modifications of MBP such as deimination and phosphorylation could serve to alter the nature of its interactions with other proteins such as CaM and thereby shift equilibria subtly. This phenomenon could perhaps occur during early myelinogenesis when the proportions of deiminated MBP are significantly increased (Moscarello et al. 1994; Palma et al. 1997). It is also possible that some of the many other developmentally regulated Golli (genes of the oligodendrocyte lineage) isoforms of MBP (Givogri et al. 2000, 2001) might bind CaM in vivo, although work in our laboratory has already discounted this idea for J37 (Kaur et al. 2003).
Conclusions
MBP that is minimally posttranslationally modified (natural component C1) or unmodified (recombinant) interacted with CaM to form equimolar complexes in a Ca2+-dependent manner that could be modeled by a single-binding site equation. Deimination of MBP to six citrullinyl residues per molecule (natural component C8), or quasi-deimination to six glutaminyl residues per molecule, altered the nature of the interaction significantly, although it was still Ca2+-dependent. The association of MBP-Cit6 with Ca2+-CaM could be described as involving a second binding site, perhaps one that became exposed due to deimination perturbing the tertiary structure of MBP, or to multiple conformational states of the deiminated MBP. The environment of the sole tryptophanyl residue of MBP was probed with acrylamide, which quenched its fluorescence in both static and dynamic modes. The quenching constants thus obtained were the same for both the deiminated and unmodified proteins. When an equimolar amount of CaM was present, however, there were clear differences between the unmodified and deiminated proteins. The fluorescence lifetime measurements of this residue confirmed that the quenching was primarily dynamic and showed differences between the unmodified and deiminated proteins. The comparable behavior of the naturally deiminated MBP (hMBP-Cit6) and its recombinant analog (rmMBP-qCit6) with respect to CaM-binding supports further use of the latter protein to study the effects of deimination. The MBP:Ca2+-CaM is potentially physiologically relevant, and posttranslational modifications such as deimination alter the nature of the association significantly.
Materials and methods
Materials
Electrophoresis grade acrylamide, ultrapure TRIS base, and ultrapure Na2EDTA were purchased from ICN Biochemicals. Electrophoresis grade sodium dodecyl sulphate (SDS) was purchased from Bio-Rad Laboratories. The Ni2+-NTA agarose beads were obtained from Qiagen. All other chemicals were reagent grade and purchased from either Fisher Scientific or Sigma-Aldrich.
Protein purification
The purification of Leu-Glu-His6-tagged rmMBP-Cit0 and rmMBP-qCit6 by nickel-chelation chromatography, and assessment of purity by SDS-PAGE, reversed-phase HPLC, and electrospray ionization mass spectrometry were performed as previously described (Bates et al. 2000, 2002). The natural human charge isomers hMBP-Cit0 and hMBP-Cit6 were obtained from autopsy material (from a patient with chronic MS) as previously described (Wood et al. 1996). These proteins were a gift from Dr. Denise Wood and Dr. Mario Moscarello (Hospital for Sick Children, Toronto, Ontario, Canada). For use as an occasional control and for DLS experiments, the least-modified C1 component of the natural 18.5-kD isoform from bovine brain (bMBP/C1, or bMBP-Cit0) was purified as previously described (Beniac et al. 1997). Purified bovine brain CaM was purchased from Calbiochem.
Protein concentrations were determined here by measuring the absorbance at 280 nm, a parameter that we have calibrated by amino acid analysis. The values of the extinction coefficients used were 0.590 L/g/cm (bMBP-Cit0), 0.623 L/g/cm (rmMBP-Cit0), 0.627 L/g/cm (rmMBP-qCit6), 0.586 L/g/cm (hMBP-Cit0), 0.586 L/g/cm (hMBP-Cit6), and 0.152 L/g/cm (CaM). These values (in 6.0 M guanidine hydrochloride and 0.02 M phosphate at pH 6.5) were calculated on the basis of the amino acid sequences by using the ProtParam software tool available at the Web site http://www.expasy.ch.
Interactions with Ca2+-CaM via Trp fluorescence emission spectroscopy
We measured the changes in the intrinsic fluorescence emission spectra of the single Trp residue in each MBP species (Fig. 1▶) upon titrating with CaM (which does not contain Trp) in 50 mM Tris-HCl (pH 7.4), 250 mM NaCl, and 1 mM CaCl2. Using an Alphascan-2 spectrofluorimeter (Photon Technology International) at ambient temperature (22°C) with slits set at 2 nm, excitation was at 295 nm, and emission was scanned from 300 to 400 nm in 2-nm steps for collection of spectra. Data were acquired by using the Felix (version 1.4) software of the Alphascan-2 spectrofluorimeter. Upon titration with CaM, emissions were measured here at 340 nm (in contrast, in our previous work with bMBP-Cit0 and rmMBP-Cit0, emissions were measured at 330 nm, and the concentration of Ca2+ was 5 mM; Libich and Harauz 2002a). Fluorescence emission data were corrected for dilution, scattering, and the inner filter effect using
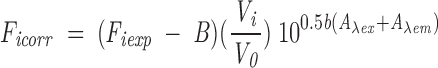 |
(1) |
where Ficorr and Fiexp are the corrected and experimentally determined fluorescence intensities at any given titration point, respectively; B is the background fluorescence intensity; V0 is the initial sample volume; Vi is the sample volume at any given titration point (thus, Vi/V0 is the dilution factor); b is the path length in centimeters; and Aλex and Aλem are the absorbances of the sample at the excitation and emission wavelengths, respectively (Liu and Sharom 1996).
Determination of dissociation constants
For the formation of a 1:1 complex of MBP:CaM, the association constant is
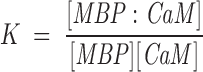 |
(2) |
where [MBP] and [CaM] are the concentrations of free (unreacted) MBP and CaM, respectively. The dissociation constant is simply Kd = 1/K. Because we knew from previous work that the dissociation constant is of the order of magnitude of these concentrations (Libich and Harauz 2002a), and because we had to work under conditions in which these concentrations were comparable (∼2 μM), we corrected for unbound protein using the quadratic expression
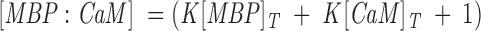 |
(3) |
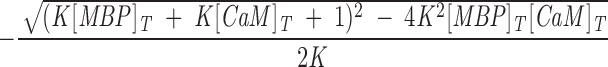 |
where [MBP]T and [CaM]T are the total MBP and CaM concentrations, respectively (Heyduk and Lee 1990; Winzor and Sawyer 1995; Ninfa and Ballou 1998). The total fluorescence signal FT arose from both free and bound MBP, and was:
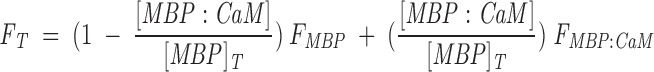 |
(4) |
Here, corrected fluorescence data were regressed by using the SigmaPlot computer program (SPSS) and Equation 4 and by treating K (= 1/Kd), FMBP:CaM, and [MBP]T as fit parameters and [CaM]T as an independent variable. The value of FMBP was obtained at zero CaM concentration.
To ensure that the MBP:CaM interaction was in fact a Ca2+-dependent process under these conditions, control experiments were performed in the presence of chelating agents (8 mM EDTA and 2 mM EGTA) instead of 1 mM CaCl2.
Dynamic light scattering
DLS was performed by using a Coherent Laser Group Model 532 DPSS Nd:Yag laser at a wavelength of 532 nm. A four-window quartz cuvette with a path length of 0.5 cm (Hellma) and a sample volume of 300 μL were used. Detection was accomplished by using a photomultiplier tube at θ = 90 degrees connected to a Brookhaven digital autocorrelator running the 9KDLSW control program (Brookhaven Instruments Corporation). Glans-Thompson filters were placed between the laser and the sample, and between the sample and the photomultiplier tube, to permit transmission of vertically polarized light only. Delay times of 5 μs were used during data collection, and a calculated baseline was used for normalization. Samples were prepared a day in advance and allowed to sit overnight at 4°C to allow air bubbles to come out of solution, and a computational dust filter was applied to avoid data artefacts. Data collection was performed at room temperature (∼23°C) after allowing adequate time for sample warming, and data analysis was performed with Brookhaven software.
Preliminary experiments were performed with samples of 0.1 mg/mL bMBP-Cit0 in 20 mM HEPES-NaOH (pH 7.4), 200 mM NaCl, and 1 mM CaCl2. Subsequent experiments were performed with samples of 0.1 mg/mL bMBP-Cit0 alone, 0.1 mg/mL CaM alone, and a mixture of 0.1 mg/mL bMBP-Cit0 and 0.1 mg/mL CaM, all in 20 mM MES-NaOH (pH 6.0), 200 mM NaCl, and 1 mM CaCl2. The mean diameter of each particle population was taken to be that corresponding to the highest (central) peak of the size histogram.
CD spectroscopy
CD spectroscopy was performed at room temperature as previously described (Bates et al. 2000, 2002). All spectra were collected by using a Jasco J-600 spectropolarimeter (Japan Spectroscopic Co.). The MBP (rmMBP-Cit0, rmMBP-qCit6, hMBP-Cit0, or hMBP-Cit6) was at a concentration of 10 μM (0.2 mg/mL) in fluorescence buffer (50 mM Tris-HCl at pH 7.4, 250 mM NaCl, and 1 mM CaCl2), and was analyzed alone and in the presence of an equimolar amount of CaM. The background signal from buffer was first subtracted from the protein signals. The data were then smoothed by using an inverse-square algorithm in the SigmaPlot (SPSS) computer program for presentation.
Accessibility of Trp residue via acrylamide quenching: Steady-state measurements
The molecular environment of Trp in each MBP species was probed by measuring the fluorescence emission at 340 nm in aqueous solution and by titrating with the collisional quencher acrylamide. A 5-M stock solution of the natural collisional quencher acrylamide was added in 5-μL aliquots to 500 μL of 2.0 μM MBP in buffer. The buffer conditions were 50 mM Tris-HCl (pH 7.4), 250 mM NaCl, and 1 mM CaCl2. The data were fitted to the following nonlinear form of the Stern-Volmer equation
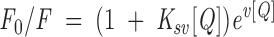 |
(5) |
where F0 and F are the fluorescence intensities at 340 nm in the absence and presence of acrylamide, respectively, and [Q] is the concentration of the acrylamide quencher in M (Nowak and Berman 1991; Lakowicz 1999). The Stern-Volmer quenching constant is Ksv (M−1), representing the dynamic component (Laws and Contino 1992). The parameter V (also in units of M−1) describes a hypothetical "sphere of action" in which the quencher and fluorophore coexist at the moment of excitation, and represents the static component. The soluble Trp analog NATA (N-acetyltryptophanamide) was used as the reference.
The acrylamide quenching measurements were also performed for each MBP species in the same buffer conditions as above, but in the presence of Ca2+ and an equimolar amount of CaM.
Fluorescence lifetime measurements
Fluorescence lifetime measurements of Trp in each MBP species, upon titration with acrylamide, were measured with a fluorescence lifetime instrument (model C-720, Photon Technology International Inc.) using a proprietary stroboscopic detection technique (James et al. 1992; Liu et al. 2000). The system used a GL-330 pulsed nitrogen laser pumping a GL-302 high-resolution dye laser (Photon Technology International Inc.). The dye laser output at 590 nm was frequency-doubled to 295 nm with a GL-103 frequency doubler coupled to an MP-1 sample compartment via fiber optics. The emission was observed at 90° relative to the excitation via an M-101 emission monochromator and a stroboscopic detector equipped with a Hamamatsu 1527 photomultiplier. Fluorescence decays were analyzed with the Felix 32 analysis package using a discrete one- to four-exponential fitting program. The weighted-average lifetimes were calculated from the results of multiexponential fits by using the expression
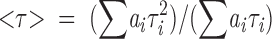 |
(6) |
where ai and τi represented preexponential factors and lifetimes, respectively. (An alternative treatment uses the expression ≤τ> = (∑aiτi)/(∑ai).) If quenching is purely collisional, then a dynamic Stern-Volmer constant can be calculated from the following equation,
 |
(7) |
which was fitted here to the data using SigmaPlot, where again [Q] is the concentration of acrylamide, ≤τ> is the weighted-average lifetime, and ≤τ0> is the weighted-average lifetime in the absence of quencher.
Finally, another set of fluorescence lifetime measurements of the tryptophanyl residue of each MBP species, upon titration with acrylamide, was obtained for each MBP species in the same buffer conditions as above, but in the presence of Ca2+ and an equimolar amount of CaM.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (G.H.), the Natural Sciences and Engineering Research Council of Canada (F.R.H., G.H.), and the Multiple Sclerosis Society of Canada (G.H.). We are grateful to Dr. Denise Wood and Dr. Mario Moscarello for the gift of human MBP charge isomers C1 and C8 (hMBP-Cit0 and hMBP-Cit6), to Dr. Dawn Larson for advice on the site-directed mutagenesis, and to Dr. Frances Sharom and Dr. Rod Merrill for advice on the fluorescence experiments. The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0303603.
References
- Anthony, J. and Moscarello, M.A. 1971a. A conformational change induced in the basic encephalitogen by lipids. Biochim. Biophys. Acta 243 429–433. [DOI] [PubMed] [Google Scholar]
- ———. 1971b. Conformational transition of a myelin protein. FEBS Lett. 15 335–339. [DOI] [PubMed] [Google Scholar]
- Arbuzova, A., Schmitz, A.A.P., and Vergéres, G. 2002. Cross-talk unfolded: MARCKS proteins. Biochem. J. 362 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitis, D.N., Yang, W., and Boggs, J.M. 2002. Myelin proteolipid protein, basic protein, the small isoform of myelin-associated glycoprotein, and p42MAPK are associated in the Triton X-100 extract of central nervous system myelin. J. Neurosci. Res. 70 8–23. [DOI] [PubMed] [Google Scholar]
- Bates, I.R., Matharu, P., Ishiyama, N., Rochon, D., Wood, D.D., Polverini, E., Moscarello, M.A., Viner, N.J., and Harauz, G. 2000. Characterization of a recombinant murine 18.5-kDa myelin basic protein. Protein Expr. Purif. 20 285–299. [DOI] [PubMed] [Google Scholar]
- Bates, I.R., Libich, D.S., Wood, D.D., Moscarello, M.A., and Harauz, G. 2002. An Arg/Lys→Gln mutant of recombinant murine myelin basic protein as a mimic of the deiminated form implicated in multiple sclerosis. Protein Expr. Purif. 25 330–341. [DOI] [PubMed] [Google Scholar]
- Beechem, J.M. and Brand, L. 1985. Time-resolved fluorescence of proteins. Annu. Rev. Biochem. 54 43–71. [DOI] [PubMed] [Google Scholar]
- Beniac, D.R., Luckevich, M.D., Czarnota, G.J., Tompkins, T.A., Ridsdale, R.A., Ottensmeyer, F.P., Moscarello, M.A., and Harauz, G. 1997. Three-dimensional structure of myelin basic protein, I: Reconstruction via angular reconstitution of randomly oriented single particles. J. Biol. Chem. 272 4261–4268. [DOI] [PubMed] [Google Scholar]
- Beniac, D.R., Wood, D.D., Palaniyar, N., Ottensmeyer, F.P., Moscarello, M.A., and Harauz, G. 2000. Cryoelectron microscopy of protein-lipid complexes of human myelin basic protein charge isomers differing in degree of citrullination. J. Struct. Biol. 129 80–95. [DOI] [PubMed] [Google Scholar]
- Boggs, J.M. and Rangaraj, G. 2000. Interaction of lipid-bound myelin basic protein with actin filaments and calmodulin. Biochemistry 39 7799–7806. [DOI] [PubMed] [Google Scholar]
- Boggs, J.M., Rangaraj, G., Koshy, K.M., Ackerley, C., Wood, D.D., and Moscarello, M.A. 1999. Highly deiminated isoform of myelin basic protein from multiple sclerosis brain causes fragmentation of lipid vesicles. J. Neurosci. Res. 57 529–535. [PubMed] [Google Scholar]
- Calhoun, D.B., Vanderkooi, J.M., Holtom, G.R., and Englander, S.W. 1986. Protein fluorescence quenching by small molecules: Protein penetration versus solvent exposure. Proteins 1 109–115. [DOI] [PubMed] [Google Scholar]
- Cao, L., Goodin, R., Wood, D.D., Moscarello, M.A., and Whitaker, J.N. 1999. Rapid release and unusual stability of immunodominant peptide 45–89 from citrullinated myelin basic protein. Biochemistry 38 6157–6163. [DOI] [PubMed] [Google Scholar]
- Cavatorta, P., Giovanelli, S., Bobba, A., Riccio, P., Szabo, A.G., and Quagliariello, E. 1994. Myelin basic protein interaction with zinc and phosphate: Fluorescence studies on the water-soluble form of the protein. Biophys. J. 66 1174–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, K.F., Robb, N.D., and Chen, W.H. 1990. Myelin basic protein: Interaction with calmodulin and gangliosides. J. Neurosci. Res. 25 535–544. [DOI] [PubMed] [Google Scholar]
- Chen, Y. and Barkley, M.D. 1998. Toward understanding tryptophan fluorescence in proteins. Biochemistry 37 9976–9982. [DOI] [PubMed] [Google Scholar]
- Clayton, A.H.A. and Sawyer, W.H. 1999. Tryptophan rotamer distributions in amphipathic peptides at a lipid interface. Biophys. J. 76 3235–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döppenschmitt, S., Langguth, P., Regardh, C.G., Andersson, T.B., Hilgendorf, C., and Spahn-Langguth, H. 1999. Characterization of binding properties to human P-glycoprotein: Development of a [3H]verapamil radioligand-binding assay. J. Pharmacol. Exp. Ther. 288 348–357. [PubMed] [Google Scholar]
- Doyle, H.A. and Mamula, M.J. 2001. Post-translational protein modifications in antigen recognition and autoimmunity. Trends Immunol. 22 443–449. [DOI] [PubMed] [Google Scholar]
- ———. 2002. Post-translational protein modifications: New flavors in the menu of autoantigens. Curr. Opin. Rheumatol. 14 244–249. [DOI] [PubMed] [Google Scholar]
- Dyer, C.A., Phillbotte, T., Wolf, M.K., and Billings-Gagliardi, S. 1997. Regulation of cytoskeleton by myelin components: Studies on shiverer oligodendrocytes carrying an Mbp transgene. Dev. Neurosci. 19 395–409. [DOI] [PubMed] [Google Scholar]
- Finch, P.R., Wood, D.D., and Moscarello, M.A. 1971. The presence of citrulline in a myelin protein fraction. FEBS Lett. 15 145–148. [DOI] [PubMed] [Google Scholar]
- Gell, D., Kong, Y., Eaton, S.A., Weiss, M.J., and Mackay, J.P. 2002. Biophysical characterization of the α-globin binding protein α-hemoglobin stabilizing protein. J. Biol. Chem. 277 40602–40609. [DOI] [PubMed] [Google Scholar]
- Givogri, M.I., Bongarzone, E.R., and Campagnoni, A.T. 2000. New insights on the biology of myelin basic protein gene: The neural-immune connection. J. Neurosci. Res. 59 153–159. [PubMed] [Google Scholar]
- Givogri, M.I., Bongarzone, E.R., Schonmann, V., and Campagnoni, A.T. 2001. Expression and regulation of Golli products of myelin basic protein gene during in vitro development of oligodendrocytes. J. Neurosci. Res. 66 679–690. [DOI] [PubMed] [Google Scholar]
- Gomes, A.V., Barnes, J.A., and Vogel, H.J. 2000. Spectroscopic characterization of the interaction between calmodulin-dependent protein kinase I and calmodulin. Arch. Biochem. Biophys. 379 28–36. [DOI] [PubMed] [Google Scholar]
- Han, B.G., Nunomura, W., Takakuwa, Y., Mohandas, N., and Jap, B.K. 2000. Protein 4.1R core domain structure and insights into regulation of cytoskeletal organization. Nat. Struct. Biol. 7 871–875. [DOI] [PubMed] [Google Scholar]
- Harauz, G., Ishiyama, N., and Bates, I.R. 2000. Analogous structural motifs in myelin basic protein and in MARCKS. Mol. Cell. Biochem. 209 155–163. [DOI] [PubMed] [Google Scholar]
- Heyduk, T. and Lee, J.C. 1990. Application of fluorescence energy transfer and polarization to monitor Escherichia coli cAMP receptor protein and lac promoter interaction. Proc. Natl. Acad. Sci. 87 1744–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, C.M.D., Bates, I.R., White, G.F., Hallett, F.R., and Harauz, G. 2002. Effects of the osmolyte trimethylamine-N-oxide on conformation, self-association, and two-dimensional crystallization of myelin basic protein. J. Struct. Biol. 139 13–26. [DOI] [PubMed] [Google Scholar]
- Hill, T.J., Lafitte, D., Wallace, J.I., Cooper, H.J., Tsvetkov, P.O., and Derrick, P.J. 2000. Calmodulin-peptide interactions: Apocalmodulin binding to the myosin light chain kinase target-site. Biochemistry 39 7284–7290. [DOI] [PubMed] [Google Scholar]
- Hoeflich, K.P. and Ikura, M. 2002. Calmodulin in action: Diversity in target recognition and activation mechanisms. Cell 108 739–742. [DOI] [PubMed] [Google Scholar]
- Ikura, M., Clore, G.M., Gronenborn, A.M., Zhu, G., Klee, C.B., and Bax, A. 1992. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 256 632–638. [DOI] [PubMed] [Google Scholar]
- Imparl, J.M., Senshu, T., and Graves, D.J. 1995. Studies of calcineurin-calmodulin interaction: probing the role of arginine residues using peptidylarginine deiminase. Arch. Biochem. Biophys. 318 370–377. [DOI] [PubMed] [Google Scholar]
- Ishiyama, N., Bates, I.R., Hill, C.M.D., Wood, D.D., Matharu, P., Viner, N.J., Moscarello, M.A., and Harauz, G. 2001. The effects of deimination of myelin basic protein on structures formed by its interaction with phosphoinositide-containing lipid monolayers. J. Struct. Biol. 136 30–45. [DOI] [PubMed] [Google Scholar]
- James, D.R., Siemiarczuk, A., and Ware, W.R. 1992. Stroboscopic optical boxcar technique for the determination of fluorescence lifetimes. Rev. Sci. Instrum. 63 1710–1716. [Google Scholar]
- Jones, A.J. and Rumsby, M.G. 1975. The intrinsic fluorescence characteristics of the myelin basic protein. J. Neurochem. 25 565–572. [DOI] [PubMed] [Google Scholar]
- Kaur, J., Libich, D.S., Campagnoni, C.W., Wood, D.D., Moscarello, M.A., Campagnoni, A.T., and Harauz, G. 2003. Expression and properties of the recombinant murine Golli-myelin basic protein isoform J37. J. Neurosci. Res. 71 777–784. [DOI] [PubMed] [Google Scholar]
- Keniry, M.A. and Smith, R. 1979. Circular dichroic analysis of the secondary structure of myelin basic protein and derived peptides bound to detergents and to lipid vesicles. Biochim. Biophys. Acta 578 381–391. [DOI] [PubMed] [Google Scholar]
- Kim, S.J., Chowdhury, F.N., Stryjewski, W., Younathan, E.S., Russo, P.S., and Barkley, M.D. 1993. Time-resolved fluorescence of the single tryptophan of Bacillus stearothermophilus phosphofructokinase. Biophys. J. 65 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz, J.K., Flynn, P.F., Fuentes, E.J., and Wand, A.J. 2002a. Dissection of the pathway of molecular recognition by calmodulin. Biochemistry 41 2599–2608. [DOI] [PubMed] [Google Scholar]
- Kranz, J.K., Lee, E.K., Nairn, A.C., and Wand, A.J. 2002b. A direct test of the reductionist approach to structural studies of calmodulin activity: Relevance of peptide models of target proteins. J. Biol. Chem. 277 16351–16354. [DOI] [PubMed] [Google Scholar]
- Kroes, S.J., Canters, G.W., Gilardi, G., van Hoek, A., and Visser, A.J. 1998. Time-resolved fluorescence study of azurin variants: Conformational heterogeneity and tryptophan mobility. Biophys. J. 75 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz, J.R. 1999. Principles of fluorescence spectroscopy, 2nd ed. Kluwer Academic, New York, NY.
- Lamensa, J.W. and Moscarello, M.A. 1993. Deimination of human myelin basic protein by a peptidylarginine deiminase from bovine brain. J. Neurochem. 61 987–996. [DOI] [PubMed] [Google Scholar]
- Laws, W.R. and Contino, P.B. 1992. Fluorescence quenching studies: Analysis of nonlinear Stern-Volmer data. Methods Enzymol. 210 448–463. [DOI] [PubMed] [Google Scholar]
- Libich, D.S. and Harauz, G. 2002a. Interactions of the 18.5 kDa isoform of myelin basic protein with Ca2+-calmodulin: In vitro studies using fluorescence microscopy and spectroscopy. Biochem. Cell Biol. 80 395–406. [DOI] [PubMed] [Google Scholar]
- ———. 2002b. Interactions of the 18.5 kDa isoform of myelin basic protein with Ca2+-calmodulin: In vitro studies using gel shift assays. Mol. Cell. Biochem. 241 45–52. [DOI] [PubMed] [Google Scholar]
- Liebes, L.F., Zand, R., and Phillips, W.D. 1975. Solution behavior, circular dichroism and 220 MHz PMR studies of the bovine myelin basic protein. Biochim. Biophys. Acta 405 27–39. [DOI] [PubMed] [Google Scholar]
- Lintner, R.N. and Dyer, C.A. 2000. Redistribution of cholesterol in oligodendrocyte membrane sheets after activation of distinct signal transduction pathways. J. Neurosci. Res. 60 437–449. [DOI] [PubMed] [Google Scholar]
- Liu, R. and Sharom, F.J. 1996. Site-directed fluorescence labeling of P-glycoprotein on cysteine residues in the nucleotide binding domains. Biochemistry 35 11865–11873. [DOI] [PubMed] [Google Scholar]
- Liu, R., Siemiarczuk, A., and Sharom, F.J. 2000. Intrinsic fluorescence of the P-glycoprotein multidrug transporter: Sensitivity of tryptophan residues to binding of drugs and nucleotides. Biochemistry 39 14927–14938. [DOI] [PubMed] [Google Scholar]
- Mendz, G.L., Brown, L.R., and Martenson, R.E. 1990. Interactions of myelin basic protein with mixed dodecylphosphocholine/palmitoyllysophosphatidic acid micelles. Biochemistry 29 2304–2311. [DOI] [PubMed] [Google Scholar]
- Mendz, G.L., Miller, D.J., and Ralston, G.B. 1995. Interactions of myelin basic protein with palmitoyllysophosphatidylcholine: Characterization of the complexes and conformations of the protein. Eur. Biophys. J. 24 39–53. [DOI] [PubMed] [Google Scholar]
- Moscarello, M.A. 1997. Myelin basic protein, the "executive" molecule of the myelin membrane. In Cell biology and pathology of myelin: Evolving biological concepts and therapeutic approaches, (eds. B.H.J. Juurlink et al.), pp. 13–25. Plenum Press, New York.
- Moscarello, M.A., Wood, D.D., Ackerley, C., and Boulias, C. 1994. Myelin in multiple sclerosis is developmentally immature. J. Clin. Invest. 94 146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskaitis, J.E. and Campagnoni, A.T. 1986. A comparison of the dodecyl sulfate-induced precipitation of the myelin basic protein with other water-soluble proteins. Neurochem. Res. 11 299–315. [DOI] [PubMed] [Google Scholar]
- Murase, T. and Iio, T. 2002. Static and kinetic studies of complex formations between calmodulin and mastoparanX. Biochemistry 41 1618–1629. [DOI] [PubMed] [Google Scholar]
- Ninfa, A.J. and Ballou, D.P. 1998. Ligand binding. In Fundamental laboratory approaches for biochemistry and biotechnology, pp. 247–271. Fitzgerald Science Press, Bethesda, MD.
- Noonan, D.J., Lou, D., Griffith, N., and Vanaman, T.C. 2002. A calmodulin binding site in the tuberous sclerosis 2 gene product is essential for regulation of transcription events and is altered by mutations linked to tuberous sclerosis and lymphangioleiomyomatosis. Arch. Biochem. Biophys. 398 132–140. [DOI] [PubMed] [Google Scholar]
- Nowak, M.W. and Berman, H.A. 1991. Fluorescence studies on the interactions of myelin basic protein in electrolyte solutions. Biochemistry 30 7642–7651. [DOI] [PubMed] [Google Scholar]
- Osawa, M., Tokumitsu, H., Swindells, M.B., Kurihara, H., Orita, M., Shibanuma, T., Furuya, T., and Ikura, M. 1999. A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin–dependent kinase kinase. Nat. Struct. Biol. 6 819–824. [DOI] [PubMed] [Google Scholar]
- Palma, A.E., Owh, P., Fredric, C., Readhead, C., and Moscarello, M.A. 1997. Characterization of myelin basic protein charge microheterogeneity in developing mouse brain and in the transgenic shiverer mutant. J. Neurochem. 69 1753–1762. [DOI] [PubMed] [Google Scholar]
- Papish, A.L., Tari, L.W., and Vogel, H.J. 2002. Dynamic light scattering study of calmodulin-target peptide complexes. Biophys. J. 83 1455–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polverini, E., Fasano, A., Zito, F., Riccio, P., and Cavatorta, P. 1999. Conformation of bovine myelin basic protein purified with bound lipids. Eur. Biophys. J. 28 351–355. [DOI] [PubMed] [Google Scholar]
- Porumb, T., Crivici, A., Blackshear, P.J., and Ikura, M. 1997. Calcium binding and conformational properties of calmodulin complexed with peptides derived from myristoylated alanine-rich C kinase substrate (MARCKS) and MARCKS-related protein (MRP). Eur. Biophys. J. 25 239–247. [DOI] [PubMed] [Google Scholar]
- Pritzker, L.B., Joshi, S., Gowan, J.J., Harauz, G., and Moscarello, M.A. 2000a. Deimination of myelin basic protein, 1: Effect of deimination of arginyl residues of myelin basic protein on its structure and susceptibility to digestion by cathepsin D. Biochemistry 39 5374–5381. [DOI] [PubMed] [Google Scholar]
- Pritzker, L.B., Joshi, S., Harauz, G., and Moscarello, M.A. 2000b. Deimination of myelin basic protein, 2: Effect of methylation of MBP on its deimination by peptidylarginine deiminase. Biochemistry 39 5382–5388. [DOI] [PubMed] [Google Scholar]
- Rhoads, A.R. and Friedberg, F. 1997. Sequence motifs for calmodulin recognition. FASEB J. 11 331–340. [DOI] [PubMed] [Google Scholar]
- Romsicki, Y. and Sharom, F.J. 1999. The membrane lipid environment modulates drug interactions with the P-glycoprotein multidrug transporter. Biochemistry 38 6887–6896. [DOI] [PubMed] [Google Scholar]
- Schleiff, E., Schmitz, A., McIlhinney, R.A., Manenti, S., and Vergéres, G. 1996. Myristoylation does not modulate the properties of MARCKS-related protein (MRP) in solution. J. Biol. Chem. 271 26794–26802. [DOI] [PubMed] [Google Scholar]
- Schumacher, M.A., Rivard, A.F., Bachinger, H.P., and Adelman, J.P. 2001. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature 410 1120–1124. [DOI] [PubMed] [Google Scholar]
- Seidel, C.A.M., Schulz, A., and Sauer, M.H.M. 1996. Nucleobase-specific quenching of fluorescent dyes, 1: Nucleobase one-electron redox potentials and their correlation with static and dynamic quenching efficiencies. J. Phys. Chem. 100 5541–5553. [Google Scholar]
- Smith, R. 1992. The basic protein of CNS myelin: Its structure and ligand binding. J. Neurochem. 59 1589–1608. [DOI] [PubMed] [Google Scholar]
- Staugaitis, S.M., Colman, D.R., and Pedraza, L. 1996. Membrane adhesion and other functions for the myelin basic proteins. Bioessays 18 13–18. [DOI] [PubMed] [Google Scholar]
- Tarcsa, E., Marekov, L.N., Mei, G., Melino, G., Lee, S.C., and Steinert, P.M. 1996. Protein unfolding by peptidylarginine deiminase: Substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J. Biol. Chem. 271 30709–30716. [DOI] [PubMed] [Google Scholar]
- Tompkins, T.A. and Moscarello, M.A. 1991. A 57-kDa phosphatidylinositol-specific phospholipase C from bovine brain. J. Biol. Chem. 266 4228–4236. [PubMed] [Google Scholar]
- ———. 1993. Stimulation of bovine brain phospholipase C activity by myelin basic protein requires arginyl residues in peptide linkage. Arch. Biochem. Biophys. 302 476–483. [DOI] [PubMed] [Google Scholar]
- Vadas, E.B., Melançon, P., Braun, P.E., and Galley, W.C. 1981. Phosphorescence studies of the interaction of myelin basic protein with phosphatidylserine vesicles. Biochemistry 20 3110–3116. [DOI] [PubMed] [Google Scholar]
- Weiss, J.N. 1997. The Hill equation revisited: Uses and misuses. FASEB J. 11 835–841. [PubMed] [Google Scholar]
- Wells, T.A., Nakazawa, M., Manabe, K., and Song, P.S. 1994. A conformational change associated with the phototransformation of Pisum phytochrome A as probed by fluorescence quenching. Biochemistry 33 708–712. [DOI] [PubMed] [Google Scholar]
- Whitaker, J.N. and Mitchell, G.W. 1996. A possible role for altered myelin basic protein in multiple sclerosis. Ann. Neurol. 40 3–4. [DOI] [PubMed] [Google Scholar]
- Winzor, D.J. and Sawyer, W.H. 1995. Quantitative characterization of ligand binding. Wiley-Liss, New York.
- Wood, D.D. and Moscarello, M.A. 1997. Molecular biology of the glia: Components of myelin: The implication of post-translational changes for demyelinating disease. In The molecular biology of multiple sclerosis, (ed. W. Russell), pp. 37–54. Wiley, New York.
- Wood, D.D., Bilbao, J.M., O’Connors, P., and Moscarello, M.A. 1996. Acute multiple sclerosis (Marburg type) is associated with developmentally immature myelin basic protein. Ann. Neurol. 40 18–24. [DOI] [PubMed] [Google Scholar]
- Yap, K.L., Kim, J., Truong, K., Sherman, M., Yuan, T., and Ikura, M. 2000. Calmodulin target database. J. Struct. Funct. Genomics 1 8–14. [DOI] [PubMed] [Google Scholar]
- Yuan, T., Mietzner, T.A., Montelaro, R.C., and Vogel, H.J. 1995. Characterization of the calmodulin binding domain of SIV transmembrane glycoprotein by NMR and CD spectroscopy. Biochemistry 34 10690–10696. [DOI] [PubMed] [Google Scholar]
- Yuan, T., Tencza, S., Mietzner, T.A., Montelaro, R.C., and Vogel, H.J. 2001. Calmodulin binding properties of peptide analogues and fragments of the calmodulin-binding domain of simian immunodeficiency virus transmembrane glycoprotein 41. Biopolymers 58 50–62. [DOI] [PubMed] [Google Scholar]
- Zand, R., Li, M.X., Jin, X., and Lubman, D. 1998. Determination of the sites of posttranslational modifications in the charge isomers of bovine myelin basic protein by capillary electrophoresis-mass spectroscopy. Biochemistry 37 2441–2449. [DOI] [PubMed] [Google Scholar]
- Zhan, Q.Q., Wong, S.S., and Wang, C.L. 1991. A calmodulin-binding peptide of caldesmon. J. Biol. Chem. 266 21810–21814. [PubMed] [Google Scholar]
- Zuiderweg, E.R. 2002. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry 41 1–7. [DOI] [PubMed] [Google Scholar]