Abstract
A method to predict lipoprotein signal peptides in Gram-negative Eubacteria, LipoP, has been developed. The hidden Markov model (HMM) was able to distinguish between lipoproteins (SPaseII-cleaved proteins), SPaseI-cleaved proteins, cytoplasmic proteins, and transmembrane proteins. This predictor was able to predict 96.8% of the lipoproteins correctly with only 0.3% false positives in a set of SPaseI-cleaved, cytoplasmic, and transmembrane proteins. The results obtained were significantly better than those of previously developed methods. Even though Gram-positive lipoprotein signal peptides differ from Gram-negatives, the HMM was able to identify 92.9% of the lipoproteins included in a Gram-positive test set. A genome search was carried out for 12 Gram-negative genomes and one Gram-positive genome. The results for Escherichia coli K12 were compared with new experimental data, and the predictions by the HMM agree well with the experimentally verified lipoproteins. A neural network-based predictor was developed for comparison, and it gave very similar results. LipoP is available as a Web server at www.cbs.dtu.dk/services/LipoP/.
Keywords: Signal peptides, lipoprotein prediction, HMM, neural networks
Bacterial lipoproteins consist of a large group of proteins with many different functions. The characteristic feature of all lipoproteins is a signal sequence in the N-terminal end, followed by a cysteine (Hayashi and Wu 1990). The signal sequence is cleaved by signal peptidase II (SPaseII), also called lipoprotein signal peptidase (Lsp). These lipoprotein signal peptides are quite similar to the signal peptides of secreted proteins, which are cleaved by signal peptidase I (SPaseI). So far, a few hundred putative lipoproteins in Gram-negative Eubacteria have been annotated in SWISS-PROT (Bairoch and Apweiler 2000).
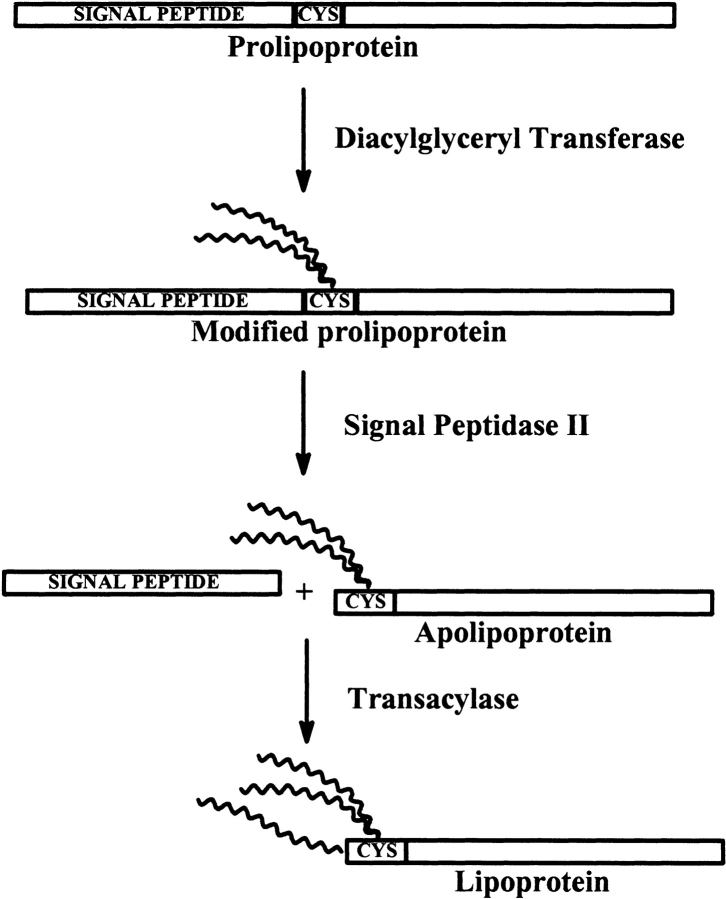
Biosynthesis of lipoproteins in Gram-negative and Gram-positive bacteria consists of three steps, as shown in Figure 1 ▶: transfer of a diacylglyceride to the cysteine sulphydryl group of the unmodified prolipoprotein; cleavage of the signal peptide by signal peptidase II, forming an apolipoprotein; and, finally, acylation of the α-amino group of the N-terminal cysteine of the apolipoprotein (Sankaran and Wu 1994). Before the processing of the prolipoprotein, which takes place on the periplasmic side of the inner membrane, the prolipoprotein is exported through the inner membrane by the general secretory pathway that is also used by secretory proteins processed by SPaseI (Hayashi and Wu 1990). In Gram-negative bacteria, the lipoproteins are anchored to either the inner or the outer membrane, and a single amino acid in position +2 is proposed to determine the final destination of the lipoproteins (Yamaguchi et al. 1988; Seydel et al. 1999). For more details about biosynthesis and export of lipoproteins, see Braun and Wu (1994).
Figure 1.
Biosynthesis of a lipoprotein. Lipids are attached to cysteine. Peptides are shown to the left and to the right of the cysteine residue. Catalytic enzymes are written beside reaction arrows.
The signal sequence can be divided into three regions: an n-region, an h-region, and a c-region. The n-region is characterized by presence of the positive amino acids lysine and/or arginine, the h-region consists of hydrophobic amino acids, and the c-region has a characteristic region of four amino acids around the cleavage site that is very well conserved, a so-called lipobox. The most conserved amino acids in the lipobox are a leucine in position −3 from the cleavage site, an alanine in position −2, and a glycine or an alanine in position −1. The cysteine at position +1 is required: LA(G,A) ↓ C (von Heijne 1989). The consensus for the lipoprotein signal sequence has previously been characterized further, so it could be used for lipoprotein predictions. One example is the consensus made by von Heijne, (LVI)(ASTG)(GA) ↓ C, requiring only one match to the first two positions. This pattern was able to discriminate between all lipoprotein signal peptides and SPaseI-cleaved signal peptides known at the time (von Heijne 1989). The lipoprotein predictor in PSORT (Nakai and Kanehisa 1991) integrates the von Heijne consensus sequence in its predictions. Another example is the Prosite pattern PS00013 {DERK} (6)(LIVMFWSTAG)(2)(LIVMFYSTAGCQ) (AGS) ↓ C, where {DERK}(6) means that none of the four amino acids are allowed in the first six positions (position −10 to −5 relative to the cleavage site). The pattern has two additional rules: The cysteine must be between position 15 and 35, and at least one lysine or arginine must be in one of the first seven positions of the signal peptide (Falquet et al. 2002). More recently, a new regular expression was made for Gram-positive bacteria (Sutcliffe and Harrington 2002).
The lipoprotein signal peptide has been compared with the SPaseI-cleaved signal peptides. The lipoprotein signal peptides have a similar n-region, but the h-regions of lipoprotein signal peptides are shorter and the SPaseI-cleaved signal peptides have a polar c-region before the cleavage site (Klein et al. 1988; von Heijne 1989). For lipoproteins, as well as for the SpaseI-cleaved proteins, the n- and h-regions are required for the translocation of the uncleaved protein precursor through the inner membrane. The c-region is necessary for the recognition of the cleavage site by the signal peptidase (von Heijne 1990).
Methods for prediction of SPaseI-cleaved signal peptides have been around for some time (Nakai and Kanehisa 1991; Nielsen et al. 1997). The performance of these methods is generally quite good, but it is a problem to discriminate SPaseI-cleaved signal peptides from SPaseII-cleaved signals and N-terminal transmembrane helices (Nielsen et al. 1997; Nielsen and Krogh 1998). Similarly, methods for predicting transmembrane helices often, by mistake, predict signal peptides as membrane helices (for example, see Krogh et al. 2001).
Here we present a method to predict lipoproteins in Gram-negative bacteria and their signal peptide cleavage site based on a hidden Markov model (HMM) or a neural network. Both methods are significantly better than the above-mentioned existing methods. The HMM is trained on both SPaseI-cleaved proteins, lipoproteins, and cytoplasmic and transmembrane proteins, and it is able to classify an N-terminal protein sequence as a lipoprotein signal peptides, a SPaseI-cleaved signal peptide, or a protein without a signal sequence (cytoplasmic or transmembrane) with very low error rates. The HMM is also able to predict the cleavage site in both SPaseI- and SPaseII-cleaved signal peptides.
Results
Protein sets for training and testing was extracted from SWISS-PROT as described in Materials and Methods. They consisted of lipoproteins, SPaseI-cleaved proteins, cytoplasmic proteins from the two Gram-negative phyllums Proteobacteria and Spirochetes (order: Spirochaetales), and transmembrane proteins from phyllums Proteobacteria and Gracilicutes.
Analysis of signal peptides
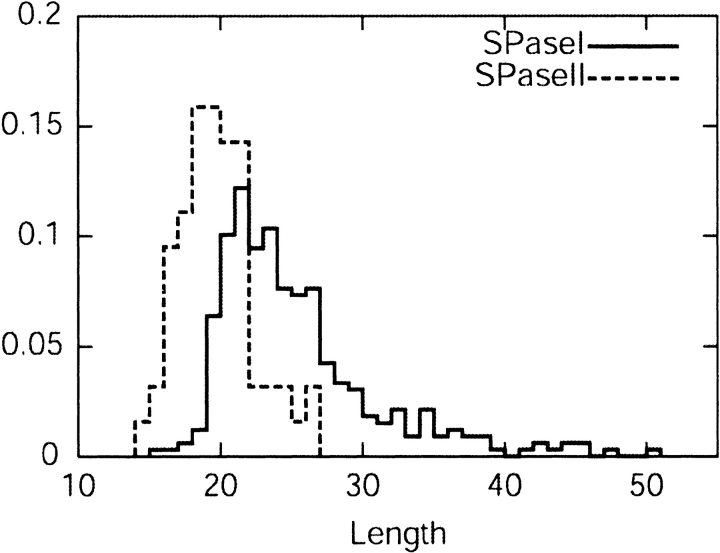
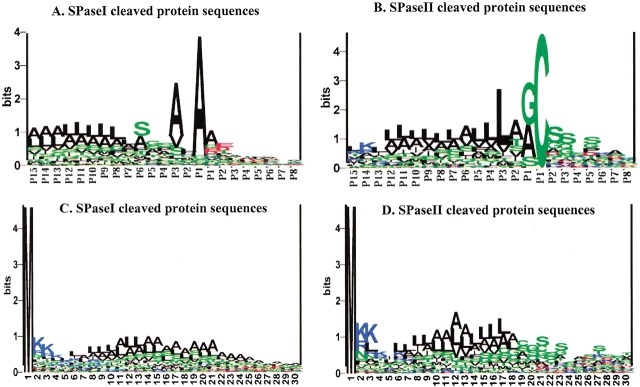
The length distributions of the two kinds of signal peptides are shown in Figure 2 ▶. The mean length of lipoprotein signal peptide is found to be 19.3, and for the SPaseI-cleaved signal peptide, it is 24.9. Sequence logos (Schneider and Stephens 1990) for the regions close to the cleavage sites (Fig. 3A,B ▶) show that the cleavage site consensus differs in amino acid distribution, which corresponds well with the fact that the signal peptides are cleaved by different proteolytic enzymes. The lipoproteins must have a cysteine after the cleavage site, whereas the SPaseI-cleaved signal peptide can have several different amino acids in the first position after the cleavage site. The signal parts to the left of the cleavage site differ as well. Figure 3, A and B ▶, indicates that the hydrophobic region is closer to the cleavage site for the lipoproteins than for the SPaseI-cleaved signal peptides. The SPaseI-cleaved signal peptides have a polar region right before the cleavage site (mostly serine). Figure 3, C and D ▶, shows the sequence logos of the first 30 amino acids for SPaseI- and SPaseII-cleaved proteins. SPaseI and SPaseII signal peptides both have some positive amino acids in the beginning of the sequence followed by a hydrophobic region with a similar amino acid distribution after that, and the similarity between the sequences corresponds well with the fact that all signal peptides are recognized by the same secretory enzymes. Figure 3, C and B ▶, also shows that the hydrophobic region of the lipoproteins is shorter than the one for secretory proteins, as expected. Cytoplasmic proteins do not have a preference for a particular amino acid in any positions besides the first methionine.
Figure 2.
Length distribution for lipoprotein signal peptides and for SPaseI-cleaved signal peptides.
Figure 3.
Sequence logos of cleavage sites for SPaseI-cleaved proteins (A)and lipoproteins (B) aligned at the cleavage sites (cleavage is between positions −1 and 1). Sequence logos of the 30 N-terminal residues for SPaseI-cleaved protein precursors (C) and lipoprotein precursors (D). A logo displays the amino acid conservation at each position as the information content measured in bits (Schneider and Stephens 1990). Black indicates hydrophobic amino acid (AA); green, neutral/polar AA; blue, positive AA; and red, negative AA.
Neural networks
The logos (Fig. 3 ▶) showed that the lipoprotein cleavage site consensus is quite different from the one for SPaseI-cleaved proteins, whereas the rest of the signal peptides are quite similar and are therefore a weaker discrimination factor. Therefore, we chose to base the prediction of lipoproteins with neural networks on whether a cysteine belonged to a lipoprotein cleavage site or not. The neural network training was thus carried out by using the lipoprotein cleavage site in lipoproteins as positive examples and all remaining cysteines from lipoproteins, SpaseI-cleaved proteins, and cytoplasmic proteins as negative examples. The neural network was designed to classify whether a cysteine in the center of a symmetric window was a lipoprotein cleavage site or not as in Nielsen et al. (1997). The number of hidden neurons was varied from zero to four, and the size of the symmetric input window was varied from 27 to 33.
The neural networks were evaluated by their performance on the test data sets by cross-validation, and the correlation coefficient were calculated (Matthews 1975). Based on the correlation coefficients and the number of lipoproteins predicted, the best network was chosen and the optimal parameters were estimated.
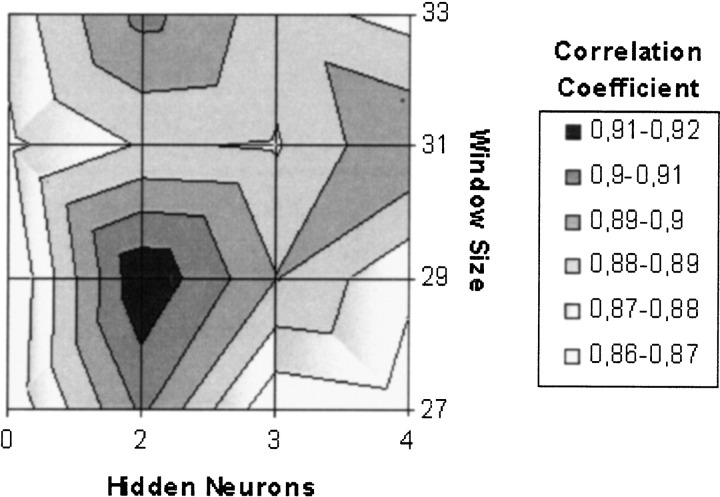
Judging from the correlation coefficients, the best neural network prediction (Fig. 4 ▶) was obtained for neural networks with the window size 29 and two hidden neurons, which was chosen as optimal parameters. For this neural network, there were 61 true positives (96.8% of all positives) and eight false positives (1.1% of all negative). Some of the other neural networks with high correlation coefficients had less false negatives but also less true positives. The network with these optimized parameters was used in all the following analysis. With this neural network, none of the transmembrane proteins were predicted as lipoproteins.
Figure 4.
Correlation coefficient as a function of window size and number of hidden neurons.
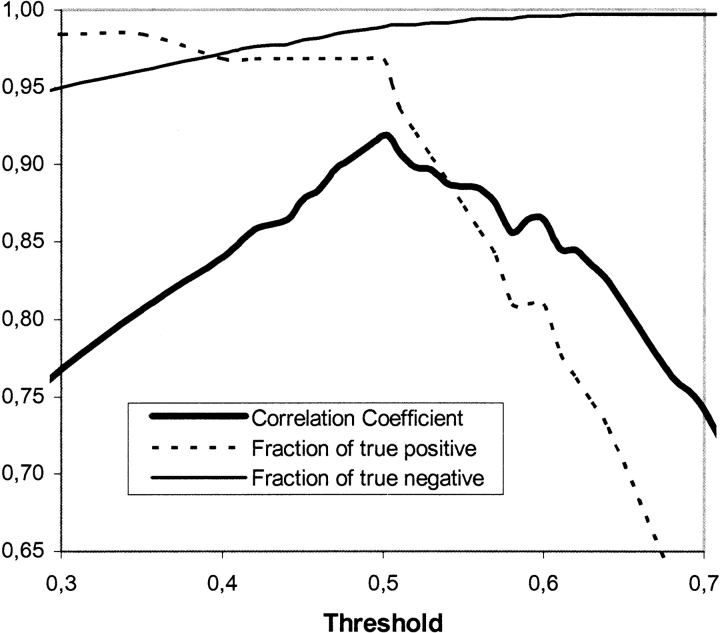
The fractions of true positive and true negative and the correlation coefficient are dependent on the threshold for the output neurons, and when the number of true positive predictions increase, it is consequently difficult to avoid an increase in false-positive predictions as well. Figure 5 ▶ illustrates this. When the threshold is raised, the number of true positives decreases remarkably, whereas the number of false positive remains constant. The opposite happens when the threshold is lowered, and the correlation coefficient has a visible maximum at threshold 0.5. If the number of false positives has to be reduced significantly, the number of true positives decreases even more. When the threshold is raised, the probability of the predicted lipoprotein actually being a lipoprotein increases. For example, with a threshold of 0.85, all the predicted lipoproteins are true positives. Therefore, the variation of the threshold can be used, if the relative number of false positive needs to be lowered, even though the number of true positives hereby decreases.
Figure 5.
Correlation coefficient and fraction of true positives and true negatives as a function of the threshold.
HMMs
The classification system was made as an HMM with four branches. From a “begin” state of the model, there are transitions to each of these branches or submodels:
SPaseI-cleaved signal peptides
The submodel for signal peptides is shown in Figure 6 ▶. It has states modeling the n-region, the h-region, and the c-region. It also models part of the protein after the cleavage site. The signal peptide model is very similar to the one in Nielsen and Krogh (1998), but is simplified a little. Initially, the same model as in Nielsen and Krogh (1998) was used, but after estimation, states and transitions with very small probabilities were eliminated.
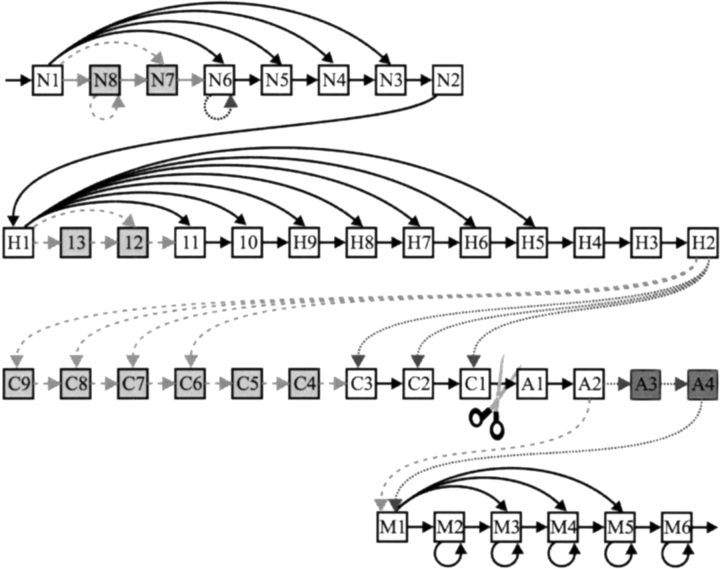
Figure 6.
The architecture of the SPaseI and SPaseII models. N-states model the n-region; H-states model the h-region; C-states and A-states model the regions before and after the cleavage site, respectively; and M-states model the remaining residues. All N-states except N1 are tied, all H-states are tied, states C7–C9 are tied, and all M-states are tied. Dashed transitions and light gray states are present only in the model of SpaseI-cleaved signal peptides, and dotted transitions and dark gray states are present only in the model of lipoproteins.
SPaseII-cleaved signal peptides (lipoproteins)
The layout of the model for lipoprotein signal peptides is essentially the same as the one for signal peptides, as shown in Figure 6 ▶. The differences from the signal peptide model were again arrived at by observing which states were very unlikely to be used after initially estimating a larger model.
N-terminal transmembrane helices
N-terminal transmembrane helices are often mistaken as signal peptides, and vice versa (Krogh et al. 2001). Therefore, a submodel for N-terminal transmembrane helices was included. It is essentially a part of the TMHMM model (Krogh et al. 2001), in which just one membrane helix can be modeled. The intention with this part of the model is primarily to limit the number of false positives from the signal peptide predictions and not to predict whether a protein has an N-terminal transmembrane helix or not.
Cytoplasmic proteins
This submodel consists of two states: a state for the first amino acid and a state for the rest with a transition to itself.
Only the first 70 amino acids were used for both training and testing. The first three branches have a six-state submodel for modeling the length distribution and amino acid composition of the last part of the sequence in the mature protein. The whole model was estimated from the data and tested with cross-validation as the neural network (see Methods).
To classify proteins into each of the four classes presented by the submodels, the posterior probabilities of the sequence given each branch were used. These probabilities were divided by the probability of the protein according to a null model. The logarithm of the ratio (the log-odds) was used as the score for each class. We used the submodel for cytoplasmic proteins as the null model, so the score for cytoplasmic is always the same for any protein. To predict the class of a protein, we chose the highest scoring branch. Table 1 shows the number of predictions in each class in the cross-validation versus the correct classification (a “confusion matrix”).
Table 1.
Results of the prediction by the HMM
| Predicted class | |||||
| Correct class | SPaseI | SPaseII | Cytoplasmic | TMH | Total |
| SPaseI | 309 | 2 | 14 | 3 | 328 |
| SPaseII | 2 | 61 | 0 | 0 | 63 |
| Cytoplasmic | 5 | 1 | 382 | 0 | 388 |
| TMH | 8 | 0 | 21 | 142 | 171 |
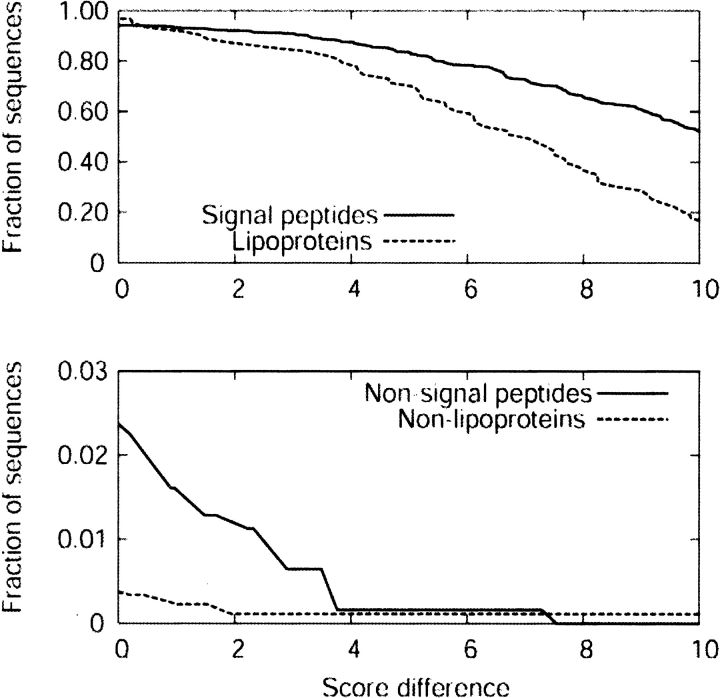
The score difference between the predicted class and the second highest score can be used as a measure of confidence in the prediction. Figure 7 ▶ shows the fraction of correctly predicted signal peptides and lipoproteins, as well as the fraction of sequences wrongly classified in one of those classes as a function of this score difference. For the signal peptides, one can increase specificity significantly at a moderate cost in sensitivity by setting a cut-off between two and four in score difference. To a lesser extent, the same is true for lipoproteins.
Figure 7.
HMM performance as a function of score difference. (Top) The fraction of correct predictions as a function of score difference. (Bottom) The fraction of sequences wrongly predicted as signal peptides or lipoproteins.
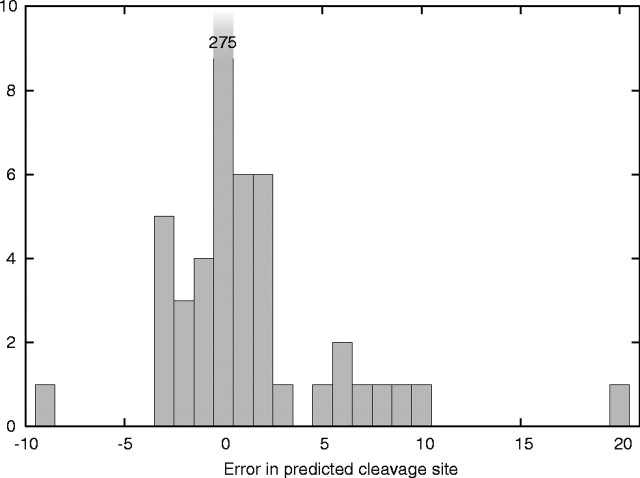
For the prediction of cleavage sites, we used the log-odds based on the posterior probability of the state immediately after the cleavage site, but otherwise normalized as above. This score can immediately be turned into a probability of a cleavage site given the model by subtracting the score for the relevant branch and exponentiating. Usually several positions have a cleavage site score above a threshold of, say, zero in log-odds, but we always chose the highest scoring as the predicted site. Of the 61 lipoproteins correctly classified, all except one had the correct cleavage site predicted. The exception is NLPD_PVPSEAE (Lipoprotein nlpD/lppB homolog, Precursor, from Pseudomonas aeruginosa) in which the predicted cleavage site is 15 amino acids after the annotated one. Of the 309 correctly predicted signal peptides (SPaseI), 275 had the cleavage site correctly predicted, corresponding to 11% error rate in the precise location of the cleavage site or a 16% error rate as a fraction of the total number of signal peptides (328). This is at about the same level of performance as SignalP (Nielsen and Krogh 1998). Most of the predicted sites are within ±5 amino acids from the correct site, as seen in Figure 8 ▶.
Figure 8.
Histogram of cleavage site prediction errors.
Discussion
To compare the results from the neural network and the HMM with existing tools, the PROSITE consensus pattern with additional rules and the von Heijne consensus pattern (von Heijne 1989) were tested on the same data sets.
The von Heijne consensus pattern predicted 54 of the 63 lipoproteins correctly, but also gave a total of 74 false positives. Forty-four of these were SpaseI-cleaved signal peptides, which the consensus pattern should be able to distinguish from lipoprotein signal peptides. This is not surprising, because many lipoproteins and SPaseI-cleaved proteins have been annotated since 1989. The Prosite pattern predicted 56 of the 63 lipoproteins correctly and it came up with only 14 false positives, significantly better than the von Heijne consensus pattern. Still, the HMM and the neural network were both significantly better, as the number of false positives was almost twice as much for the Prosite pattern prediction compared with the neural network, and even more when compared with the HMM.
When comparing the two new predictors, the HMM seemed superior. By using the HMM, the same number of lipoproteins were predicted correctly as with the neural network. However, less false positives were predicted with the HMM. Varying the threshold for the neural network, the rate of false positives could be decreased, but at the cost of true positives. Actually, raising the threshold for the output neuron so that the number of false positives was decreased to the same number as for the HMM, the number of true positives was decreased to as little as 51. The worse performance of the neural network could well be due to a much larger number of free parameters compared with the relatively small number of lipoproteins used for training. It is possible that the neural net could have been further improved by, for example, using an asymmetric input window as in SignalP.
Table 2 summarizes the comparison. Because the HMM gave the best results, it was used for further investigations.
Table 2.
Comparison of the HMM and neural network prediction with other available lipoprotein prediction methods
| Correct predictions of Lipoprotein | False predictions of Lipoproteins | ||||
| Number | Lipoproteins | Number | Nonlipoproteins | Correlation coefficient | |
| Prosite pattern | 56 | 88.9% | 14 (11a/2b/1c) | 1.6% | 0.83 |
| von Heijne consensus | 54 | 85.7% | 74 (44/20/10) | 8.3% | 0.56 |
| Neural network | 61 | 96.8% | 8 (6/2/0) | 0.9% | 0.92 |
| HMM | 61 | 96.8% | 3 (2/1/0) | 0.3% | 0.96 |
a SPaseI cleaved proteins.
b Cytoplasmic proteins.
c Transmembrane proteins.
It has previously been discussed whether the lipoproteins should be considered as positive examples for SignalP (Nielsen et al. 1997). SignalP is only trained on SpaseI-cleaved proteins, considering the signal score and the cleavage score. Applying SignalP to lipoproteins the signal score is very high, but the cleavage score is low as expected. Depending on the combined score values, most lipoproteins were predicted as having a signal peptide, but the cleavage site was rarely predicted at the correct position.
The amino acid in position +2 relative to the cleavage site is believed to determine whether the protein is attached to the inner or outer membrane of Gram-negative bacteria. Traditionally, it was thought that an aspartic acid in this position directs the protein to the inner membrane, and all other amino acids direct it to the outer membrane. However, it has been shown that the situation is not quite so simple (Seydel et al. 1999). We have not been able to find sufficient experimental data to include this sorting signal into the model, and instead the server provided at www.cbs.dtu.dk/services/LipoP/ simply reports which amino acid is in the +2 position to help users judge for themselves.
Prediction of Gram-positive lipoproteins
Because lipoproteins from Gram-negative and Gram-positive Eubacteria are resembling each other in the consensus sequence close to the lipid attachment site (Sutcliffe and Russell 1995), we also tested the HMM on Gram-positive lipoproteins.
A small data set consisting of 28 lipoproteins from Gram-positive Eubacteria was extracted from SWISS-PROT by using the same criteria as for the Gram-negative lipoproteins. Twenty-six of these lipoproteins were correctly predicted by the HMM to be lipoproteins, whereas the last two proteins were predicted as transmembrane proteins. Because of the limited number of available sequences, the data set was not similarity reduced, and these two sequences not predicted to be lipoproteins by the HMM were actually homologous cytochrome C oxidase polypeptide II precursors from two different Bacillus species, B. firmus and B. subtilis (COX2_PVBACFI and QOX2_PVBACSU). As these are annotated in SWISS-PROT as having several potential transmembrane helices, as well as having a lipoprotein signal peptide, the HMM prediction is actually not wrong. The SPaseII scores were still relatively high (2.92 and 5.70) compared with TMH scores (6.95 and 9.22). It should be noted that several examples of integral membrane proteins with cleavable lipoprotein signal peptide has been shown to exist (Pyrowolakis et al. 1998; Bengtsson et al. 1999; Sakamoto et al. 1999).
After the completion of the above data set, it came to our attention that another set of 33 experimentally verified Gram-positive lipoproteins was used by Sutcliffe and Harrington (2002). Twelve of these sequences are also in our data set. We have tested our method on 31 of the sequences (LppC and MBL from Streptococcus equi were not found). Four sequences were wrongly classified, but three of them had the correct cleavage site predicted in a suboptimal prediction of lipoprotein (see www.binf.ku.dk/krogh/LipoP/). Two cytochrome C oxidases (QOX2_PVBACSU and Q93HZ4) were predicted as transmembrane, one protein was predicted as an ordinary signal peptide (SODC_PVMYCTU), and one as cytosolic (KAPB_PVBACSU).
Genome search
Lipoproteins were predicted in the complete proteomes of 13 microbial genomes from GenBank. Because the above results indicated that the HMM also was capable of predicting Gram-positive lipoproteins, the genome of the industrially very important B. subtilis was included for testing. Table 3 lists the number of proteins predicted as lipoproteins by the HMM model. The number of predicted lipoproteins annotated as such is listed for both GenBank and SWISS-PROT. Many of the proteins included in the whole genome data sets from GenBank cannot be found in SWISS-PROT. Therefore, the number of predicted lipoproteins, which can be found in SWISS-PROT, is included for comparison.
Table 3.
Proteins predicted as lipoproteins for the 13 genomes
| Out of the number of predicted lipoproteins | ||||
| Organism | Number of predicted lipoproteins by HMM | Annotated as lipoproteins in GenBank | Found in SWISS-PROT | Annotated as lipoproteins in SWISS-PROT |
| Agrobacterium tumefaciens str. C58 | 47 | 2 | 3 | 2 |
| Bacillus subtilis | 101 | 3 | 50 | 32 |
| Borrelia burgdorferi | 113 | 12 | 14 | 9 |
| Campylocater jejuni | 47 | 29 | 6 | 6 |
| Escherichia coli K12 | 101 | 16 | 101 | 63 |
| Haemophilus influenzae Rd | 48 | 13 | 48 | 19 |
| Helicobacter pylori 26695 | 37 | 5 | 8 | 3 |
| Neisseria meningitidis serogroup A | 72 | 28 | 6 | 5 |
| Pseudomonas aeruginosa | 186 | 8 | 13 | 10 |
| Salmonella typhi | 116 | 69 | 12 | 7 |
| Salmonella typhimurium | 110 | 63 | 19 | 15 |
| Treponema pallidum | 31 | 2 | 22 | 12 |
| Vibrio cholerae | 82 | 17 | 10 | 10 |
The predicted lipoproteins for Escherichia coli strain K12 are listed in Table 4 and sorted according to descending differences in SPaseII and SPaseI scores. A more thorough list of prediction results for all the analyzed genomes can be found at www.binf.ku.dk/krogh/LipoP/. The lipoproteins predicted for the E. coli strain K12 were compared with new experimental data (S. Matsuyama et al., unpubl.). Table 4 indicates which of the predicted lipoproteins have been experimentally confirmed by the sensitivity to globomycin and/or lipid-modification. As can be seen, 26 of the predicted lipoproteins have not yet been experimentally verified. When looking at the SWISS-PROT annotation, 17 of these 26 predictions are annotated as hypothetical proteins. The remaining nine predictions are all annotated as something else. We would, however, from the test results, expect 0.3% of the nonlipoproteins to be predicted falsely as lipoproteins, which corresponds to ~13 false positives for the E. coli K12 genome with ~4000 annotated proteins. This corresponds well with the previous annotations. Fortunately, all the lipoproteins included in the training set were experimentally verified.
Table 4.
Lipoproteins predicted for the Escherichia coli K12 genome
| SWISSPROT entry | SPaseII–SPaseI score | Predicted lipid attachment site | Experimentally verifieda |
| YFHM_ECOLI | 15.770 | 18 | − |
| YFBK_ECOLI | 13.390 | 19 | − |
| ACFD_ECOLI | 13.220 | 24 | − |
| YAIW_ECOLI | 13.010 | 21 | − |
| MLTA_ECOLI | 12.940 | 21 | + |
| YIIG_ECOLI | 12.460 | 22 | + |
| RLPB_ECOLIb | 12.140 | 19 | + |
| YFIB_ECOLI | 12.070 | 19 | + |
| YIFL_ECOLI | 11.890 | 20 | − |
| YCFL_ECOLI | 11.760 | 19 | − |
| NLPB_ECOLIb | 11.520 | 26 | + |
| NLPA_ECOLIb | 11.360 | 24 | + |
| YHIU_ECOLI | 11.060 | 21 | + |
| YIAD_ECOLI | 10.880 | 21 | + |
| MULI_ECOLI | 10.770 | 21 | + |
| YFIO_ECOLI | 10.680 | 20 | + |
| MLTB_ECOLIb | 10.660 | 19 | + |
| YCFM_ECOLI | 10.520 | 20 | + |
| YBJR_ECOLI | 10.250 | 17 | + |
| SLYB_ECOLI | 10.190 | 18 | + |
| YCEB_ECOLI | 9.875 | 19 | + |
| YEHR_ECOLI | 9.793 | 27 | + |
| YCJN_ECOLI | 9.705 | 21 | + |
| NLPI_ECOLIb | 9.677 | 19 | + |
| YBHC_ECOLI | 9.570 | 22 | + |
| YQHH_ECOLI | 9.422 | 20 | + |
| YCDR_ECOLI | 9.138 | 21 | + |
| PAL_ECOLI | 9.039 | 22 | + |
| YAFT_ECOLI | 8.961 | 19 | + |
| NLPD_ECOLIb | 8.943 | 26 | + |
| YAEC_ECOLI | 8.699 | 23 | + |
| YHFL_ECOLI | 8.644 | 20 | + |
| YEAY_ECOLI | 8.481 | 23 | + |
| APBE_ECOLI | 8.469 | 20 | + |
| SLP_ECOLIb | 8.097 | 30 | + |
| YGER_ECOLI | 8.042 | 34 | + |
| RLPA_ECOLIb | 7.906 | 18 | + |
| YGHG_ECOLI | 7.802 | 25 | + |
| ACRA_ECOLIb | 7.696 | 25 | + |
| ACRE_ECOLI | 7.696 | 24 | + |
| HSLJ_ECOLI | 7.680 | 17 | + |
| FLGH_ECOLI | 7.581 | 22 | + |
| NLPC_ECOLI | 7.428 | 16 | + |
| LOLB_ECOLIb | 7.363 | 22 | + |
| YBFN_ECOLI | 7.188 | 17 | + |
| BLC_ECOLIb | 7.034 | 19 | + |
| MLTD_ECOLI | 6.735 | 16 | + |
| YBAY_ECOLI | 6.537 | 19 | + |
| OSME_ECOLIb | 6.535 | 21 | + |
| VACJ_ECOLI | 6.469 | 18 | + |
| RCSF_ECOLI | 6.450 | 16 | + |
| YBBC_ECOLI | 6.358 | 18 | − |
| YJEI_ECOLI | 6.146 | 30 | + |
| YGDR_ECOLI | 5.750 | 20 | + |
| BORD_ECOLI | 5.519 | 17 | + |
| YDCL_ECOLI | 5.493 | 21 | + |
| SPR_ECOLI | 5.441 | 27 | + |
| YCAL_ECOLI | 5.318 | 28 | + |
| YFGH_ECOLI | 5.271 | 22 | + |
| YJAH_ECOLI | 4.807 | 31 | − |
| CUTF_ECOLIb | 4.759 | 21 | + |
| YRAM_ECOLI | 4.703 | 27 | − |
| OSMB_ECOLIb | 4.544 | 24 | + |
| YOAF_ECOLI | 4.476 | 17 | + |
| WZA_ECOLI | 4.375 | 21 | + |
| YDDW_ECOLI | 4.167 | 28 | − |
| YGDI_ECOLI | 4.107 | 21 | + |
| YCCZ_ECOLI | 3.873 | 21 | + |
| YMCC_ECOLI | 3.867 | 16 | + |
| YBET_ECOLI | 3.850 | 19 | − |
| NRFG_ECOLI | 3.780 | 22 | − |
| YFEY_ECOLI | 3.476 | 18 | + |
| MLTC_ECOLI | 3.345 | 18 | + |
| YRAP_ECOLI | 3.248 | 19 | + |
| CSGG_ECOLI | 3.121 | 16 | + |
| YNFC_ECOLI | 2.941 | 29 | + |
| YNBE_ECOLI | 2.896 | 17 | + |
| CUSC_ECOLI | 2.754 | 18 | + |
| YFGL_ECOLI | 2.544 | 20 | + |
| YBFP_ECOLI | 2.535 | 23 | + |
| YFHG_ECOLI | 2.522 | 26 | − |
| YHDV_ECOLI | 2.447 | 17 | + |
| LEP_ECOLI | 2.253 | 21 | − |
| GUN_ECOLI | 2.058 | 23 | − |
| YCEK_ECOLI | 2.041 | 16 | + |
| YJBF_ECOLI | 1.952 | 26 | + |
| PANE_ECOLI | 1.927 | 20 | − |
| YFIL_ECOLI | 1.884 | 30 | + |
| YEDD_ECOLI | 1.802 | 16 | + |
| YECR_ECOLI | 1.530 | 16 | + |
| FLIL_ECOLI | 1.291 | 27 | − |
| YIHN_ECOLI | 1.282 | 16 | − |
| DCRB_ECOLI | 1.123 | 38 | − |
| KEFA_ECOLI | 0.949 | 27 | − |
| YJBH_ECOLI | 0.801 | 18 | − |
| YDEK_ECOLI | 0.797 | 19 | − |
| YBJP_ECOLI | 0.641 | 19 | + |
| VISC_ECOLI | 0.435 | 20 | − |
| SRLD_ECOLI | 0.364 | 19 | − |
| YPDI_ECOLI | 0.131 | 19 | − |
| YJCP_ECOLI | 0.095 | 24 | − |
a According to S. Matsuyama et al., unpubl.
b Sequences included in training set.
It seemed that the HMM prediction missed 15 of the experimentally verified lipoproteins from E. coli (data not shown). Two of the lipoproteins were, however, encoded by plasmid genes and therefore not included in the E. coli K12 genome. By using the sequences from SWISS-PROT, they were both predicted correctly as lipoproteins. Also, by using the SWISS-PROT sequences instead of the GenBank sequences, seven of the remaining proteins were predicted as lipoproteins. Because the protein sequences given in GenBank in these cases were longer than the ones given in SWISS-PROT, it is therefore very likely that the position of the start codon in the GenBank sequence is incorrectly annotated. One additional protein was annotated as both potential transmembrane and lipoprotein in SWISS-PROT (CYOA_PVECOLI, ubiquinol oxidase polypeptide II precursor), and as with the Gram-positive case, the SPaseII score was relatively high (6.64) compared with the TMH score (7.87). All the above considered, only five of the 90 experimentally verified lipoproteins were actually not predicted as such by the HMM model.
In the Gram-positive B. subtilis, 101 annotated proteins were predicted as lipoproteins. For comparison, Tjasma et al. (1999) found 114 probable lipoproteins by a SignalP search combined with a lipobox search and a Blast similarity search. Sutcliffe and Harrington (2002) found 67 lipoproteins (61 probable and six proven) lipoproteins by a regular expression called G+LPP, and Gonnet and Lisacek (2002) found 65 lipoproteins predicted by another refined regular expression.
Conclusion
A method for lipoprotein prediction, LipoP, was developed. Both an HMM and a neural network were significantly better at predicting lipoproteins than were any of the existing methods discussed in this article. The HMM method was chosen for the remainder of the analysis, mainly because it distinguishes between lipoproteins, SPaseI-cleaved signal peptides, cytoplasmic proteins, and proteins with N-terminal transmembrane helices. However, when handling proteins, which are both lipoproteins and have transmembrane regions, the HMM, in some cases, misses the lipoprotein signal peptide.
The method was used to predict lipoproteins in 12 Gram-negative bacteria. When comparing a genome search of E. coli with new experimental data, most of the experimentally verified lipoproteins were correctly predicted as lipoproteins (94.6%). This verification of the lipoproteins predicted in E. coli might be an indication of how well the HMM performs on genome data in general. Even though the HMM is trained on proteins from Gram-negative bacteria, it also seems to be able to predict Gram-positive lipoproteins. This feature was used to make a genome search of the Gram-positive bacteria, B. subtilis.
The LipoP server is accessible at www.cbs.dtu.dk/services/LipoP/. Genome predictions and other material are accessible at www.binf.ku.dk/krogh/LipoP/.
Materials and methods
Data sets
A data set consisting of Gram-negative lipoproteins and SPaseI and cytoplasmic proteins was created. The sequences were extracted from SWISS-PROT (release 40) by using keywords and comments and by including only proteins from organisms belonging to the two phyllums Proteobacteria and Spirochetes (order: Spirochaetales).
Only a very limited number of lipoproteins with known signal length and lipid attachment site for Gram-negative Bacteria could be retrieved. Therefore, also lipoproteins annotated as probable for signal length and lipid attachment site, as well as lipoproteins annotated as potential in only one of these categories, but certain lipoproteins in the other were allowed in the data set. Hereby, we were able to extract 99 lipoproteins. More sequences where available for Gram-negative SPaseI-cleaved proteins and for Gram-negative cytoplasmic proteins; thus all proteins with annotations such as probable and potential were excluded from these data sets, creating two parts of the data set consisting of 528 SPaseI-cleaved proteins and 1026 cytoplasmic proteins, respectively. In these sets, the first amino acid after the cleavage site was labeled.
The combined data set was then homology reduced to limit biasing so it could be used for testing with cross-validation. Because we were primarily interested in the signal part of the sequence, only the first 30 amino acids were taken into consideration for the lipoproteins and the first 60 amino acids for the SPaseI-cleaved proteins and the cytoplasmic proteins in the similarity reduction. To generate a nonredundant data set, we searched each sequence in the data set against all the other sequences by using BLASTP (Altschul et al. 1997) and a Blosum62 score matrix (Henikoff and Henikoff 1992). By using a threshold of 10−6 on the expectation score, we subsequently generated a maximal nonredundant version of the data set using the Hobohm-2 algorithm (Hobohm et al. 1992). Finally, the data set consisted of 63 nonhomologous lipoproteins (Table 5), 328 SPaseI-cleaved proteins, and 388 cytoplasmic proteins.
Table 5.
Lipoproteins included in homology reduced data set
| 17KD_RICPR ACRA_ECOLI ANIA_NEIGO BLC_ECOLI BLC_VIBCH BMPA_BORAF BMPB_BORGA BMPC_BORBU BMPD_BORBU COML_NEIGO CUTF_ECOLI CYCR_RHOVI GLPQ_HAEIN GUN_BURSO H8_NEIMC HBPA_HAEIN HFD1_HAEIN HLPA_HAEIN LOLB_ECOLI LP20_HELPY LPPB_HAESO |
LPPL_PSEAE LYS4_ECOLI MLTA_VIBCH MLTB_ECOLI MP17_FRATU MULI_MORMO MULI_PROMI MULI_PSEAE NLPA_ECOLI NLPB_ECOLI NLPD_ECOLI NLPD_PSEAE NLPI_ECOLI OMLA_ACTPL OMLA_PSEAE OSA1_BORBU OSB1_BORBU OSMB_ECOLI OSME_ECOLI OUTS_ERWCH P22_BORBU |
PAL_HAEIN PCP_HAEIN PMEB_ERWCH PULA_KLEPN PULS_KLEPN RLPA_ECOLI RLPB_ECOLI SLP_ECOLI SMPA_TREHY TA15_TREPA TA17_TREPA TA47_TREPA TBB1_NEIMB TMPA_TREPA TMPA_TREPH TMPC_TREPA TRT3_ECOLI VACJ_SHIFL VM07_BORHE VM17_BORHE VM21_BORHE |
A data set of N-terminal transmembrane segments was created from the set of 160 membrane proteins used in Krogh et al. (2001). Sixty-eight of them were from the above-mentioned phyllums, and they were extracted. Because the original data set was already similarity reduced, no more reduction was done. To obtain a reasonable number of N-terminal membrane helices for training and testing the methods, a set of “fake” N-terminal sequences was created. Finding all the TM helices starting on the cytoplasmic side, the following was done for each sequence. If it was the first TM helix in the protein, up to 40 amino acids upstream were included (or as many as there were). Otherwise, half the upstream amino acids on the cytoplasmic side or up to 40 were included. The first amino acid was replaced by a methionine. The sequences were cut off at a total length of 70 amino acids, excluding those that were short (C-terminal TM helices). The set of “constructed” TM peptides ended up containing 171 sequences.
For testing the methods, the data were divided into 63 sets. Each set contained exactly one lipoprotein. The other sets were distributed equally and randomly among the 63 sets. In the cross-validation procedure, the HMM or neural network was trained on 62 of these sets and tested on the one that was left out. This was repeated 63 times, so that all sets were used for testing once, and finally, the test results were averaged. This is a standard method for obtaining unbiased test results when the amount of data is limited.
A data set consisting of Gram-positive lipoproteins was made for testing purposes only. The extraction of proteins was done in the same way as the data set for Gram-negative lipoproteins, but the data set was not homology-reduced because the Gram-positive lipoproteins would only be used for testing.
Recently, 90 lipoproteins from E. coli have been experimentally verified by S. Matsuyama et al. (unpubl.). These were used as a base for comparison of the results from the genome search carried out for E. coli.
Neural networks
The neural network training was carried out by using the lipoprotein cleavage site on lipoproteins as positive examples and all remaining “C”s from lipoproteins, SpaseI-cleaved proteins and cytoplasmic proteins as negative examples. Thus, the neural networks were trained only on cysteines, and backward propagation was used under the training. The number of hidden neurons was varied from zero to four, and the size of the symmetric windows was varied from 27 to 33. The neural networks were evaluated by their performance on the test data sets. The test data from the 63 cross-validations were added together, and the correlation coefficient were calculated (Matthews 1975). In this way, all proteins in the entire data set were included in the calculation, and none of them were tested on the network they were trained on. By considering the correlation coefficient and the number of lipoproteins predicted, the best network was chosen and the optimal parameters were estimated.
The training set for the neural networks consisted of Gram-negative lipoproteins and SPaseI and cytoplasmic proteins data sets, whereas the transmembrane data set was used only for testing. The neural network was trained on the first 100 amino acids of each sequence. For testing, only the first 50 amino acids of each sequence were considered.
HMMs
The four branches or submodels already described were denoted SPaseI, SPaseII, TMH, and CYT. The first state of each branch was given probability 1 out of 20 for all amino acids. This is because the first amino acid always is methionine, so there is no information in this amino acid. The advantage of this scheme is that the model can deal with a wrongly assigned first amino acid (which happens sometimes when the start codon of a gene is not ATG).
The probability for entering each of the branches was not estimated from the data. These entry probabilities reflect the prior probabilities (in a Bayesian statistical sense) that a randomly chosen protein belongs to each of the four classes. They also determine the number of predictions of each class, so by changing them, one can, for instance, increase the number of predictions from a certain class. They were set by trial and error so as to get reasonable prediction levels for the classes, but mostly focused on the performance on lipoproteins. Equivalently, one could fix the four entry probabilities to, for example, one out of four and then instead of choosing the highest scoring branch for prediction, one could have class-specific cut-offs on the log-odds score. In our final model, we have these probabilities: P(SPaseI) = 0.08, P(SPaseII) = 0.02, P(TMH)=0.03, and P(CYT) = 0.87. The system is not very sensitive to these parameters.
The model was trained using the Baum-Welch procedure for labeled sequences (Krogh 1997; Durbin et al. 1998; Krogh and Riis 1999). The sequences were labeled according to which of the three classes it belonged to, and the cleavage site was labeled for signal sequences. This ensures that a submodel is trained on the correct set of proteins, and that cleavage sites are correctly positioned during training. Only the first 70 amino acids of each protein were used for training and testing.
We used the submodel for cytoplasmic proteins as the null model, so the score for cytoplasmic is always equal to log[P(CYT)] = −0.1393 (the natural logarithm is used).
Genome search
Data sets were extracted from the GenBank genomic library (Benson et al. 2002). The extracted genomes and the corresponding GenBank files are listed in Table 6. The number of sequences included in each genome file is listed as well.
Table 6.
List of organisms used for the genome search
| Organism | # Sequences in genome | Genbank file |
| Agrobacterium tumefaciens str. C58 (U. Washington) | 5402 | AE008688 (circular chromosome) AE008689 (linear chromosome) AE008687 (plasmid AT) AE008690 (plasmid TI) |
| Bacillus subtilis | 4112 | NC_000964 |
| Borrelia burgdorferi w/plasmids | 1596 | NC_001318 (chromosome) Plasmids: NC_001904 (cp9), NC_001903 (cp26), NC_000948 (cp32-1), NC_000949 (cp32-3), NC_000950 (cp32-4), NC_000951 (cp32-6), NC_000952 (cp32-7), NC_000953 (cp32-8), NC_000954 (cp32-9), NC_000957 (lp5), NC_001849 (lp17), NC_000955 (lp21), NC_001850 (lp25), NC_001851 (lp28-1), NC_001852 (lp28-2), NC_001853 (lp28-3), NC_001854 (lp28-4), NC_001855 (lp36), NC_001856 (lp38), NC_001857 (lp54), NC_000956 (lp56) |
| Campylocater jejuni | 1634 | AL111168 |
| Escherichia coli K12 | 4279 | NC_000913 |
| Haemophilus influenzae Rd | 1714 | NC_000907 |
| Helicobacter pylori 26695 | 1576 | NC_000915 |
| Neisseria meningitidis serogroup A strain Z2491 | 2065 | AL157959 |
| Pseudomonas aeruginosa | 5567 | NC_002516 |
| Salmonella enterica subsp. enterica serovar Typhi | 4600 | NC_003198 |
| Salmonella typhimurium LT2 | 4451 | NC_003197 |
| Treponema pallidum | 1036 | NC_000919 |
| Vibrio cholerae | 3828 | AE003852 (chromosome I) AE003853 (chromosome II) |
The predictions were carried out on the whole genome data sets extracted from GenBank. The proteins predicted as lipoproteins by the HMM were compared with previous annotations in GenBank as well as in SWISS-PROT. To find the corresponding SWISS-PROT entry, the gene names were extracted from the GenBank file (*.gbk) for each predicted lipoprotein, and by comparing these with a SWISS-PROT file (*.sprot) for each microorganism, it was possible to extract SWISS-PROT entry names for gene products that were available in SWISS-PROT. By using “function” and “product” annotation in the GenBank files and “keyword” annotation in the SWISS-PROT files, the prediction was compared with the existing GenBank and SWISS-PROT annotations.
Acknowledgments
We thank Hajime Tokuda for experimental results prior to publication and Lars Juhl Jensen for his programming advice. This work was sponsored by a grant to the Center for Biological Sequence Analysis (S.B.) from the Danish National Research Foundation. A.K. was supported by EU grant no. QLRI-CT-2001-00015.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0303703.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch, A. and Apweiler, R. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson, J., Tjalsma, H., Rivolta, C., and Hederstedt, L. 1999. Subunit II of Bacillus subtilis cytochrome c oxidase is a lipoprotein. J. Bacteriol. 181 685–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., Rapp, B.A., and Wheeler, D.L. 2002. GenBank. Nucleic Acids Res. 30 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, V. and Wu, H.C. 1994. Lipoproteins, structure, function, biosynthesis, and a model for protein export. In Bacterial cell wall (eds. J.M. Ghuysen and R. Hakenbeck), pp. 319–341. Elsevier, Amsterdam, The Netherlands.
- Durbin, R.M., Eddy, S.R., Krogh, A., and Mitchison, G. 1998. Biological sequence analysis. Cambridge University Press, Cambridge, UK.
- Falquet, L., Pagni, M., Bucher, P., Hulo, N., Sigrist, C.J., Hofmann, K., and Bairoch, A. 2002. The PROSITE database: Its status in 2002. Nucleic Acids Res. 30 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonnet, P. and Lisacek, F. 2002. Probabilistic alignment of motifs with sequences. Bioinformatics 18 1091–1101. [DOI] [PubMed] [Google Scholar]
- Hayashi, S. and Wu, H.C. 1990. Lipoproteins in bacteria. J. Bioenerg. Biomembr. 22 451–471. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. and Henikoff, J.G. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. 89 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobohm, U., Scharf, M., Schneider, R., and Sander, C. 1992. Selection of representative protein data sets. Protein Sci. 1 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, P., Somorjai, R.L., and Lau, P.C. 1988. Distinctive properties of signal sequences from bacterial lipoproteins. Protein Eng. 2 15–20. [DOI] [PubMed] [Google Scholar]
- Krogh, A. 1997. Two methods for improving performance of a HMM and their application for gene finding. In Proceedings of the Fifth International Conference on Intelligent Systems for Molecular Biology (eds. T. Gaasterland et al.), pp. 179–186. AAAI Press, Menlo Park, CA. [PubMed]
- Krogh, A. and Riis, S.K. 1999. Hidden neural networks. Neural Comput. 11 541–563. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L.L. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305 567–580. [DOI] [PubMed] [Google Scholar]
- Matthews, B.W. 1975. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta 405 442–451. [DOI] [PubMed] [Google Scholar]
- Nakai, K. and Kanehisa, M. 1991. Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins 11 95–110. [DOI] [PubMed] [Google Scholar]
- Nielsen, H. and Krogh, A. 1998. Prediction of signal peptides and signal anchors by a hidden Markov model. In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology (eds. J. Glasgow et al.), pp. 122–130. AAAI Press, Menlo Park, CA. [PubMed]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10 1–6. [DOI] [PubMed] [Google Scholar]
- Pyrowolakis, G., Hofmann, D., and Herrmann, R. 1998. The subunit b of the F0F1-type ATPase of the bacterium Mycoplasma pneumoniae is a lipoprotein. J. Biol. Chem. 273 24792–24796. [DOI] [PubMed] [Google Scholar]
- Sakamoto, J., Shibata, T., Mine, T., Miyahara, R., Torigoe, T., Noguchi, S., Matsushita, K., and Sone, N. 2001. Cytochrome c oxidase contains an extra charged amino acid cluster in a new type of respiratory chain in the amino-acid-producing Gram-positive bacterium Corynebacterium glutamicum. Microbiology 147 2865–2871. [DOI] [PubMed] [Google Scholar]
- Sankaran, K. and Wu, H.C. 1994. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269 19701–19706. [PubMed] [Google Scholar]
- Schneider, T.D. and Stephens, R.M. 1990. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydel, A., Gounon, P., and Pugsley, A.P. 1999. Testing the “+2 rule” for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34 810–821. [DOI] [PubMed] [Google Scholar]
- Sutcliffe, I.C. and Harrington, D.J. 2002. Pattern searches for the identification of putative lipoprotein genes in Gram-positive bacterial genomes. Microbiology 148 2065–2077. [DOI] [PubMed] [Google Scholar]
- Sutcliffe, I.C. and Russell, R.R. 1995. Lipoproteins of Gram-positive bacteria. J. Bacteriol. 177 1123–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma, H., Kontinen, V.P., Pragai, Z., Wu, H., Meima, R., Venema, G., Bron, S., Sarvas, M., and van Dijl, J.M. 1999. The role of lipoprotein processing by signal peptidase II in the Gram-positive eubacterium Bacillus subtilis: Signal peptidase II is required for the efficient secretion of α-amylase, a non-lipoprotein. J. Biol. Chem. 274 1698–1707. [DOI] [PubMed] [Google Scholar]
- von Heijne, G. 1989. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 2 531–534. [DOI] [PubMed] [Google Scholar]
- ———. 1990. The signal peptide. J. Membr. Biol. 115 195–201. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, K., Yu, F., and Inouye, M. 1988. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53 423–432. [DOI] [PubMed] [Google Scholar]