Abstract
The use of spectroscopy in the study of fatty acids binding to bovine β-lactoglobulin (BLG) appears to be a difficult task, as these acid compounds, assumed as the protein natural ligands, do not exhibit favorable optical response such as, for example, absorption or fluorescence. Therefore, the BLG fatty-acid equilibrium has been tackled by exploiting the competition between fatty acids and ANS, a widely used fluorescent hydrophobic probe, whose binding sites on the protein have been characterized recently. Two lifetime decays of the ANS–BLG complex have been found; the longer one has been attributed to the internal binding site and the shorter one to the external site. At increasing fatty acids concentration, the fractional weight associated with ANS bound to the internal site drops, in agreement with a model describing the competition of the dye with fatty acids, whereas the external site occupancy appears to be unaffected by the fatty acids binding to BLG. This model is supported by docking studies. An estimate of the acid-binding affinities for BLG has been obtained by implementing the fitting of the bound ANS intensities with a competitive binding model. A relevant dependence has been found upon the solution pH, in the range from 6 to 8, which correlates with the calyx accessibility modulated by the conformation of the EF loop. Fatty acids with longer aliphatic chains (palmitate and laurate) are found to display larger affinities for the protein and the interaction free energy nicely correlates with the number of contacts inside the protein calyx, in agreement with docking simulations.
Keywords: Fatty acid, β-lactoglobulin, ANS, time resolved fluorescence, binding, docking
Bovine β-lactoglobulin (BLG), the major component of ruminant milk whey, is a 162 residues soluble protein belonging to the lipocalin family. BLG proper functionality has not been fully clarified yet, despite the great number of studies performed. However, its presence in the milk of several mammals has suggested a nutritional role, supported also by its ability to interact with a great variety of hydrophobic ligands, such as retinol (Futterman and Heller 1972; Dufour et al. 1990; Dufour and Haertlé 1991; Cho et al. 1994; Narayan and Berliner 1997; Lange et al. 1998), fatty acids and triglycerides (Dufour et al. 1990; Frapin et al. 1993; Narayan and Berliner 1997; Wang et al. 1997a,b; Qin et al. 1998b; Wu et al. 1999; Zsila et al. 2002). The lipocalin family includes, among others, transport proteins such as the retinol-binding protein, the odorant-binding protein, and the major urinary protein, which all share the common structural feature of a β-barrel calyx, built from eight antiparallel β sheets, arranged as an ideal site for hydrophobic ligands (Brownlow et al. 1997). Among the above-cited ligands, fatty acids are the most abundant endogenous ligands of BLG, thus suggesting the importance of the study of their interaction with the protein. Some obstacles, however, are encountered in the direct study of their binding to the protein, as these acid compounds do not exhibit a convenient spectroscopic response (such as absorption, circular dichroism, or fluorescence). Measurements aimed at the determination of the fatty-acid-binding affinities have been performed by equilibrium dialysis or by an equilibrium partition method using labeled fatty acids (Spector and Fletcher 1970; Spector 1975; Anel et al. 1989; Pérez et al. 1992). Otherwise indirect procedures could be used following, for instance, how the response of other chromophores, intrinsic or bound to the protein, is affected by the presence of fatty acids. Several studies have been reported on changes of intrinsic (Tryptophan) fluorescence (Narayan and Berliner 1997, 1998; Wang et al. 1997b; Lange et al. 1998) or spectral shifts in the fluorescence emission of covalently bound probes (Richieri et al. 1992). However, the variations are often as small as 10% at most, thereby making difficult quantitative estimates of the binding constant. Structural studies of protein fatty-acid complexes, performed with X rays (Qin et al. 1998b; Wu et al. 1999) and NMR spectroscopy (Ragona et al. 2000) have given indications of the location of binding sites and of the protein residues that are involved in the interaction.
The present work deals with a novel investigation of the BLG fatty-acid binding, which exploits the competition between fatty acids and ANS, a widely used fluorescent hydrophobic probe whose binding sites to BLG have been characterized recently (Collini et al. 2000). We have chosen to follow the modification of the ANS fluorescence upon fatty-acid addition to a BLG–ANS solution in order to obtain information on (1) the preferred site for fatty-acid binding, once the locations of the ANS sites are known, and (2) the strength of the fatty-acid–BLG interaction. When performing a competition study, the probe ligand must be chosen in such a way that its binding affinity for the protein is large enough to ensure the observation of a meaningful fluorescent response of the bound dye, but also sufficiently low to allow dye displacement by the fatty acids. ANS has been chosen, as its binding characteristics fulfill the above requirements, its interaction constant lying around 103 M−1, depending upon solution pH, but nevertheless, lower than the values reported for the endogenous ligands of BLG.
The binding parameters were derived under different solution conditions, in the pH range from 6–8. This neutral-to-alkaline region is particularly interesting, as the charges on the protein modulate the accessibility to its hydrophobic calyx, an ideal site for small hydrophobic ligands. In fact, the EF loop, sensitive to pH, bends over the calyx entrance in a closed conformation when in acid solutions, whereas, at pH >7.5, it moves away from the calyx (Tanford transition), assuming an open conformation that favors ligand binding (Tanford et al. 1959; Qin et al. 1998a).
To investigate the nature of the fatty acid interaction with BLG, we have studied saturated fatty acids with different chain lengths: palmitic acid, lauric acid, and caprylic acid, which share a similar chemical structure, CH3(CH2)nCOOH, but possess aliphatic chains of different lengths (n = 14, 10, 6; see Fig. 1 ▶).
Figure 1.
Chemical structure of the investigated ligands; fatty acids and ANS (1-8-anilinonaphthalene sulfonate).
Time-resolved fluorescence measurements yielded ANS fluorescence decay parameters, lifetimes, and fractional intensities values, which have been fitted using a competition model in order to obtain the association constants for fatty-acid BLG interactions.
Fluorescence results have been accompanied by docking simulations performed using the program GRID.
Results and Discussion
Steady-state fluorescence measurements of the ANS–BLG complex in the presence of fatty acids have been performed in order to identify the suitable range of protein and probe concentration causing the largest fluorescence change upon fatty acid addition. The useful concentrations ranged from 5 to 10 μM for both ANS and BLG, depending upon the pH of the solution, which in turn affects the ANS-binding constant to BLG (Collini et al. 2000). To obtain efficient competition, the concentration of fatty acids was varied from 0 to 12 μM.
Fluorescence lifetime measurements performed with the three different fatty acids in the pH range described above have been fitted to a three-exponential decay, in which the lifetime of free ANS in solutions has been fixed to τF = 0.26 nsec. ANS bound to BLG displays two different lifetimes (~14 nsec and ~3 nsec) whose values are only slightly dependent on pH and ionic strength, in agreement with previous results (D’Alfonso et al. 1999; Collini et al. 2000). These lifetimes have been attributed to two different BLG-binding sites; the shorter lifetime corresponds to a site partially exposed to the solvent and located on a surface hydrophobic patch (Ragona et al. 1997; Fogolari at al. 1998). The longer one corresponds to a binding site shielded from the solvent and located inside protein calyx, in which the probe feels a nonpolar environment, thus decaying with a longer lifetime value.
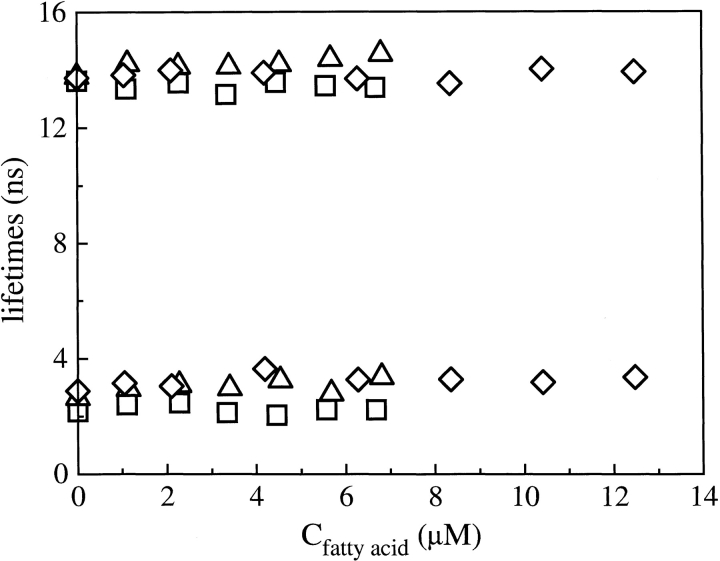
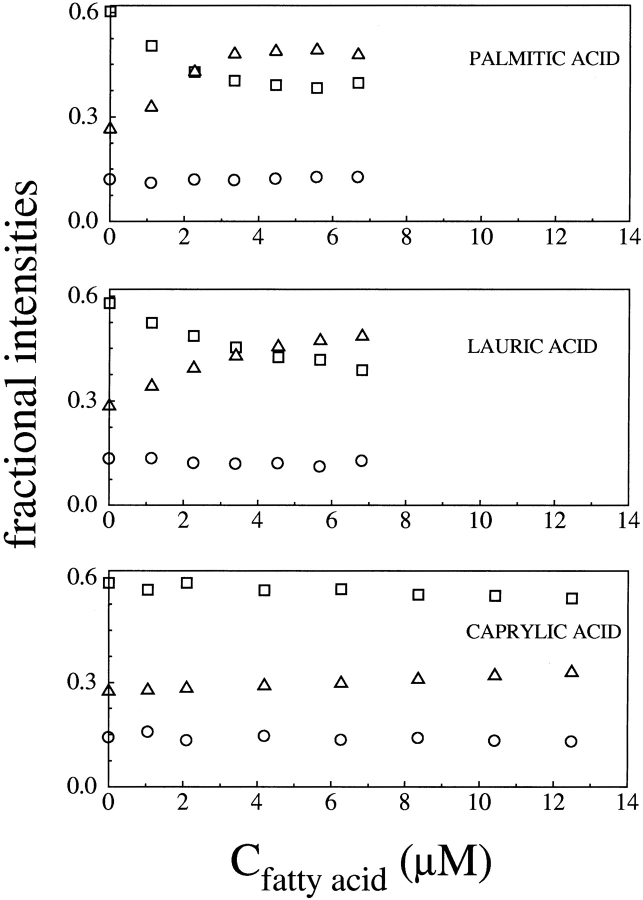
Fluorescence lifetime values of bound ANS are essentially constant, within the experimental error affecting the two exponential analysis, under all of the examined conditions, and they are independent of fatty-acid concentration. In Figure 2 ▶, for example, are shown the fluorescence lifetimes of bound ANS versus fatty-acid concentration for the titrations performed at pH 8.3 with palmitic, lauric, and caprylic acid, whereas Figure 3 ▶ reports the corresponding fractional intensities. The observation that the lifetimes of bound ANS are unaffected by fatty acids suggests that the presence of fatty acid does not appreciably alter the polarity of ANS-binding sites. It is clear from Figure 3 ▶ that the fractional intensities associated with the bound ANS longer lifetime, f1, and with the free ANS, fF, change at increasing fatty-acid concentration, whereas the fractional intensity associated with the bound ANS shorter lifetime, f2, remains essentially constant. The extent of these changes appear to be related to the specific fatty acid under study, being largest for palmitic acid, and also to the solution pH, being larger at the higher pH values.
Figure 2.
Fluorescence lifetimes of ANS bound to BLG versus fatty-acid concentration at pH 8.3. (□) Palmitic acid; (▵) lauric acid; (⋄) caprylic acid.
Figure 3.
Fractional intensities of ANS decay in the presence of BLG versus fatty-acid concentration at pH 8.3. Different symbols refer to different fractional intensities. (□) f1, corresponding to the longer bound ANS lifetime; (○) f2, corresponding to the shorter bound ANS lifetime; (▵) f3, corresponding to the free ANS lifetime.
To derive from the fractional intensities quantitative information on how the ANS bound concentration changes upon fatty-acid addition, one can introduce the ratio of the two bound ANS fractional intensities, f1/f2. In fact, as reported in Materials and Methods (Equation 6), the fractional intensities depend upon the lifetime, the corresponding dye concentration, and an unknown normalizing factor, which can cancel out when taking the ratio f1/f2:
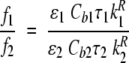 |
(1) |
The indexes 1 and 2 refer to BLG internal and external sites for ANS, respectively, as stated above. In view of the fact that the extinction coefficient of bound ANS was reported to be almost the same for the two sites (D’Alfonso et al. 1999), and the ANS lifetimes are found to be constant, it is reasonable to assume that the radiative lifetimes do not change (Robinson et al. 1978; Lakowicz 1999. The fractional intensity f2 is constant as well, thus suggesting that the concentration of ANS bound to the external site does not appreciably change upon fatty-acid binding.
Equation 1 can be rewritten by including all the constant terms in a proportionality factor as:
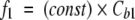 |
(2) |
In this way, it is possible to relate the experimental fractional intensity to the concentration of ANS bound to the internal site. It is worth noting that the decrease of f1 at increasing fatty-acid concentration is a strong indication that fatty acids compete with ANS only for the internal binding site. This finding is in good agreement with X-ray and NMR data on the complexes of BLG with dodecanoic and palmitic acids (Qin et al. 1998b; Wu et al. 1999; Ragona et al. 2000).
The aim of the following analysis is the determination of the fatty-acid-binding constant to BLG in terms of the known ANS-binding constant (Collini et al. 2000), by using a competitive model only for the BLG internal site. The concentration of ANS bound to the external site can be taken into account in the analysis as a renormalization factor of the total ANS concentration; nonetheless, this correction affects the value of the free ANS concentration within the experimental precision of its determination.
When both ANS and fatty acids are present in BLG solution, the following equilibria are present,
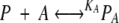 |
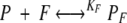 |
in which P, A, and F represent the free protein, ANS, and fatty-acid concentrations, PA and PF are the concentrations of protein with bound ANS or fatty acid, and KA and KF are the ANS and fatty-acid-binding constants, respectively. Then P0, A0, and F0 represent the total protein, ANS, and fatty-acid concentrations, respectively, and can be expressed as follows:
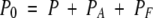 |
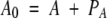 |
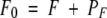 |
The concentration of ANS bound to the protein, PA, is the sum of ANS bound to both the external and the internal site of BLG:
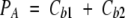 |
in which the indexes 1 and 2 refer to the internal and external BLG-binding sites, respectively. Because the two ANS-binding affinities (K1 and K2) are known, and Cb2 is constant, it is possible to define the new total dye concentration, A0* as:
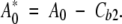 |
In this way, an equation relating the unknown fatty-acid-binding constant, KF, to the concentration of ANS bound to the internal site, Cb1, can be derived:
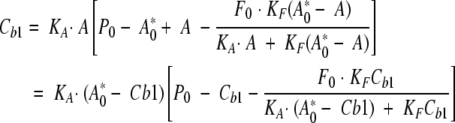 |
(3) |
By solving Equation 3 for Cb1, a cubic expression is obtained in Cb1, according to:
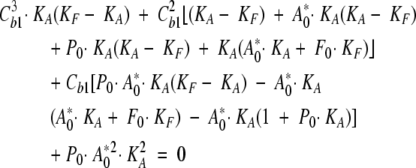 |
(4) |
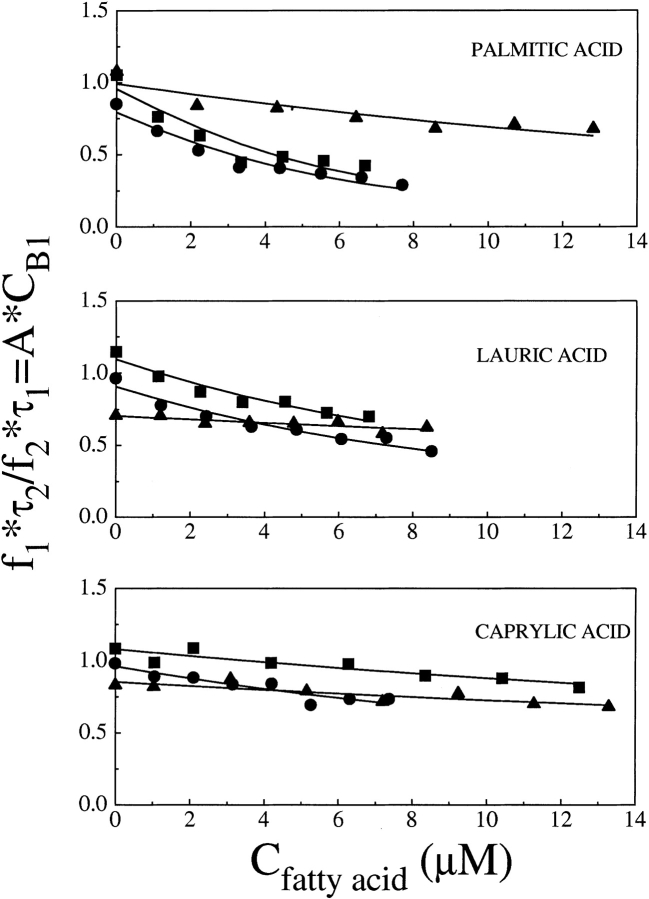
The amount of ANS initially bound to the external site (Cb2) can be neglected in the expression of the total protein concentration P0, as it induces a change in P0 of ~0.5%, at most. Similarly, the variation of A0 due to the free ANS initially bound in the internal site (at most equal to Cb1) can be neglected in the computation of both Cb1 and Cb2, because the maximum induced A0 variation would be of about ≈0.0002 μM, corresponding to a negligible Δf1≈0.7%. Therefore, the fractional intensity associated to the longer lifetime component can be fitted by Equation 2. However, it has been often observed that fit parameters, in a three-exponential decomposition of ANS lifetime decay, are not completely independent from each other. For this reason, instead of using the values of f1 obtained from the fit, we use the ratio f1τ2/f2τ1 = const’ × Cb1, thereby greatly reducing the noise on the derived parameters. The values of the fatty-acid association constant, KF, are shown in Table 1. The good agreement between the data and the fitting model is shown in Figure 4 ▶, in which f1τ2/f2τ1 values and the fitting functions are shown versus fatty-acid concentration. It was not possible to determine the binding constant for caprylic acid, at pH 6.2 and pH 7.2, as the change in the fractional intensity of ANS bound to the internal site is too small; in this case, only an upper limit estimate can be given.
Table 1.
Binding affinities of the fatty acid studied in the different experimental conditions examined
| KF (*102) M−1 | pH 6 | pH 7 | pH 8 |
| Palmitic acid | 660 ± 30 | 4700 ± 100 | 5050 ± 200 |
| Lauric acid | 235 ± 20 | 1630 ± 160 | 1350 ± 140 |
| Caprylic acid | <3 | <7 | 255 ± 50 |
Figure 4.
Ratio f1τ2/f2τ1 = const’ × Cb1 versus fatty-acid concentration at the different investigated pH values: (▪) pH 8.3; (•) pH 7.2; (▴) pH 6.2. Broken lines refer to the fitting functions as defined in the text, Equations 4–6.
A few values for the association constant of palmitic acid to BLG at different pHs and ionic strengths are reported in the literature. Association constants of palmitic and lauric acids, reported in an earlier work (Spector and Fletcher 1970) are in good agreement with our data and display a similar pH dependence. A larger (about fourfold) association constant was reported recently by Narayan and Berliner (1998). Binding constants were also reported for different fatty acids, such as oleic (KF ≅ 0.4× 105 M−1) and stearic (KF ≅ 4× 105 M−1; Spector and Fletcher 1970) and for cholesterol (KF ≅ 3× 107 M−1; Wang et al. 1997b) and have been summarized in Table 2 of the review by Sawyer and Kontopidis (2000).
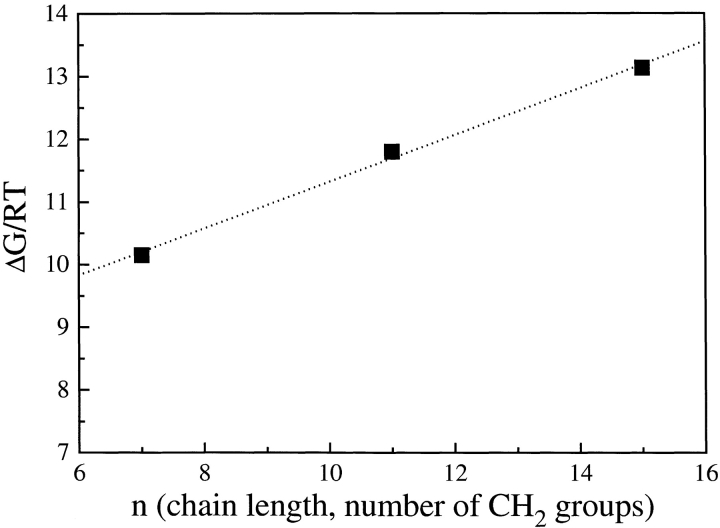
From the analysis of the association constants reported in Table 1, it is clear that near the Tanford transition, in the pH range 6.2–8.3, the strength of the interaction increases with pH, that is, it depends on the opening of the EF loop, regardless of the length of the fatty-acid chain. A strong dependence of the binding constant upon the fatty-acid chain length is also observed, with larger binding affinities for the longer palmitic acid. When the free energies (ΔG) for the binding of fatty acids to BLG are derived from the equilibrium constants at pH 8.3, in which the largest fractional intensity changes allowed for the determination of all the interaction constants, a linear relation of ΔG versus the number of methylene groups of the alkylic fatty-acid chain is found, as shown in Figure 5 ▶. This free energy can be decomposed in a leading term (ΔG0), which might depend upon the electrostatic interaction between the fatty-acid carboxylate and BLG lysines located at the calyx entrance (Wu et al. 1999), plus a contribution arising from the number of hydrophobic contacts between fatty-acid methyl and methylene groups (nmet) and BLG residues within the calyx ΔG = ΔG0 + ψ nmet. Data fitting yields ΔG0 = 7.6 RT and ψ = 0.37 RT per methylene with nmet = 7, 11, 15 for caprylic, lauric, and palmitic acid, respectively.
Figure 5.
Free energy difference for fatty-acid binding to BLG at pH 8.3 versus the number of methylene groups in the fatty-acid chain.
To complement experimental fluorescence data with an analysis, at a molecular level, of the binding specificity, the three fatty acids and the ANS probe were docked within BLG by use of the GRID-docking program (Goodford 1985, Kastenholz et al. 2000). The GRID method has been developed for determining energetically favorable binding sites for small chemical groups (probes) on a target molecule (protein). The probe groups are small chemical entities, such as carboxy oxygens, water molecules, and hydrophobic probes, which are moved through a regular grid of points around the target molecule in order to calculate, at each point of the grid, an interaction energy, thus generating a molecular interaction field (MIF). The ligand molecule is represented as a collection of GRID probes, and MIFs calculated for each probe are used to define the ligand position with respect to the target and to estimate the binding energy of the ligand at its binding site. The docked solutions are ranked on the basis of the total energy of interaction with the target, evaluated by GRID forcefield, to estimate the binding affinity of each solution.
Palmitic, lauric, and caprylic acids have been docked to the BLG structure obtained for the apo-protein at pH 7.1 (PDB code 1bsy), with the EF loop in the open conformation, as it was shown (Ragona et al. 2000) that this is a prerequisite for fatty-acid binding to BLG. At low pH, when the EF loop is in the closed conformation, the protein is unable to bind fatty acids.
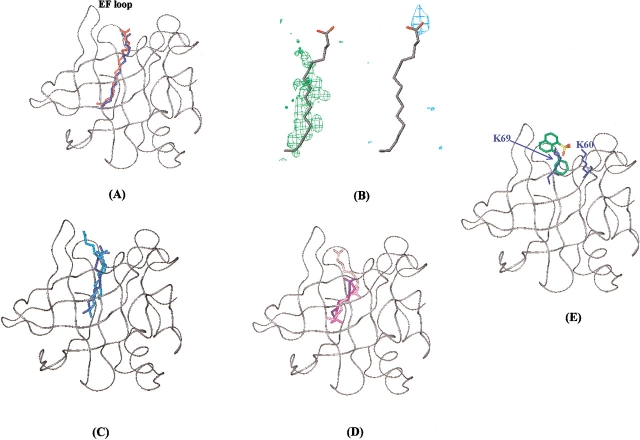
The X-ray structure of BLG complexed with palmitic acid (Wu et al. 1999) indicated, in agreement with NMR data (Ragona et al. 2000), that the fatty acid lies within the central cavity of the protein, with the methyl end buried deeply within the protein and the carboxyl end, protruding outside BLG open end. The GRID docking program generates a single solution for palmitic acid, with an interaction energy of −14.8 Kcal/mole. To assess the quality of the docking procedure, we compared the coordinates of the docked palmitic acid with the ones of the crystal structure (PDB code 1b0o; Fig. 6A ▶); a very good agreement is observed for the positioning of both the aliphatic chain and the carboxyl tail. The inspection of MIFs, generated for hydrophobic and carboxyl oxygen probe, graphically represented as three-dimensional contours around the target molecule, describing the probe attractive interaction regions, indicates that palmitic-acid docking solution maximizes both electrostatic and hydrophobic interactions (Fig. 6B ▶). This indicates that both of these interactions are relevant for selectivity toward this ligand.
Figure 6.
Docking solutions obtained from GRID program for interaction of BLG with palmitic, lauric, caprylic acids, and ANS. (A) Superposition of predicted (blue) and experimentally observed (red) positions of palmitic acid within BLG. BLG backbone (PDB code 1bsy) is shown in gray in a ribbon representation. (B) Graphical representation of MIF in the region of palmitic acid binding site. Palmitic acid docking solution is shown in gray. The green and blue contours indicate regions of favorable interaction energies for hydrophobic (energy level = −0.5 Kcal/mole) and carboxyl oxygen (energy level = −6.2 Kcal/mole) probes, respectively. (C) Docking solutions obtained for lauric acid. (D) Docking solutions obtained for caprylic acid. Darker colors correspond to lower energy solutions. (E) Docking solution obtained for ANS. ANS molecule is colored by atom type. K60 and K69 side-chains are displayed in blue.
Three different solutions have been generated by GRID for lauric acid, the ligand is always located in the internal cavity with the carboxyl tail toward BLG open end (Fig. 6C ▶). A careful analysis of the three predicted positions with respect to MIFs, indicated that the aliphatic chain always lies along BLG “hydrophobic spine,” but only in the second docking solution, the carboxyl group is close to a favorable electrostatic interaction region. Moreover, this solution displays the highest similarity with the previously determined X-ray structure of 12-bromododecanoic acid complexed to BLG (Qin et al. 1998b).
The comparison of palmitic acid-binding energy (−14.8 Kcal/mole), with the most reliable docked solution obtained for lauric acid (−11.9 Kcal/mole), points to a lower affinity for this shorter fatty acid, in agreement with the fluorescence data showing a decrease of binding affinity with the shortening of the fatty-acid chain.
Finally, the shorter caprylic acid, docked to BLG, afforded four solutions (Fig. 6D ▶) in which the aliphatic chains fit, for different extensions, the favorable hydrophobic interaction regions. The energy values obtained for the four solutions range from −13 to −9 Kcal/mole; however, the location of the carboxyl tail never fits the favorable electrostatic interaction regions. This observation suggests a very low-binding specificity of caprylic acid to BLG, possibly due to its minor sterical hindrance, conferring a large conformational freedom within a cavity that preferentially hosts the endogenous longer-chain fatty acids (Perez et al. 1989).
Concerning the GRID docking simulations relative to the BLG–ANS complex, three solutions were generated with energy values ranging from −6.2 to −5.5 Kcal/mole. The best docking solution, selected on the basis of MIFs analysis, presents ANS laying at the entrance of the cavity, with the aniline aromatic group pointing within the internal cavity and the negatively charged sulphonate group, in close contact with K60 and K69, nicely fitting the favorable electrostatic interaction field generated by GRID (Fig. 6E ▶). The obtained energy values suggest a lower affinity of ANS with respect to fatty acids.
Conclusions
The competition of ANS with fatty acids for a site on BLG, as detected here by time-resolved fluorescence of the dye during titration, has led to establish the strength of the protein to acids affinity. In particular, the fatty-acid-binding affinities values, which were shown to increase with pH in the range 6.2–8.3, independent of the acid-chain length, confirm the major role of the binding played by the EF loop conformation. Furthermore, palmitic acid, exhibiting the longest aliphatic chain, was shown to display the highest affinity for BLG, and a linear dependence of the binding-free energy versus fatty-acid chain length was observed, in agreement with the results of docking simulations performed using GRID. Both fluorescence data and docking simulations thus suggest the leading role of the electrostatic interactions in modulating the binding interaction energies. Docking solutions for the short caprylic acid, consistent with the very low affinity measured at all the tested pHs, were characterized by a large variability of carboxylate locations, and did not fit the favorable electrostatic interaction regions.
The docking solution obtained for ANS is in nice agreement with fluorescence data, indicating that the ANS probe lays within the protein (Collini et al. 2000). The comparison of the GRID energy figures of the investigated complexes indicate that the ANS-binding energy values are lower than those obtained for fatty acids, in agreement with fluorescence lifetime competition data, which suggest that fatty acids displace ANS.
It is worth mentioning that the higher specificity displayed by BLG for palmitic acid correlates well with biochemical data on the composition of fatty acids bound to BLG isolated from milk, in which palmitic acid is 30 times more abundant than lauric acid (Perez et al. 1989). This figure should be compared with that reported for cow milk fatty-acid content showing only an eightfold amount of palmitic acid, with respect to lauric acid (Belitz and Gosch 1999). The larger amount of palmitic acid complexed to BLG can be justified confidently by the stronger affinity of the protein for this ligand.
Materials and methods
Sample preparation
Lyophilized bovine β-lactoglobulin, genetic variant B, (lot. 11K7032, Sigma-Aldrich Inc.) was dissolved in the proper buffer at the desired concentration determined photometrically using the molar extinction coefficient at the absorption peak, ɛ(280) = 17600 cm−1 M−1. Palmitic acid, lauric acid, and caprylic acid have been purchased from Sigma Chemical Co. and dissolved (or diluted in the case of caprylic acid) in ethanol at a concentration of ~1 mM (stock solutions). ANS (8-anilino-1-naphthalenesulfonic acid) ammonium salt has been purchased from Fluka Chemical Co. Dye concentrations were determined photometrically using ɛ(350) = 5000 cm−1 M−1. Fresh BLG–ANS solutions were prepared before each measurement by adding to a BLG solution proper aliquots of a stock solution of ANS dissolved in the desired buffer, obtaining final probe and protein concentrations of 5–10 μM, depending upon the particular experiment, as explained in the Results section.
Phosphates buffers used in the titration experiments were as follows: (1) 0.010 M KH2PO4-Na2HPO4 at pH 6.2; (2) 0.010 M KH2PO4-Na2HPO4 at pH 7.2; (3) 0.010 M KH2PO4-Na2HPO4-NaOH at pH 8.3. All of the reagents used in sample preparation were of analytical grade.
Fluorescence measurements
Fluorescence titration experiments with fatty acids were performed at 25°C by adding small aliquots (typically 2μL) of the fatty-acid stock solutions to a BLG–ANS solution, reaching a concentration ratio fatty acid:BLG >1:1 in the presence of, at most, 1% w/w ethanol. Such a small fatty-acid total added volume allows us to consider the protein and ANS concentrations as constants during the titration; the concentration values used were 5 or 10 μM, according to the solution pH value. Each titration has been performed at least twice.
Steady-state spectra were acquired on a Cary Eclipse (Varian Inc.) spectrofluorometer, recording the fluorescence signal between 380 and 650 nm after excitation at 363 nm.
Dynamic fluorescence measurements were performed with a frequency-modulated phase fluorometer (Digital K2, I.S.S.). The excitation was accomplished by the 363.7-nm line of an Argon ion laser at 30 mW power (2025, Spectra Physics). For further details, see Collini et al. 1995. Digital data acquisition and storage was provided by the ISS-A2D ACD card inserted in a personal computer. For each data set, at least 15 logarithmically spaced frequencies were used in the range 2–220 MHz with a cross-correlation frequency of 400 Hz. Each lifetime measurement has been repeated at least twice in order to obtain an estimate of the errors affecting the results. Phase angles and modulation ratios accuracy were of 0.2° and 0.004, respectively. Lifetime measurements have been performed under the magic angle conditions and a long pass filter at 435 nm (Andover Co.) was used to cut Rayleigh and Raman scattering. A solution of dimethyl-popop [1,4-bis(4-methyl-5-phenyloxazol-2-yl)benzene] in ethanol was used as a reference sample of known lifetime (τ = 1.45 nsec). Data fitting was accomplished by means of a least square minimization procedures based on the Marquardt algorithm. Fluorescence lifetimes were analyzed in terms of sums of discrete exponential components, with the lifetimes values τi and their corresponding fractional intensities fi as unknown parameters, according to the equations:
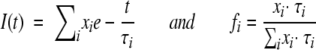 |
(5) |
in which xi represent the pre-exponential factors, which are proportional to the concentration (C), to the molar extinction coefficient (ɛ) and to the radiative constant (kR) of the corresponding emitting chromophore, leading to:
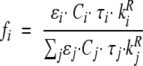 |
(6) |
Circular dichroism measurements
To test whether the BLG conformation was affected by the addition of ethanol or palmitic acid, circular dichroism (CD) experiments were performed with a Jasco (Easton) J-500A spectropolarimeter both in the near (350–250 nm) and in the far (250–190 nm) UV, by use of 1- and 0.2-cm path length cuvettes, 40- and 4-μM protein concentrations, ethanol concentration about 1% w/w, and 12-μM palmitic acid samples. In all of the tested experiments, the presence of a small quantity of ethanol or of the fatty acids used did not induce any modification in BLG ellipticity.
GRID calculations
BLG X-ray coordinates, obtained for the apo-protein at pH 7.1 (PDB code 1bsy), were used for the docking program. Crystallographic water molecules were removed. The coordinates of the palmitic, lauric, and caprylic acids were derived from the coordinates of palmitic acid bound to BLG (PDB code 1b0o). The fatty-acid carboxyl group was considered deprotonated, bearing a net negative charge of −1. ANS molecule was built with InsightII (Accelrys), and minimized with CVFF forcefield (200 steps steepest descent and 2000 steps of conjugate gradient).
The calculation was performed with version 20 of the GRID software (Molecular Discovery Ltd). The protein was considered rigid, and hydrogens were added with the program GRIN (part of GRID package). The docking search was performed on the whole protein. All GRID input parameters retained their default values. The GRID probes used were DRY (for hydrophobic interactions), carboxy oxygen, water, and neutral hydrogen atom. The calculated molecular interaction fields (MIF) were inspected with Gview (part of GRID package). The docking results were visualized with Gview and InsightII (Accelrys).
Acknowledgments
We thank Valentina Guatteo for her help during measurements and Silvio Mecucci for his support with the GRID program. This work has been partly supported by the National Institute for the Physic of Matter (INFM), PAIS-FOLGIN. H.M., L.R, and M.C. acknowledge Ministero dell’Istruzione, delle Università e della Ricerca (M.I.U.R.) ex 40% 2002 and Fondo per gli Investimenti della ricerca di Base (F.I.R.B.) 2001 grants.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
BLG, bovine β-lactoglobulin
NMR, nuclear magnetic resonance
ANS, 1-8-anilinonaphthalene sulfonate
DMSO, dimethylsulfoxide
MIF, molecular interaction field
PDB, Protein Data Bank
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0304403.
References
- Anel, A., Calvo, M., Naval, J., Iturralde, M., Alava, M.A., and Piñeiro, A. 1989. Interaction of rat α-fetoprotein and albumin with polyunsaturated and other fatty acids: Determination of apparent association. FEBS Lett. 250 22–24. [DOI] [PubMed] [Google Scholar]
- Belitz, H.D. and Gosch, W. 1999. Food chemistry, p. 387. ed. Springer-Verlag, Heidelberg, Germany.
- Brownlow, S., Cabral, J.H.M., Cooper, R., Flower, D.R., Yewdall, S.J., Polikarpov, I., North, A.C.T., and Sawyer, L. 1997. Bovine β-lactoglobulin at 1.8 Å resolution—Still an enigmatic lipocalin. Structure 5 481–495. [DOI] [PubMed] [Google Scholar]
- Cho, Y., Batt, C.A., and Sawyer, L. 1994. Probing the retinol-binding site of bovine β-lactoglobulin. J. Biol. Chem. 269 11102–11107. [PubMed] [Google Scholar]
- Collini, M., Chirico, G., Baldini, G., and Bianchi, M.E. 1995. Conformation of short DNA fragments by modulated fluorescence polarization anisotropy. Biopolymers 36 211–225. [DOI] [PubMed] [Google Scholar]
- Collini, M., D’Alfonso, L., and Baldini, G. 2000. New insight on β-lactoglobulin binding sites by 1-anilinonaphthalene-8-sulfonate fluorescence decay. Protein Sci. 9 1968–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alfonso, L., Collini, M., and Baldini, G. 1999. Evidence of heterogeneous 1-anilinonaphthalene-8-sulfonate binding to β-lactoglobulin from fluorescence spectroscopy. Biochim. Biophys. Acta 1432 194–202. [DOI] [PubMed] [Google Scholar]
- Dufour, E. and Haertlé, T. 1991. Binding of retinoids and β-carotene to β-lactoglobulin. Influence of protein modifications. Biochim. Biophys. Acta 1079 316–320. [DOI] [PubMed] [Google Scholar]
- Dufour, E., Marden, M.C., and Haertlé, T. 1990. β-Lactoglobulin binds retinol and protoporphyrin IX at two different binding sites. FEBS Lett. 277 223–226. [DOI] [PubMed] [Google Scholar]
- Fogolari, F., Ragona, L., Zetta, L., Romagnoli, S., De Kruif, K.G., and Molinari, H. 1998. Monomeric bovine β-lactoglobulin adopts a β-barrel fold at pH 2. FEBS Lett. 436 149–154. [DOI] [PubMed] [Google Scholar]
- Frapin, D., Dufour, E., and Haertle, T. 1993. Probing the fatty acid binding site of β-lactoglobulins. J. Protein. Chem. 12 443–449. [DOI] [PubMed] [Google Scholar]
- Futterman, S. and Heller, J. 1972. The enhancement of fluorescence and the decreased susceptibility to enzymatic oxidation of retinol complexed with bovine serum albumin, β-lactoglobulin, and the retinol-binding protein of human plasma. J. Biol. Chem. 247 5168–5172. [PubMed] [Google Scholar]
- Goodford, P.J. 1985. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 28 849–857. [DOI] [PubMed] [Google Scholar]
- Kastenholz, M.A., Pastor, M., Cruciani, G., Haaksma, E.E., and Fox, T. 2000. GRID/CPCA: A new computational tool to design selective ligands. J. Med. Chem. 43 3033–3044. [DOI] [PubMed] [Google Scholar]
- Lakowicz, J.R. 1999. Principles of fluorescence spectroscopy, 2nd ed. Kluver Academic/Plenum Publishers, New York.
- Lange, D.C., Kothari, R., Patel, R.C., and Patel, S.C. 1998. Retinol and retinoic acid bind to a surface cleft in bovine β-lactoglobulin: A method of binding site determination using fluorescence resonance energy transfer. Biophys. Chem. 74 45–51. [DOI] [PubMed] [Google Scholar]
- Narayan, M. and Berliner, L.J. 1997. Fatty acids and retinoids bind independently and simultaneously to β-lactoglobulin. Biochemistry 36 1906–1911. [DOI] [PubMed] [Google Scholar]
- ———. 1998. Mapping fatty acid binding to β-lactoglobulin: Ligand binding is restricted by modification of Cys 121. Protein Sci. 7 150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, M.D., Diaz de Villegas, C., Sanchez, L., Aranda, P., Ena, J.M., and Calvo, M. 1989. Interaction of fatty acids with β-lactoglobulin and albumin from ruminant milk. J. Biochem. 106 1094–1097. [DOI] [PubMed] [Google Scholar]
- Perez, M.D., Sanchez, L., Aranda, P., Ena, J.M., Oria, R., and Calvo, M. 1992. Effect of β-lactoglobulin on the activity of pregastric lipase. A possible role for this protein in ruminant milk. Biochim. Biophys. Acta 1123 151–155. [DOI] [PubMed] [Google Scholar]
- Qin, B.Y., Bewley, M.C., Creamer, L.K., Baker, H.M., Baker, E.N., and Bewley, G.B. 1998a. Structural basis of the Tanford transition of bovine β-lactoglobulin. Biochemistry 37 14014–14023. [DOI] [PubMed] [Google Scholar]
- Qin, B.Y., Creamer, L.K., Baker, E.N., and Jameson, G.B. 1998b. 12-bromododecanoic acid binds inside the calyx of bovine β-lactoglobulin. FEBS Lett. 438 272–278. [DOI] [PubMed] [Google Scholar]
- Ragona, L., Pusterla, F., Zetta, L., Monaco, H.L., and Molinari, H. 1997. Identification of a conserved hydrophobic cluster in partially folded bovine β-lactoglobulin at pH 2. Fold. Des. 2 281–290. [DOI] [PubMed] [Google Scholar]
- Ragona, L., Fogolari, F., Zetta, L., Pérez, D.M., Puyol, P., De Kruif, K., Löhr, F., Rüterjans, H., and Molinari, H. 2000. Bovine β-lactoglobulin: Interaction studies with palmitic acid. Protein Sci. 9 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richieri, G.V., Ogata, R.T., and Kleinfeld, A.M. 1992. A fluorescently labeled intestinal fatty acid binding protein. Interactions with fatty acids and its use in monitoring free fatty acids. J. Biol. Chem. 267 23495–23501. [PubMed] [Google Scholar]
- Robinson, G.W., Robbins, R.J., Fleming, G.R., Morris, J.M., Knight, A.E.W., and Morrison, R.J.S. 1978. Picosecond studies of the fluorescence probe molecule 8-anilino-1-naphthalenesulfonic acid. J. Am. Chem. Soc. 100 7145–7150. [Google Scholar]
- Sawyer, L. and Kontopidis, G. 2000. The core lipocalin, bovine β-lactoglobulin. Biochim. Biophys. Acta 1482 136–148. [DOI] [PubMed] [Google Scholar]
- Spector, A.A. 1975. Fatty acid binding to plasma albumin. J. Lipid Res. 16 165–179. [PubMed] [Google Scholar]
- Spector, A.A. and Fletcher, J.E. 1970. Binding of long chain fatty acids to β-lactoglobulin. Lipids 5 403–411. [DOI] [PubMed] [Google Scholar]
- Tanford, C., Bunville, L.G., and Nozaki, Y. 1959. The reversible transformation of β-lactoglobulin at pH 7.5. J. Am. Chem. Soc. 81 4032–4035. [Google Scholar]
- Wang, Q., Allen, J.C., and Swaisgood, H.E. 1997a. Binding of retinoids to β-lactoglobulin isolated by bioselective adsorption. J. Dairy Sci. 80 1047–1053. [DOI] [PubMed] [Google Scholar]
- ———. 1997b. Binding of vitamin D and cholesterol to β-lactoglobulin. J. Dairy Sci. 80 1054–1059. [DOI] [PubMed] [Google Scholar]
- Wu, S., Perez, M.D., Puyol, P., and Sawyer, L. 1999. β-Lactoglobulin binds palmitate within its central cavity. J. Biol. Chem. 274 170–174. [DOI] [PubMed] [Google Scholar]
- Zsila, F., Imre, T., Szabo, P.T., Bikadi, Z., and Simonyi, M. 2002. Induced chirality upon binding of cis-parinaric acid to bovine β-lactoglobulin: Spectroscopic characterization of the complex. FEBS Lett. 520 81–87. [DOI] [PubMed] [Google Scholar]