Abstract
The identification of protein biochemical functions based on their three-dimensional structures is strongly required in the post-genome-sequencing era. We have developed a new method to identify and predict protein biochemical functions using the similarity information of molecular surface geometries and electrostatic potentials on the surfaces. Our prediction system consists of a similarity search method based on a clique search algorithm and the molecular surface database eF-site (electrostatic surface of functional-site in proteins). Using this system, functional sites similar to those of phosphoenoylpyruvate carboxy kinase were detected in several mononucleotide-binding proteins, which have different folds. We also applied our method to a hypothetical protein, MJ0226 from Methanococcus jannaschii, and detected the mononucleotide binding site from the similarity to other proteins having different folds.
Keywords: Protein function prediction, three-dimensional structure, molecular surface, electrostatic potential, clique search algorithm
With the progress of genome projects, more than 60 genomic sequences have already been provided. However, large fractions of the gene products have not been annotated, and even when annotations were assumed, not all of them might be reliable due to the inherent ambiguity in functional inference using conventional methods based mainly on sequence homology (Devos and Valencia 2000; Wilson et al. 2000; Aloy et al. 2001; Teichmann et al. 2001). To extend our understanding of genomes in light of biological consequences, we should overcome this difficulty and try to identify the functions of gene products encoded on the genomic sequences.
The protein function is considered to have two different aspects: a biological function and a biochemical function. From the viewpoint of molecular biology, the former function could be described in terms of protein–protein interactions, whereas the latter should be directly related to each protein structure. Our goal in the present study was to develop a reliable method to predict the biochemical functions of proteins from their three-dimensional (3D) structures.
Our approach is to search for similar substructures of known proteins against hypothetical proteins. Currently, it is well known that fold-level similarity can give us only a limited amount of information regarding the hypothetical proteins’ function (Thornton et al. 2000; Todd et al. 2002). Thus, our method should be able to detect the similarities among proteins that have different folds. For that purpose, we focused our attention on the local structures of the functional sites in proteins.
In particular, we describe the protein structure based on the molecular surfaces of the proteins and the electrostatic potential on the surface, in order to discriminate the physicochemical differences in the local surface.
Several approaches have been proposed based on molecular surface geometry (Rosen et al. 1998; Exner et al. 2002a,b; Heifetz et al. 2002; Venkatachalam et al. 2003). In these methods, for the purpose of reducing the computation time of the similarity search, the number of vertices describing the molecular surface is reduced, or the search space is limited by using a grid-based search algorithm. These reductions might cause trouble such that the same molecule would not be identified when it is put on a different coordinate system. Thus, we did not reduce the surface points and developed a new method based on a clique search algorithm (Kinoshita et al. 2002).
For the system to predict a protein’s biochemical function from its 3D structure, a database for protein functional sites and a similarity search method against the database are required. For the functional site database, we constructed a database called eF-site (electrostatic surface of functional-site in proteins), which is available on the Web at http://ef-site.protein.osaka-u.ac.jp/eF-site. In the database, the molecular surfaces of many protein molecules with their electrostatic potentials on the surfaces are contained. The information about the functional residues is also stored. Each molecular surface was generated by Connolly’s algorithm (Connolly 1983), and the electrostatic potential was calculated by numerically solving Poisson-Boltzmann equations with a precise continuum model (Nakamura and Nishida 1987). We have already registered more than 7000 entries in the eF-site database, and 1684 entries were selected as the representative functional surfaces according to the local surface similarity, as described below. It should be noted that the redundancy, which we eliminated to select a set of representatives, is not based on the homology but on the atomic configuration of the binding site (Kinoshita et al. 1999). Thus, some homologs could be selected as different representatives according to the different binding conditions, such as those crystallized with different ligands and/or with point mutations. These representative entries were used for the following studies.
Our search method is an application of the graph theorem using the particular descriptors for the surface geometry and the physicochemical properties of a protein’s molecular surface. In fact, there are a set of curvatures at each vertex forming the molecular surface (Fig. 1 ▶) and the electrostatic potential at the vertex. From a mathematical viewpoint, the geometric feature of each vertex at a local surface can be described by two principal curvatures. Among the various principal curvatures, a combination of the maximum and minimum curvatures was found to be the best descriptor which is more sensitive than the conventional combination of the mean curvature and Gaussian curvatures. Thus, during graph construction, the nodes were assumed when these descriptors as the properties of each vertex were similar between the two molecular surfaces. The edges between these nodes were then connected when the mutual distances between the vertices were also similar (Fig. 1A ▶). This procedure for forming a nondirected graph was found to be much more effective at identifying the similar local molecular surface than our previous algorithm (Kinoshita et al. 2002), which was more sensitive to local structural changes than the current method, and so even a small structural change or structural difference would significantly alter the description of molecular surfaces. Finally, the largest clique was found for the graph using the conventional exhaustive search algorithm (Bron and Kerbosch 1973).
Figure 1.
Search algorithm and calculation of curvature. (A) The molecular surface was calculated using Connolly’s algorithm (Connolly 1983) and is represented by a set of triangular meshes. Properties such as electrostatic potential were also considered and were added at each vertex. Our search method is based on the clique search algorithm known in the mathematical field of the graph theorem. A graph is made from nodes and edges. Here, each node is considered as a pair of vertices with similar electrostatic potentials (V) and curvatures (Å−1) at the vertices. All possible combinations of the nodes were then examined. Similarity was defined as absolute differences in the electrostatic potential and curvatures less than 0.04 and 0.2, respectively. With this definition of the nodes, each pair of nodes specifies the pairs of vertices, one from Surface A and the other from Surface B. When the pairs of vertices have similar mutual distances, an edge between the pairs of nodes is drawn. Similarity was defined as an absolute difference in the mutual distance less than 1.5 Å. We then have a graph for a pair of surfaces to be compared. In this graph, we focus our attention on the special subgraph called “clique.” Clique is a subgraph all of whose nodes are connected by edges. In a clique, the corresponding vertices then have a similar spatial arrangement. Thus, the problem when searching for a similar subsurface is to find the largest clique in the graph. To search for the largest clique from a given graph, we used an established algorithm by Bron and Kerbosch (1973). (B) Curvature at a vertex P was calculated by considering the normal cross-section at point P. The normal cross-section is a crossing line between the normal plane and the molecular surface. The cross-section was approximated with a second-order polynomial, and the curvature was determined from its coefficient. According to the freedom of the normal planes around a normal vector at point P, a set of curvatures was calculated for each normal plane obtained by rotating the plane around the normal vector. We obtained the maximum and minimum curvatures by rotating the normal plane from 0° to 180° with a step of 5°, and used them to describe the surface geometry at each vertex.
With this search method and the eF-site database, we first examined whether our method could detect any similarity beyond the fold level. As an example, we studied the functional similarity, which was recently noted (Tari et al. 1996) among phosphoenoylpyruvate carboxy kinase (PCK; SCOP: c.19; Lo Conte et al. 2002) and “some p-loop containing triphosphate hydrolase fold (SCOP: c.37).” In these cases, protein folds are quite different, as seen in the different SCOP classification number and the different β-sheet topologies.
By using the whole surface of PCK (PDB: 1ayl) as a query, a similarity search was carried out against the representative eF-site entries. The search results for PCK are shown in Figure 2 ▶. The three proteins homologous to PCK and two proteins with different SCOP folds (Fig. 2B,C,E ▶) were detected with a relatively high similarity score (Z-score ≥3.0), and were followed by F1 ATPase (PDB: 1e1q, SCOP: c.37.1.1, Z-score = 2.8), CheA (PDB: 1i5b, SCOP: d.122.1.3, Z-score = 2.8), and some others with mononucleotide binding proteins. The two nonhomologous entries with high Z-scores are ferredoxin-NADP+-oxidoreductase (FNR) complexed with adenosine-2′-5′-diphosphate (PDB: 1fnc, SCOP: c.25.1.1, Z-score = 3.3) and tryptophanyl-tRNA synthetase (TrpRS) complexed with tryptophanyl-5′ AMP (PDB: 1i6k, SCOP: c.26.1.1, Z-score = 3.1), both of which bind mononucleotides as well as PCK. In both cases, similarities were partially observed around the ligand binding site. The local surface geometry and property are similar around the adenosine base and sugar binding part in the former case (Fig. 2D ▶), and around the phosphate binding part in the latter case (Fig. 2F ▶).
Figure 2.
(A) Search results for PCK against the 1684 representative entries in the eF-site database. The similarity score was normalized by the Z-score. (B) Ribbon and surface models of PCK (PDB: 1ayl). Ribbon view is colored from blue to red via green from N terminus to C terminus of the protein. Surface model was colored according to the electrostatic potential at the surface, where blue and red indicate the positive and the negative potentials, respectively. The ligand molecule is represented by a ball-and-stick model in both models to indicate the ligand binding site. (C) Same as B for FNR (PDB: 1fnc). (D) Close-up view for the corresponding subsurface between PCK and FNR detected by the similarity search. The ball-and-stick models indicate the bound ligands in both complexed models. (E) Same as B for TrpRS (PDB: 1i6k). (F) Same as D for PCK and TrpRS. B–F were drawn using MOLSCRIPT (Kraulis 1991) and Raster3d (Merritt and Murphy 1994).
The relationship between their biochemical functions and the observed similarities is not clear. However, it is interesting that the similar detected surfaces are somewhat related to their biochemical functions. For the pair with completely different biochemical functions of PCK and FNR, a similarity was observed around the common adenosine base. In contrast, for the latter pair of PCK and TrpRS, both of which use the phosphate for their enzymatic reactions, a similarity was detected only around the phosphate binding sites. As a consequence, recognition of the adenine moiety is common in the former pair, and the enzymatic reaction to break a phosphodiester bond is common in the latter pair. Both the molecular recognition and enzymatic reaction are parts of the chemical function of proteins, and we may expect that the subsurface similarity corresponds with the partially similar chemical functions. Other similarity searches to identify the functional site surfaces, such as for the serine proteases, were also successfully performed, as well as in our previous study (Kinoshita et al. 2002).
These successful results encouraged us to apply our method to some hypothetical proteins as a structural genomics research. Here, we show a result for the MJ0226 free form (PDB: 1b78) from Methanococcus jannaschii because their ligand was experimentally identified. When the 3D structure of this protein was first determined, the fold was new (Hwang et al. 1999), and the overall similarity in the fold level did not work to annotate its function. However, a partial similarity to several mononucleotide related proteins was observed by Hwang et al. (1999). Based on their observation, they predicted the mononucleotide binding ability of this protein and proved it directly by solving the complex with the ATP analog, which was then registered with the PDB as 2mjp. This is an excellent example of the structure-based function prediction.
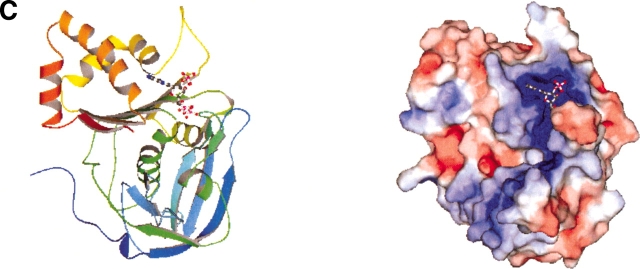
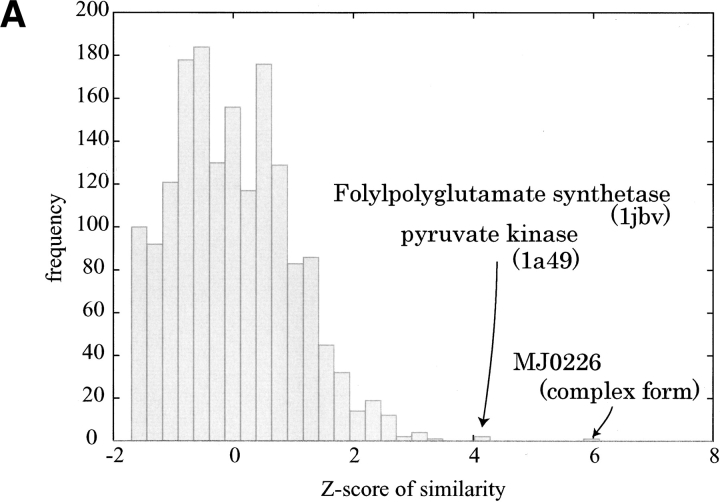
We calculated the molecular surface of the free form of this protein, MJ0226 (1b78), and a similarity search was done against the same representative eF-site entries used above. As shown in Figure 3A ▶, the first-ranked entry (Z-score = 5.9) is the complexed form of the same protein, and so it is trivial. The interesting results can be seen in the second- and third-ranked entries: the folylpolyglutamate synthetase (FPGS) complexed with an ATP analog (PDB: 1jbv, Z-score = 4.2) and pyruvate kinase (PK) complexed with an ATP (PDB: 1a49, Z-score = 4.1), respectively.
Figure 3.

(A) Same as Figure 2A ▶ for MJ0226 free form. (B) Same as Figure 2B ▶ for MJ0226 free form (PDB: 1b78). (C) Same as B for FPGS (PDB: 1jbv). (D) Same as Figure 2D ▶ but between the MJ0226 free form and FPGS. It should be noted that ATP shown in the close view of MJ0226 was taken from the complex form according to the superimposition between the complex form and the free form. (E) Same as B for PK (PDB: 1a49). (F) Same as D but between MJ0226 and PK1b78. B–F were drawn using MOLSCRIPT (Kraulis 1991) and Raster3d (Merritt and Murphy 1994).
The folds of these detected entries are completely different from that of MJ0226 (SCOP: c.51.4.1 for MJ0226, c.72.2.2 for FPGS, and c.49.1.1 for PK), and the entire molecular surfaces are also different (Fig. 3B,C,E ▶). In contrast, the local subsurfaces around the binding sites are very similar, as shown in Figure 3D,F ▶. We used the whole surface of the free form of MJ0226, and so the query contained no ligand molecules. The location of the ATP analog was modeled by superimposing the molecular surfaces of the free and the complexed forms of MJ0226, and they are shown in Figure 3D,F ▶. As can be seen in these complexed models and the complex structures we detected, our method successfully detected the binding site of the mononucleotide of this hypothetical protein. However, the binding mode of the mononucleotide is somewhat different from that of FPGS and that of PK (Fig. 3D,F ▶), which might indicate the different biochemical functions between this hypothetical protein and the detected proteins. These precise binding modes may be further discriminated by examining the atomic configurations in detail around the functional site (Wallance et al. 1997; Kinoshita et al. 1999).
Several other applications to the proteins without any functional information are now underway with the collaboration of X-ray crystallographers. Some of the successful results, including the conserved protein TT1542 from Thermus thermophilus HB8, will be published in the near future.
In summary, using the current search system for the molecular surface similarity, we successfully predicted what the ligand is and where it binds to this hypothetical protein, even though there are no known proteins with similar folds or homologous sequences in the databases. However, identifying the precise binding mode of the ligand is still a difficult task for prediction.
Acknowledgments
This work was supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) to K.K. Development of the eF-site database has been supported by a grant-in-aid from the Institute for Bioinformatics Research and Development, Japan Science and Technology Corporation (BIRD-JST) to H.N.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0368703.
References
- Aloy, P., Querol, E., Aviles, F.X., and Sternberg, M.J. 2001. Automated structure-based prediction of functional sites in proteins: Applications to assessing the validity of inheriting protein function from homology in genome annotation and to protein docking. J. Mol. Biol. 311 395–408. [DOI] [PubMed] [Google Scholar]
- Bron, C. and Kerbosch, J. 1973. Algorithm 457—Finding all cliques of an undirected graph. Commun. ACM 16 575–577. [Google Scholar]
- Connolly, M.L. 1983. Solvent-accessible surfaces of proteins and nucleic acids. Science 221 709–713. [DOI] [PubMed] [Google Scholar]
- Devos, D. and Valencia, A. 2000. Practical limits of function prediction. Proteins 41 98–107. [PubMed] [Google Scholar]
- Exner, T.E., Keil, M., and Brickmann, J. 2002a. Pattern recognition strategies for molecular surfaces. I. Pattern generation using fuzzy set theory. J. Comput. Chem. 23 1176–1187. [DOI] [PubMed] [Google Scholar]
- ———. 2002b. Pattern recognition strategies for molecular surfaces. II. Surface complementarity. J. Comput. Chem. 23 1188–1197. [DOI] [PubMed] [Google Scholar]
- Heifetz, A., Katchalski-Katzir, E., and Eisenstein, M. 2002. Electrostatics in protein–protein docking. Protein Sci. 11 571–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, K.Y., Chung, J.H., Kim, S.-H., Han, Y.S., and Cho, Y. 1999. Structure-based identification of a novel NTPase from Methanococcus jannaschii. Nat. Struct. Biol. 6 691–696. [DOI] [PubMed] [Google Scholar]
- Kinoshita, K., Sadanami, K., Kidera, A., and Go, N. 1999. Structural motif of phosphate-binding site common to various protein superfamilies: All-against-all structural comparison of protein-mononucleotide complexes. Protein Eng. 12 11–14. [DOI] [PubMed] [Google Scholar]
- Kinoshita, K., Furui, J., and Nakamura, H. 2002. Identification of protein functions from a molecular surface database, eF-site. J. Struct. Funct. Genomics 2 9–22. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of proteins structures. J. Appl. Cryst. 24 946–950. [Google Scholar]
- Lo Conte, L., Brenner, S.E., Hubbard, T.J., Chothia, C., and Murzin, A.G. 2002. SCOP database in 2002: Refinements accommodate structural genomics. Nucleic Acids Res. 30 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt, E.A. and Murphy, M.E.P. 1994. Raster3D Version 2.0, a program for photorealistic molecular graphics. Acta Crystallogr. D 50 869–873. [DOI] [PubMed] [Google Scholar]
- Nakamura, H. and Nishida, S. 1987. Numerical calculations of electrostatic potentials of protein-solvent systems by the self consistent boundary method. J. Phys. Soc. Jpn. 56 1609–1622. [Google Scholar]
- Rosen, M., Lin, S.L., Wolfson, H., and Nussinov, R. 1998. Molecular shape comparisons in searches for active sites and functional similarity. Protein Eng. 11 263–277. [DOI] [PubMed] [Google Scholar]
- Tari, L.W., Matte, A., Pugazhenthi, U., Goldie, H., and Delbaere, L.T. 1996. Snapshot of an enzyme reaction intermediate in the structure of the ATP-Mg2+-oxalate ternary complex of Escherichia coli PEP carboxykinase. Nat. Struct. Biol. 3 355–63. [DOI] [PubMed] [Google Scholar]
- Teichmann, S.A., Murzin, A.G., and Chothia, C. 2001. Determination of protein function, evolution and interactions by structural genomics. Curr. Opin. Struct. Biol. 11 354–363. [DOI] [PubMed] [Google Scholar]
- Thornton, J.M., Todd, A.E., Milburn, D., Borkakoti, N., and Orengo, C.A. 2000. From structure to function: Approaches and limitations. Nat. Struct. Biol. 7 991–994. [DOI] [PubMed] [Google Scholar]
- Todd, A.E., Orengo, C.A., and Thornton, J.M. 2002. Sequence and structural differences between enzyme and nonenzyme homologs. Structure 10 1435–1451. [DOI] [PubMed] [Google Scholar]
- Venkatachalam, C.M., Jiang, X., Oldfield, T., and Waldman, M. 2003. LigandFit: A novel method for the shape-directed rapid docking of ligands to protein active sites. J. Mol. Graph. Model 21 289–307. [DOI] [PubMed] [Google Scholar]
- Wallance, A.C., Borkakoti, N., and Thornton, J.M. 1997. TESS: A geometric hashing algorithm for deriving 3D coordinate templates for searching structural databases. Application to enzyme active sites. Protein Sci. 6 2308–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, C.A., Kreychman, J., and Gerstein, M. 2000. Assessing annotation transfer for genomics: Quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J. Mol. Biol. 297 233–249. [DOI] [PubMed] [Google Scholar]