Abstract
Herein we present a computational technique for generating helix-membrane protein folds matching a predefined set of distance constraints, such as those obtained from NMR NOE, chemical cross-linking, dipolar EPR, and FRET experiments. The purpose of the technique is to provide initial structures for local conformational searches based on either energetic considerations or ad-hoc scoring criteria. In order to properly screen the conformational space, the technique generates an exhaustive list of conformations within a specified root-mean-square deviation (RMSD) where the helices are positioned in order to match the provided distances. Our results indicate that the number of structures decreases exponentially as the number of distances increases, and increases exponentially as the errors associated with the distances increases. We also found the number of solutions to be smaller when all the distances share one helix in common, compared to the case where the distances connect helices in a daisy-chain manner. We found that for 7 helices, at least 15 distances with errors up to 8 Å are needed to produce a number of solutions that is not too large to be processed by local search refinement procedures. Finally, without energetic considerations, our enumeration technique retrieved the transmembrane domains of Bacteriorhodopsin (PDB entry1c3w), Halorhodopsin (1e12), Rhodopsin (1f88), Aquaporin-1 (1fqy), Glycerol uptake facilitator protein (1fx8), Sensory Rhodopsin (1jgj), and a subunit of Fumarate reductase flavoprotein (1qlaC) with Cα level RMSDs of 3.0 Å, 2.3 Å, 3.2 Å, 4.6 Å, 6.0 Å, 3.7 Å, and 4.4 Å, respectively.
Keywords: Protein folds, helix packing, transmembrane helices, distance constraints
Unlike soluble proteins, only a few membrane protein structures have been solved using conventional methods such as NMR and crystallography. Considering the pace at which membrane protein structures are elucidated experimentally, membrane protein fold recognition based on structural homology does not appear to be a practical option for the near future.
Alternative membrane protein structural modeling approaches take advantage of the fact that many membrane proteins contain regions of highly hydrophobic transmembrane helical fragments (e.g., helix bundle). The structural constraints placed on transmembrane helices by the lipid bilayer limit the number of possible membrane protein folds to a point that several ab initio computational techniques have been considered (Bowie 1999; Nikiforovich et al. 2001; Vaidehi et al. 2002). These techniques decompose the membrane protein folding problem into the following steps: (1) prediction of transmembrane regions, (2) construction and optimization of individual helices, (3) assembly of the helix bundle, and (4) addition of the interhelical loops.
Several codes have been developed to predict the transmembrane regions of Step 1 (Hirokawa et al. 1998; Gromiha 1999; Nikiforovich et al. 2001; Vaidehi et al. 2002). The regions are generally determined using hydropathicity analysis. Step 2 requires energy minimization or molecular dynamics simulations (Vaidehi et al. 2002) to predict sequence-specific distortions in the helices, such as kinks induced by proline. Step 3 is the main subject of this paper. The approaches that have been taken thus far to attack the problem of assembling helix bundles are different in nature. Bowie (1999) argued that the conformational space of a membrane protein can effectively be sampled and gives a truly ab initio technique where all possible helix bundles are enumerated. However, in Bowie’s calculations (1999), the orientations of the individual helices around their respective axes are not taken into account. Nikiforovich et al. (2001) used the similarity between the X-ray structures of bacteriorhodopsin and rhodopsin to find the helix packing in the membrane plane. Specifically, the intersections between the helical axes and the plane crossing the membrane are fixed at values derived from the two X-ray structures. Vaidehi et al. (2002) oriented each helical axis of the helix bundle according to the 7.5 Å electron density map of rhodopsin. Although the two previous methods use energetic calculations and molecular simulations to further refine the helical arrangements, both techniques are potentially biased toward the structures of bacteriorhodopsin and rhodopsin and have yet to be validated for other membrane proteins. The addition of loops in Step 4 can be performed using commercially available software such as WHATIF (Vriend 1990), SCWRL (Bowers et al. 1997), and Jackal (Xiang et al. 2002), whereas other solutions that segregate between small and medium loops have also been proposed (Nikiforovich et al. 2001).
The technique presented in this paper is concerned with helix bundle assembly (Step 3). We assume that transmembrane regions have been predicted and that individual helices have been built and optimized. The method takes as input a set of helices in PDB format and a set of distances between pairs of atoms on these helices. The technique outputs all possible helix arrangements matching the input data, each solution differing from another by a predefined RMSD. The arrangements that are produced are constructed at an atomistic level and can thus be further refined using local conformational search.
Distance constraints are needed because, as we will see in the next section, the conformation space for membrane proteins is too large to perform energetic calculations on each conformation. These distances can come from a variety of experiments including chemical cross-linking, dipolar electron paramagnetic resonance (dipolar EPR), fluorescence resonance energy transfer (FRET), and NMR, to name a few. Each of these methods has advantages and disadvantages in terms of high-throughput capability, distance measurement accuracy, and structural model building. The purpose of the present article is to show the utility of a set of distance constraints for membrane protein structural modeling. For validation purposes on a set of membrane proteins for which crystal structures are available, the distances we consider here correspond to pairs of amino acids (K-K, K-D, K-E, K-C, and C-C) that could potentially be cross-linked using chemical cross-linkers. However, we stress that the method is not limited to the consideration of distances derived from cross-linking experiments, and we also demonstrate the method on the dark-adapted rhodopsin structure (1f88) using a set of disulfide mapping, cross-linking, and dipolar EPR distances gathered from the literature.
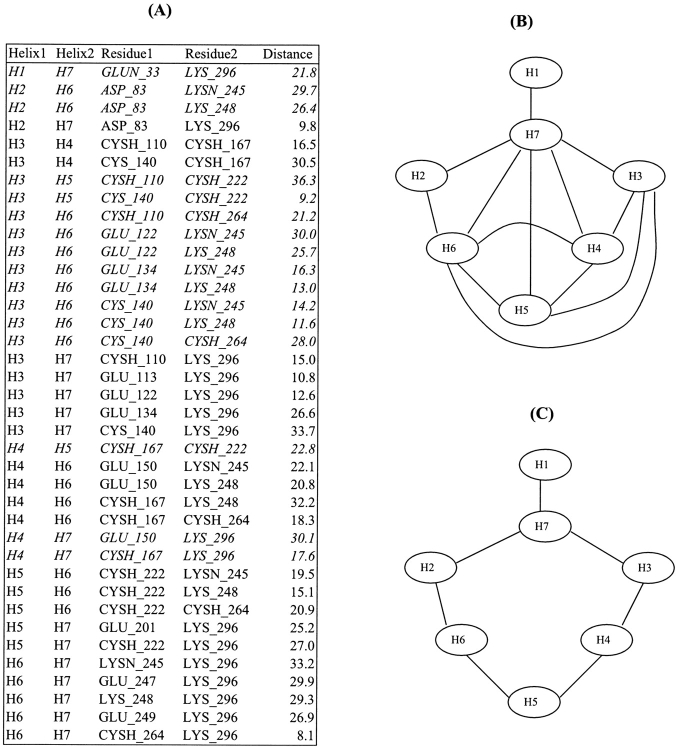
Essential to the understanding of our method is the concept of a distance graph, which was used on several occasions in this study and is illustrated in Figure 1 ▶. The distance graph for a given set of helices and interhelical distances is derived by representing each helix as a vertex and each distance as an edge connecting the two vertices corresponding to the helices between which the distance was measured: For each distance between two helices, an edge is added to the graph between the corresponding vertices. Associated with a distance graph is its radius. The radius of the distance graph is the minimum number of consecutive edges one must follow to reach all the vertices of the graph.
Figure 1.
Distance constraints and distance graph. (A) Distance list computed for rhodopsin (1f88). (B) Corresponding distance graph. An edge is drawn between two helices if a distance exists between these helices. All helices are linked to helix H7, thus the radius of the distance graph is 1. (C) Distance graph corresponding to the distances in italics in the distance list (A). The radius of this distance graph is 3, because every helix can be reached from H7 using no more than three links.
Throughout this article we also use a specific terminology to characterize increasing levels of membrane protein fold complexity. A template is a set of helices fully positioned in the membrane. Precisely, the helices are positioned in a reference system with origin in the bilayer central plane, and with z-axis pointing toward the extracellular side of the membrane and parallel to the bilayer normal. Because we do not attempt to position our helices toward other components of the membrane such as lipids, the origin is chosen arbitrarily in the bilayer central plane, and the x- and y-axes are any pair of orthogonal vectors in that plane. We refer to an unlabeled template when the sequence order of the helices is not known. In the unlabeled case, the coordinates of the helix center of mass in the bilayer reference system and the coordinates of the helix axis unit vector are used to represent each helix. A template is said to be labeled when the order of the helices is known, that is, the helix numbered i in the template corresponds to the ith membrane helix in the protein sequence. Each helix in a labeled template is represented by its sequence number and the coordinates of both its center of mass and its unit axial vector. An oriented template is a labeled template where the helices have been oriented around their helical axes. A helix in an oriented template is represented as a labeled template to which the coordinates of any backbone atom have been added. Finally, an atomistic template is a template composed of the coordinates in the bilayer reference system of all atoms of the helices. If one assumes that backbone atoms in helices are fixed, then there is only one atomistic template per oriented template, because if one knows the position of the center of mass, the axis vector, and the position of any backbone atom of a helix, then one knows the position of all backbone atoms of that helix. Consequently we will not make a distinction between oriented templates and atomistic templates.
Results and Discussion
In the following three subsections, we probe the number of membrane protein templates generated using our technique with various constraints. In the fourth subsection, we examine the ability of our technique to retrieve known membrane protein structures.
Conformational space size without distance constraints
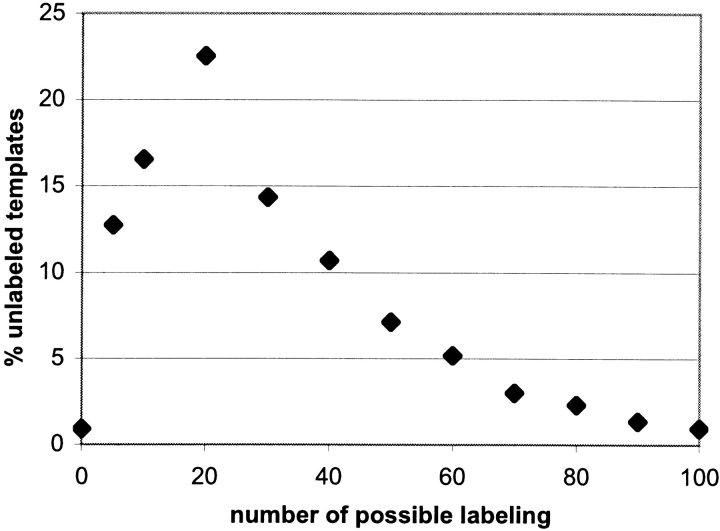
Bowie (1999) argues that, for membrane proteins of up to seven helices, unlabeled templates can be generated to cover the entire conformational space of the membrane protein folds. Precisely, he observes that conformational space is at least 80% covered by no more than 10 templates for three helices, 250 templates for four helices, 2500 templates for five helices, 25,000 templates for six helices, and 150,000 templates for seven helices. Now, for n helices there are 2n! ways to label the helices, which gives 10,080 labelings per unlabeled template for seven helices; however, this number can be drastically reduced by adding interhelix connection constraints (Bowie 1999). These constraints specify that (1) consecutive helices must be in contact, (2) the distance between the end points of consecutive helices must be in the range observed in known membrane protein structures, and (3) loops connecting helix end points cannot cross each other. Figure 2 ▶ gives the distribution of the number of labelings over a set of 50,000 unlabeled templates composed of seven helices. This distribution was obtained by running an algorithm described in Materials and Methods. The average number of labelings per template is 27.25, with a large standard deviation of 26.07. Constraint number (3) is not valid for all known membrane proteins. In fact, the structure of aquaporin-1 (1fqy) provides an example in which the loop between helices 3 and 5 and the loop between helices 5 and 6 may cross. Removing this constraint only slightly increases the number of labelings per template to 33.67. These numbers are comparable to the number of labelings, 30, reported by Bowie (1999), and a library of 150,000 unlabeled structures gives about 4.5 × 106 labeled templates. Recall that a labeled template is oriented once an arbitrary atom has been positioned for each helix in the bilayer reference system. If one imposes two orientations separated by at least Δα, then for n helices there are (360/Δα−1)n possible orientations per labeled template. For ideal helices, for example, poly-alanine helices, where the α-carbons are at a distance less than 2.5 Å from their helix axis, Δα = 60° leads to an RMSD no greater than 2.5 Å between two consecutive orientations. Thus, using Δα = 60°, a labeled library of 4.5 × 106 labeled templates with seven helices will give 78,125 possible orientations per template, and a total of 351.5 × 109 oriented or atomistic templates. Even if these templates are clustered with an RMSD of 2.5 Å, which according to Bowie leads to three times fewer structures (Bowie 1999), the number is still too large to perform energetic calculations on each template.
Figure 2.
Labeling distribution for seven-helix templates.
Conformational space size with exact distance constraints
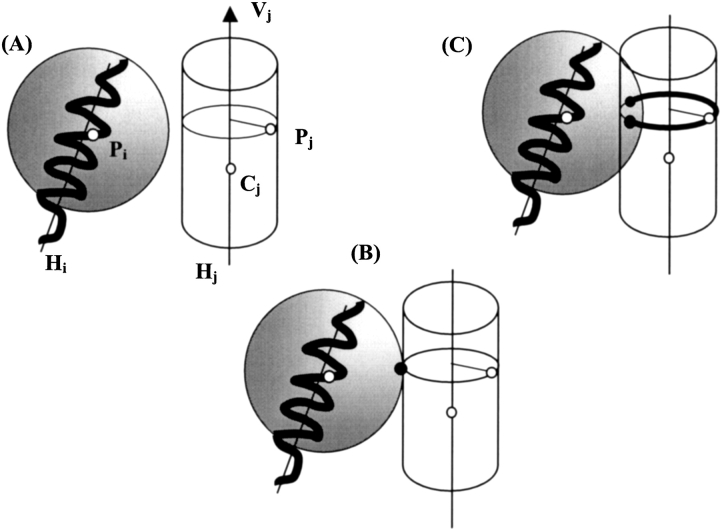
In this section, we probe the number of oriented templates when exact interhelix distances are provided. We assume we have generated all possible labeled templates and we are given a set of exact distances between backbone atoms of the helices. All backbone atoms are fully positioned relative to their respective helices, and all helices belong to a labeled template. Because each helix can freely rotate around its axis, each atom lies on a circle perpendicular to its helix axis (cf. atom Pj in Fig. 3A ▶). Now let us assume some helix Hi has already been positioned in the bilayer reference system and also assume that a distance dij is given between an atom of Hi and an atom of a second helix Hj. Given these assumptions, there are no more than two possible positions for Hj. In fact, as illustrated in Figure 3 ▶, finding the position(s) of helix Hj using distance dij from helix Hi that has already been positioned leads to no position (A), one position (B), or two positions (C), depending on the number of points obtained when intersecting a circle with a sphere. Now that helix Hj has been oriented, positioning helix Hk from Hj using distance djk should lead to no more than two positions for each Hj position, and consequently to no more than four positions from Hi. Although according to the previous section we have 360/Δα−1 possible positions for Hi, if all n−1 helices are positioned from Hi, the final number of oriented templates is bounded by 2n−1(360/Δα−1). This result is valid as long as there is no cycle in the distance graph. Indeed, as illustrated in Figure 4 ▶, when a cycle is introduced only one position remains for all helices belonging to the cycle. Although we do not rigorously prove the above statement, in all the tests carried out for this study we observed that only one position per helix permits formation of a cycle. Let n0 be the number of helices that are not linked to any other helix through a distance, and let n2 be the number of helices that are in at least one cycle in the distance graph. According to the above discussion, the total number of orientations of a given labeled template is at most 2n−n0−n2(360/Δα−1)n0. Note that this number ranges from 1 when n2 = n, that is, when the distance graph is a cyclic graph, to 26(360/Δα−1) = 320 for seven helices and Δα = 60° when the distance graph is connected but contains no cycles.
Figure 3.
Helix positioning using distance constraints. We assume that helix Hi has already been positioned and that we are provided with the distance dij between points Pi and Pj. Helix Hj is defined by its center of mass Cj and its unit vector Vj. Pj is an atom of Hj, which without distance information can be at any position on a circle induced by the possible orientations of Hj around its axis. Positioning helix Hj consists of finding a position for Pj such that Pj lies on its circle and is at distance dij from Pi, or in other words is on the sphere centered on Pi and of radius dij. (A) Circle and sphere do not intersect; all possible Pj positions are too far from Pi. The other situation without solution (not represented here) is when all possible Pj positions are too close to Pi, in which case the circle is inside the sphere. (B) The circle is tangent to the sphere; only one Pj position is at distance dij from Pi. (C) The circle crosses the sphere and the two intersection points are the two possible positions for Pj.
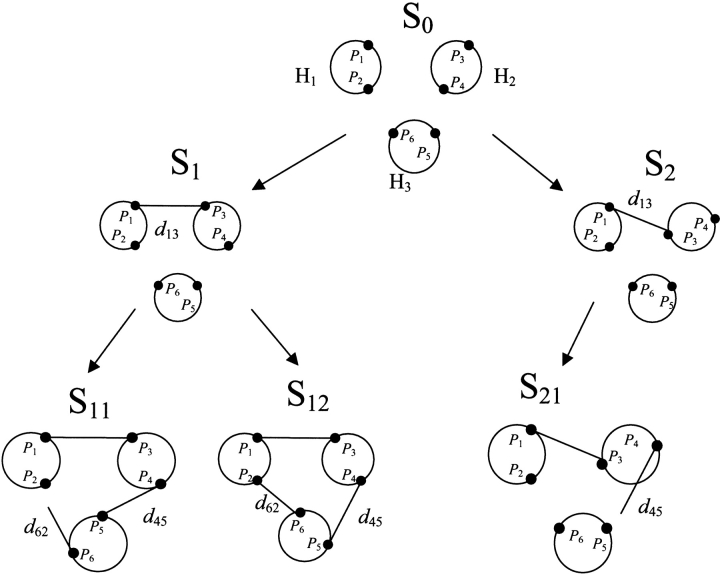
Figure 4.
Enumerating oriented templates. S0 is the initial labeled template composed on three helices viewed from the top. We assume that helix H1 has already been positioned and that distances d13 = D(P1,P3), d45 = D(P4,P5), and d62 = D(P6,P2) are provided. Helices H2 and H3 are arbitrarily positioned in S1. As explained in Figure 3 ▶, there are two positions for P3, both being at distance d13 from P1; these are represented by templates S1 and S2. Similarly there are two positions for P5 at distance d45 from P4 (templates S11 and S12). Template S11 is rejected because distance d62 is not matched; template S21 is also rejected because distance d45 is too short. Template S12 is the only one passing all distance tests.
Conformational space size with experimental distance constraints
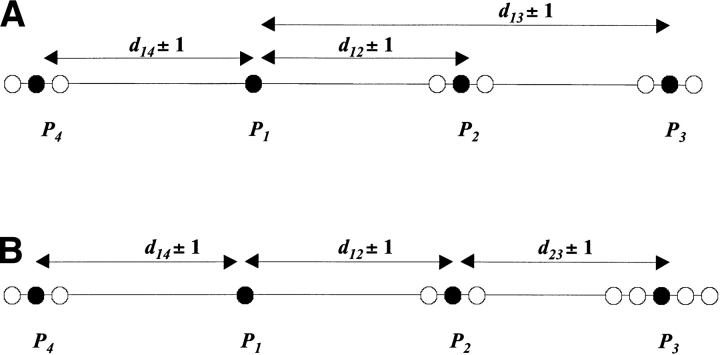
Experimental distances are never exact. For instance, NOE NMR distances are generally reported in bins of 2 Å and thus have an associated error of at least 1 Å. Cross-link distances have errors depending on the length and the flexibility of the cross-linker and the crosslinked amino acid side chains. For instance, the homobifunctional lysine-specific crosslinker BS3 (Pierce Biotechnology) yields Cα-Cα distance information in the range 5–24 Å (Young et al. 2000). Dipolar EPR distance measurements, ignoring interspin orientation, have errors of approximately 5 Å (Rabenstein and Shin 1995). Intuitively, for a given set of distances, as error increases so should the number of oriented templates matching the provided data. Less intuitive is the fact that the number of oriented templates also increases with the radius of the distance graph. In order to simplify our discussion, let us assume as illustrated in Figure 5 ▶ that we have four points, P1 through P4, embedded in a one-dimensional space. In the first case (A), distances d12, d13, and d14 are provided, each having an error of one arbitrary unit. Note that because each point is linked to P1, the radius of the distance graph is 1. We are interested in counting the number of possible configurations that are separated by one unit. Clearly, each point Pi, i=2,3,4 has three possible positions for which each configuration is separated by one unit: Pi−1, Pi, and Pi+1, where Pi = P1+d1i. The total number of configurations is thus 27. In case (B), the provided distances are d12, d23, and d14, and the radius of the distance graph is 2. For the reason mentioned above, points P2 and P4 have three possible configurations, and each P2 position leads to three possible positions for P3: P2+d23−1, P2+d23 and P2+d23+1. Now, the three positions for P2 are P1+d12−1, P1+d12, and P1+d12+1, and these lead to five final positions for P3 relative to P1: P1+d12+d23−2, P1+d12+d23–1, P1+d12+d23, P1+d12+d23+1, and P1+d12+d23+2. Consequently the total number of configurations for case (B) is 45.
Figure 5.
(A) The radius of the distance graph is 1 and the number of possible positions is 3 for points P2, P3, and P4. The total number of configurations is 27. (B) The radius of the distance graph is 2; points P1 and P4 have 3 possible positions, and point P3 has 5 positions; thus the total number of configurations is 45.
The algorithm used for enumerating oriented templates matching a set of distances with errors is detailed in Materials and Methods. This algorithm was used to probe changes in the numbers of unlabeled, labeled, and oriented templates with increasing number of distances, increasing error, and increasing radius of the distance graph. All runs were performed on a library of 150,000 unlabeled templates composed of seven helices generated using Bowie’s code, which is also briefly described in Materials and Methods. The distances were compiled for rhodopsin (PDB entry 1f88) and are listed in Figure 1 ▶. All labeled templates matching the distances were clustered with a clustering algorithm given in Materials and Methods using clustering RMSD values of 4 Å and 6 Å. Oriented templates were not clustered; however, the number of oriented templates matching the provided distances was computed on the clustered labeled templates.
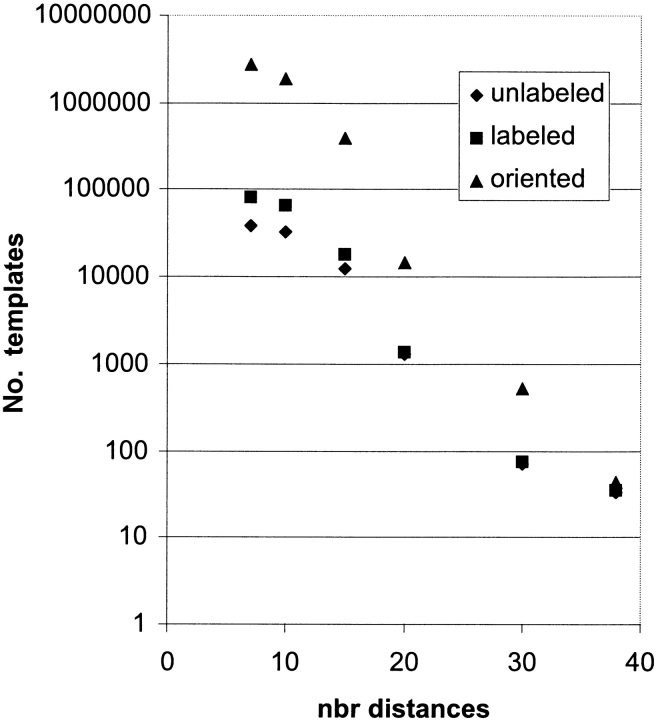
Table 1 and Figure 6 ▶ report the number of templates with number of distances ranging from 7 to 38. The 38 distances represent potential chemically cross-linkable amino acid pairs (K-K, K-D, K-E, K-C, C-C) and are listed in Figure 1 ▶. From this set, distances were removed at random, and an error of ±4 Å was attributed to each residual distance. It is clear from Figure 6 ▶ that the number of unlabeled, labeled, and oriented templates decreases exponentially as the number of distances increases. Specifically, the slopes in Figure 6 ▶ are −0.10 for unlabeled templates, −0.13 for labeled templates, and −0.22 for oriented templates. For Nd distances, the number of unlabeled templates scales as 150,000 × 0.78Nd, the number of labeled templates scales as 4.5 × 106 × 0.73Nd, and the number of oriented template scales as 351.5 × 109 × 0.59Nd. Hence, the number of oriented templates decreases slightly faster than the other template types as the number of distances increases.
Table 1.
Number of seven-helix membrane protein templates vs. number of distances
| nbr distances | Unlabeled templates | Labeled templates | Oriented templates | Labeled clustered RMSDHel = 4 Å | Labeled clustered RMSDHel = 6 Å | Oriented clustered RMSDHel = 4 Å | Oriented clustered RMSDHel = 6 Å |
| 7 | 39,606 | 84,289 | 2,671,680 | 50,189 | 8192 | 1,590,824 | 259,659 |
| 10 | 32,298 | 66,295 | 1,926,966 | 36,480 | 5372 | 1,060,347 | 156,145 |
| 15 | 12,826 | 18,212 | 394,705 | 10,738 | 1954 | 232,722 | 42,349 |
| 20 | 1274 | 1369 | 14,656 | 573 | 127 | 6134 | 1360 |
| 30 | 71 | 73 | 518 | 65 | 32 | 461 | 227 |
| 38 | 33 | 35 | 44 | 31 | 16 | 39 | 20 |
Results obtained using a distance error of ±4 Å and clustered with RMSDHel values of 4 Å and 6 Å (see Materials and Methods for definition of RMSDHel).
Figure 6.
Number of seven-helix membrane protein templates versus number of distances (data listed in Table 1).
As mentioned in the introduction, the purpose of our enumeration algorithm is to provide starting structures for further refinement with energetic calculations and molecular simulations. Local conformational search is a well-established technique in computational chemistry and has long been used for soluble protein fold recognition (Godzik et al. 1992). More recently, local search techniques have been proposed that are specific for membrane proteins. Nikiforovich et al. (2001) proposed a technique for searching the energetically favored orientation of pairs of helices. Provided that the two helices have been positioned in the bilayer reference system, the method exhaustively enumerates all possible helix rotations with an angular increment of 30°. Energy calculations are performed using the ECEPPP/2 force fields (Dunfield et al. 1978), and low-energy conformations are retained. Using this method, Nikiforovich et al. (2001) generated low-energy conformations for bacteriorhodopsin that are 3.13 Å RMSD from the crystal structure. Vaidehi et al. (2002) reported a molecular dynamics code, COARSEROT, which performs coarse-grain rotations of the helical orientations around their axis through a grid of rotation angles. Using this technique, those authors retrieved the seven-helix bundles of bacteriorhodopsin and bovine rhodopsin with RMSDs of 2.9 Å and 3.1 Å, respectively.
Now, for each labeled template, in order to fully explore the conformational space of that template, one must rotate all the helices around their axes and compute energetic parameters for each orientation. As detailed in Materials and Methods, helix orientations in an oriented template are enumerated using an angular increment of Δα = 60°; thus, the local conformational space of each oriented template can be fully explored by rotating each helix with an angle no greater than 60°. Consequently, the refinement procedures mentioned above should be able to process (360/60)7 = 279,936 oriented templates of seven helices in the same computational time it takes to run one labeled template. Using the latter number as a threshold (according to Table 1) for systems of seven helices, at least 15 distances with an error of 4 Å are needed to be able to explore the conformational space in a reasonable computational time.
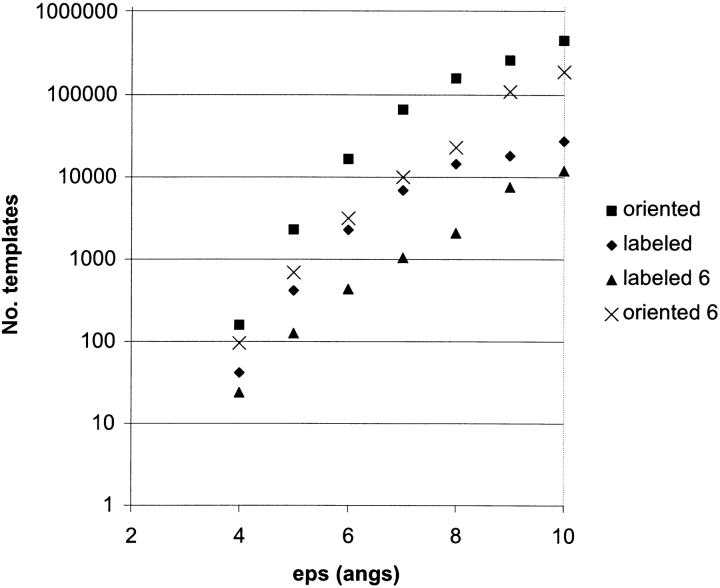
Table 2 and Figure 7 ▶ give the number of templates for the 38 distances listed in Figure 1 ▶ with increasing distance error. All template numbers increase exponentially with the distance error. The slopes computed from Table 2 are 0.62 for unlabeled and labeled templates and 0.75 for oriented templates. The scaling behavior for unlabeled and labeled templates is 0.25 × 4.2ɛ, where ɛ is the error, and 0.35 × 5.6ɛ for oriented templates. Interestingly, although the slopes are more pronounced for decreasing error than increasing number of distances (−0.62 versus −0.10 for unlabeled templates), the number of templates is not as large as one might expect, and up to a fairly large error of ±8 Å, the number of oriented templates is small enough to be manageable by local conformational refinements.
Table 2.
Number of seven-helix membrane protein templates vs. distance error
| Error (Å) | Unlabeled templates | Labeled templates | Oriented templates | Labeled clustered RMSDHel = 4 Å | Labeled clustered RMSDHel = 6 Å | Oriented clustered RMSDHel = 4 Å | Oriented clustered RMSDHel = 6 Å |
| ±4 | 33 | 35 | 44 | 31 | 16 | 39 | 20 |
| ±5 | 408 | 417 | 2314 | 318 | 126 | 1765 | 699 |
| ±6 | 2259 | 2339 | 16,575 | 1578 | 438 | 11,182 | 3104 |
| ±7 | 6468 | 6892 | 65,117 | 4551 | 1049 | 42,999 | 9911 |
| ±8 | 12,851 | 14,162 | 159,832 | 9520 | 2073 | 107,442 | 23,396 |
Results obtained using the 38 distances listed in Fig. 1 ▶ and clustered with RMSDHel values of 4 Å and 6 Å (see Materials and Methods for definition of RMSDHel).
Figure 7.
Number of seven-helix membrane protein templates versus distance error (eps [data listed in Table 2]).
The number of templates with increasing distance graph radius is given in Table 3. Although not as pronounced as with the distance error, the number of templates increases exponentially with the radius. The scaling computed on Table 3 is 500 × 2.9r, where r is the radius, for unlabeled and labeled templates, and 2500 × 4.5r for oriented templates. As a consequence of this finding, it is preferable to design experiments that will compute distances from the same reference helix rather than generating distances linking helices in a daisy-chain manner.
Table 3.
Number of seven-helix membrane protein templates vs. distance graph radius
| Radius | Unlabeled templates | Labeled templates | Oriented templates | Labeled clustered RMSDHel = 4 Å | Labeled clustered RMSDHel = 6 Å | Oriented clustered RMSDHel = 4 Å | Oriented clustered RMSDHel = 6 Å |
| 1 | 1354 | 1487 | 11510 | 1104 | 362 | 8545 | 2802 |
| 2 | 4873 | 5854 | 73,474 | 4591 | 1387 | 57,622 | 17,408 |
| 3 | 10,056 | 13,491 | 229,830 | 10,293 | 2671 | 175,350 | 45,503 |
Specific membrane proteins
In order to validate the value of the proposed method for generating structures suitable for energetic refinement, we considered seven known crystal structures with varying numbers of transmembrane helices and attempted to predict the positions of the transmembrane helices. Specifically, only those portions of the transmembrane helices completely embedded in the membrane were considered (e.g., the helix containing residues 68–79 of Glycerol uptake facilitator protein was not considered). For each structure we derived a set of distance constraints corresponding to pairs of amino acids (K-K, K-D, K-E, K-C, and C-C) that could potentially be cross-linked using chemical cross-linkers. Specifically, we considered the following cases (PDB identifier and number of distance constraints in parentheses): Bacteriorhodopsin (1c3w, 60), Halorhodopsin (1e12, 9), Rhodopsin (1f88, 38), Aquaporin-1 (1fqy, 17), Glycerol uptake facilitator protein (1fx8, 43), Sensory Rhodopsin (1jgj, 18), and a subunit of Fumarate reductase flavoprotein (1qlaC, 58). For all cases the error assigned to each distance constraint was ±4 Å, and library sizes of 300,000 unlabeled templates for seven helices (twice the size used earlier), 150,000 for six helices, and 50,000 for five helices were searched.
Table 4 reports the results for these seven cases in terms of number of distance constraints, number of solutions, and minimum RMSD among the computed structures and the structure from the PDB across all Cα atoms (Cα–RMSD) for each case (RMSD calculated using the MMTSB Tool Set; Feig et al. 2001). In general, the predicted positions of the helices in the helix assemblies were in good agreement with the PDB structures, having Cα–RMSD values ranging from 2.3 Å to 6.0 Å for the seven test cases. As discussed above, the number of solutions declines exponentially with the number of distances. For our test cases the number of solutions is heavily dependent on the number of distance constraints ranging from 8,274,078 for the structure having only nine distances to a minimum of 26 for the case with 58 constraints. Because our method involves searching a data set of potential structures for structures that satisfy a set of distance constraints, we did not expect the quality of the best structures to depend on the number of distance constraints. Table 4 supports this expectation, with very little correlation between RMSD and number of constraints. This lack of correlation is likely due in part to the fact that the quality of the computed structure is a function of the number of distance constraints, the error on each distance, and the combination of sites among which the distances were measured. Our stated goal is to provide starting structures for further refinement using either energetic constraints (force field methods) or ad hoc scoring functions based on bioinformatics data. To this end, these results clearly show that, given a set of experimental distance constraints, our method generates sets of helical positions close to those of the known structure, providing a set of candidate structures from which further structural refinements can be performed.
Table 4.
Results for seven membrane protein structures using distance constraints between potential cross-linking amino acid pairs (K-K, K-D, K-E, K-C, C-C) and for the structure of rhodopsin using a set of experimental distances
| Protein | PDB entry | Number of helicesa | Number of distances | Number of solutions for ɛ = 4 Å | Cα RMSDb (Å) |
| Fumarate reductase flavoprotein | 1qlaC | 5 | 58 | 26 | 4.4 |
| Aquaporin-1 | 1fqy | 6 | 17 | 1,154,191 | 4.6 |
| Glycerol uptake facilitator protein | 1fx8 | 6 | 43 | 208 | 6.0 |
| Bacteriorhodopsin | 1c3w | 7 | 60 | 330 | 3.0 |
| Halorhodopsin | 1e12 | 7 | 9 | 8,274,078 | 2.3 |
| Sensory Rhodopsin | 1jgj | 7 | 18 | 329,502 | 3.7 |
| Rhodopsin | 1f88 | 7 | 38 | 108 | 3.2 |
| Rhodopsin (Experimental Constraints given in Table 5) | 1f88 | 7 | 27 | 87 | 4.3 |
a Only those helices completely within the membrane were included. Specifically 1fqy helices corresponding to residues 76 to 85 and 192 to 201 were removed and for 1fx8 helices corresponding to residues 68 to 79, 125 to 133, 203 to 217, and 221 to 226 were removed.
b Minimum Cα-RMSD among all solutions (oriented templates) and the PDB entry shown in column 2.
The number of solutions reported for ±4 Å error are unclustered oriented or atomistic templates. Note that the number of solution obtained for rhodopsin (108) is different from the one found in the first row of Table 2 (44), because we doubled the size of the unlabeled templates library. The average distance error for rhodopsin in the last row is ±3.75 Å.
As a second validation we then predicted the positions of the seven transmembrane helices of dark-adapted Rhodopsin (1f88) using a set of distance constraints compiled from various experiments reported in the literature (Yeagle et al. 2001). These included dipolar EPR distances (Farrens et al. 1996; Yang et al. 1996; Albert et al. 1997; Galasco et al. 2000), disulfide mapping distances (Yu et al. 1995, 1999; Sheikh et al. 1996; Cai et al. 1997, 1999), and distances from electron cryomicroscopy (Unger and Schertler 1995). The distance constraints are shown in Table 5. The average distance error is ±3.75 Å.
Table 5.
Experimental distances used for the Rhodopsin structure
| Helix1 | Helix2 | Residue1 | Residue2 | Minimum distance | Maximum distance |
| H3 | H6 | VAL_139 | LYS_248 | 3.80 | 22.20 |
| H3 | H6 | VAL_139 | GLU_249 | 8.30 | 26.70 |
| H3 | H6 | VAL_139 | VAL_250 | 8.30 | 26.70 |
| H3 | H6 | VAL_139 | THR_251 | 3.80 | 22.20 |
| H3 | H6 | VAL_139 | ARG_252 | 8.30 | 26.70 |
| H5 | H6 | VAL_204 | PHE_276 | 3.18 | 8.18 |
| H3 | H5 | CYS_140 | CYS_222 | 1.18 | 10.18 |
| H3 | H5 | CYS_140 | GLN_225 | 4.18 | 7.18 |
| H3 | H6 | ARG_135 | VAL_250 | 2.18 | 9.18 |
| H3 | H5 | TYR_136 | CYS_222 | 4.18 | 7.18 |
| H3 | H5 | TYR_136 | GLN_225 | 3.18 | 8.18 |
| H2 | H3 | PRO_71 | GLU_134 | 7.00 | 15.00 |
| H2 | H3 | GLY_90 | PHE_116 | 5.00 | 10.00 |
| H2 | H4 | FRO_71 | ALA_153 | 4.00 | 11.00 |
| H2 | H4 | MET_86 | LEU_172 | 15.00 | 20.00 |
| H3 | H5 | TYR_136 | LEU_226 | 5.00 | 10.00 |
| H3 | H5 | LEU_125 | PRO_215 | 5.00 | 10.00 |
| H4 | H5 | HIS_152 | GLN_225 | 18.00 | 22.00 |
| H5 | H6 | LEU_216 | LEU_258 | 9.00 | 13.00 |
| H6 | H7 | MET_253 | MET_305 | 5.00 | 9.00 |
| H6 | H7 | CYS_264 | SER_298 | 5.00 | 9.00 |
| H1 | H7 | MET_39 | ILE_286 | 9.00 | 14.00 |
| H3 | H6 | GLY_114 | TYR_268 | 14.00 | 18.00 |
| H4 | H6 | PRO_171 | TYR_268 | 16.00 | 21.00 |
| H2 | H6 | ASN_73 | VAL_250 | 10.00 | 15.00 |
| H1 | H6 | THR_62 | VAL_250 | 16.00 | 20.00 |
| H1 | H6 | LEU_47 | CYS_264 | 15.00 | 20.00 |
Because the published EPR dipolar distances are between nitroxide spin labels, they do not directly correspond to distances between helical axes. To better represent the dipolar EPR distances, we used a Monte Carlo minimization-based method (Sale et al. 2002) to uncover the most probable orientations of the spin label at each of the labeled sites V139, K248, V249, V250, T251, and R252. From these orientations, the mean length of the Cα to nitroxide N vector for the six sites was approximately 9 Å. This length was used to adjust the lower and upper limits of the reported distances in order to better represent the inter-nitroxide distances as helix backbone distances. For the disulfide mapping distances, we used a Cα to Cα distance of 5.68 Å, which corresponds to two Cβ to Sγ bonds (1.82 Å) and one Sγ to Sγ bond (2.04 Å), plus and minus the reported error.
The results are shown in the last row of Table 4. Using a set of 27 distance constraints with differing amounts of error, only 87 helical bundles of the possible 300,000 × 30 × 78,125 = 0.7 × 1012 seven-helix configurations were found that matched the set of distances. This yielded structures with Cα–RMSD values ranging from 4.3 Å to 9.5 Å. These results show that even in the case of using only 27 distance constraints taken from a variety of experimental methods with differing levels of error, a reasonable number of structures suitable for further refinement can be extracted from a large data set of possible helix bundles.
Conclusions
We have demonstrated a method for generating helical bundles (oriented templates) satisfying a given set of interhelical distance constraints. Using the structure of rhodopsin (1f88) as a model, we showed that (1) the number of templates decreases exponentially as the number of distance constraints increases, (2) the number of templates increases exponentially with increasing error on the distances, (3) the number of templates increases exponentially with increasing radius of the distance graph, and (4) experiments designed to measure distances from the same reference helix are preferable to those designed to generate distances linking helices in a daisy-chain manner. We also note that the slopes derived from Tables 1 and 2 indicate that decreasing the error on the distance constraints decreases the number of solution templates at a faster rate than does increasing the number of distances. This indicates that in order to cut down the number of solutions, one may want to lower the error of a given experiment rather than adding new data points.
Conclusion (4) is particularly important to the design of experiments in which distances are derived from the interaction of external probe molecules incorporated at specific sites, such as in FRET and dipolar EPR experiments. Our findings suggest that fewer measurements from a common helix to the other helices result in a reduced set of possible structures compared to many measurements between distinct pairs of amino acids. The obvious consequence of this finding is the need for fewer experiments involving amino acid mutations and labeling. For potentially higher-throughput methods such as chemical cross-linking, this result may not assist in the initial experimental design, as cross-linking does not require a directed mutagenesis and labeling strategy. Conclusion (4) may, however, provide a guide for the selection of cross-linking reagents or sites for mutagenesis once a preliminary structural model has been constructed.
Using a set of seven helical bundles with 5–7 helices and varying numbers of distance constraints as test cases, we found that a reasonable number (in terms of starting points for further refinement) of helical bundles with helix positions near the native structure can be generated. The same conclusion can also be drawn for rhodopsin (1f88) using 27 experimental distances taken from the literature. This finding has important consequences in that further refinement using either force field or ad hoc scoring methods with local conformational search should produce reasonably accurate model structures. Furthermore, given a set of model structures, experiments can then be designed to differentiate among the models and provide data for further structural refinement.
Although the technique described here has been demonstrated on transmembrane proteins with only up to seven helices, there are no limitations on the number of helices that can be processed. Note, however, that according to Bowie (1999), the initial library of unlabeled templates has a size that increases exponentially with the number of helices. Consequently the method presented here is somewhat limited by the ability to generate a library of unlabeled templates comprising more than seven helices that is representative of the entire conformational space. One possible extension of this work is to modify Bowie’s code and incorporate distance information when generating unlabeled templates.
Another possible extension of the code is to handle not only helices but also β-strands and β-sheets. This extension of our technique is straightforward, as β-strands and β-sheets can be represented as special helices; that is, they too can be modeled with a center of mass and a unit vector. Furthermore, as with helices, if one knows the position of one atom of the β-strand or -sheet, one knows the position of all atoms. An important difference in the β-strand case is the additional constraints imposed on the structure by the interstrand hydrogen-bonding network. If pairs of adjacent strands can be determined with their respective orientations, the solution space of the corresponding template library is expected to be quite small relative to the library of an all-helical system of equivalent size.
The computer codes described next to generate labeled and oriented templates are available upon request.
Materials and methods
The computer code described next, to generate labeled and oriented templates, are available upon request.
Generating unlabeled templates
Unlabeled templates were generated using Bowie’s software, which is described in detail in a previous paper (Bowie 1999). This software generates random unlabeled templates matching specific criteria determined from known membrane protein structures (Bowie 1997). Briefly, the angle between each helix axis and the bilayer normal is random but smaller than 40°. The distance of closest approach between two neighboring helices is no greater than 13.4 Å and no smaller than 6 Å. The angle between the axes of two neighboring helices follows a distribution computed on known membrane proteins and reported by Bowie (1997). The output is a library of unlabeled templates each satisfying the above criteria. The user specifies the number of helices and the desired number of templates.
Generating labeled templates
Labeled templates were enumerated for each element of the library of unlabeled templates. Theoretically, for n helices there are 2n! labelings of the helices. However, this number can be drastically reduced if one adds the three following interhelix connection constraints: (1) The distance of closest approach between consecutive helices is in the 6–13.4 Å range; (2) the distance between the end points of consecutive helices is in the 7–22 Å range; and (3) there are no cross-over connections. The justifications for the above constraints can be found in Bowie (1999). Our enumeration algorithm finds all possible labels for each helix sequentially. The above constraints are applied on the fly by the algorithm even if the template is not yet fully labeled. Running this algorithm, we observed that most labelings are rejected before completion, and we were able to process libraries comprising several thousand templates quite efficiently in a couple of hours of CPU time on an SGI/O2 workstation.
Generating oriented or atomistic templates with distance constraints
Recall that a labeled template is oriented once an arbitrary backbone atom has been positioned in the bilayer reference system for each helix. As discussed, for n helices there are n360/Δα−1 possible orientations per labeled template, each being separated by at least one Δα angle. This number can be reduced by introducing distance constraints using the algorithm described next and illustrated in Figures 3 ▶ and 4 ▶. In order to minimize the effect of the distance graph radius (cf. Table 3 and Results and Discussion section), the first step of the algorithm consists of finding the helix that gives the smallest radius in the distance graph. If the graph is not connected (the radius is infinite), then the procedure halts, as the helices cannot be oriented due to a lack of distances. Otherwise, the algorithm then tests all possible orientations of the initial helix. Each orientation is incremented by a 10° angle until a solution is found and by a 60° angle if the previous angle led to at least one solution. The 10° angular increment is small enough to avoid missing solutions, whereas the 60° increment was chosen to provide a 2.5 Å RMSD between consecutive solutions. For each tested orientation of the initial helix, the following steps are applied until there are no more distances to be considered. (1) A helix, Hi, also named the current helix, is chosen such that this helix has already been positioned and is attached through distances to helices that have not yet been positioned. Note that if no such helix can be found then all helices have been positioned, and the algorithm then verifies that all distance constraints are satisfied and rejects the solution if there are unsatisfied distance constraints. Otherwise, (2) one chooses an unused distance dij with associated error ɛij between two atoms Pi belonging to the current helix Hi, and Pj belonging to a helix Hj. Because Hi is the current helix, Pi is fully positioned in the bilayer reference system, whereas (as illustrated in Fig. 3 ▶) Pj lies on a circle orthogonal to Vj, the axis of Hj. This circle is centered on Cj + (CjPj • Vj) Vj, where Cj is the center of mass of Hj, and has a radius |CjPj × Vj| (norm of the product between vectors CjPj and Vj). (3) The position of Pj is found by intersecting the circle on which Pj lies and the sphere centered on Pi of radius dij ± ɛij. The intersection of a sphere and a circle leads to no solution, one position, or two positions. If no solution can be found, the procedure halts and a new position of the initial helix is tested. Otherwise for each Pj position found, helix Hj is rotated clockwise and counterclockwise in 60° angular increments, and all new Pj positions that are at distances dij ± ɛij from Pi are retained. Again, 60° was chosen to provide a 2.5 Å RMSD between consecutive solutions. Finally, for each Pj position stored, steps 1 through 3 are applied until all distances have been used. Each time all distances have been processed successfully, the resulting template is a solution and is added to the library of oriented templates.
Clustering labeled templates
Labeled templates are represented by numbered helices located in the bilayer reference system through the coordinates of their center of mass and unit axial vector. Templates are clustered using a predefined RMSDHel value computed at the helix level. Precisely, RMSDHel between two templates is calculated by averaging the RMSD between the centers of mass and the two end points of the helices of the templates. Prior to performing the calculation, the templates are rotated in the bilayer plane in order to align the vectors joining the first two helices. Bowie compared RMSD values for a full atom model with RMSDHel and found that for 100 structures with RMSDHel in the range 3.9–4.1 Å, the average RMSD of the full atom models was 2.6 Å for 86% of the atoms. In the present study, we clustered labeled templates using RMSDHel of 4 Å and 6 Å.
Acknowledgments
Funding for this work was provided by Sandia National Laboratories under contract DE-AC04-94AL85000. Sandia is a multiprogram laboratory operated by Sandia Corp., a Lockheed Martin Company, for the U.S. Department of Energy (DOE)’s National Nuclear Security Administration. J.-L.F. is also pleased to acknowledge funding provided by the Mathematics, Information, and Computer Science Program of the U.S. DOE. Finally, we thank Joe Schoninger for useful comments.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0305003.
References
- Albert, A.D., Watts, A., Spooner, P., Grobner, G., Young, J., and Yeagle, P.L. 1997. A solid state NMR characterization of the substrate binding specificity and dynamics for the L-fucose-H+ membrane transport protein of E. coli. Biochim. Biophys. Acta 1328 74–82. [DOI] [PubMed] [Google Scholar]
- Bowers, M., Cohen, F.E., and Dunbrack Jr., R.L. 1997. Side chain prediction from a backbone-dependent rotamer library: A new tool for homology modeling. J. Mol. Biol. 267 1268–1282. [DOI] [PubMed] [Google Scholar]
- Bowie, J.U. 1997. Helix packing in membrane proteins. J. Mol. Biol 272 780–789. [DOI] [PubMed] [Google Scholar]
- ———. 1999. Helix-bundle membrane protein fold templates. Protein Sci. 8 2711–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, K., Langen, R., Hubbell, W.L., and Khorana, H.G. 1997. Structure and function in rhodopsin: Topology of the C-terminal RT polypeptide chain in relation to the cytoplasmic loops. Proc. Natl. Acad. Sci. 94 14267–14272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, K., Klein-Seetharaman, J., Hwa, J., Hubbell, W.L., and Khorana, H.G. 1999. Structure and function in rhodopsin. Effects of disulfide cross-links in the cytoplasmic face of rhodopsin on transducin activation and phosphorylation by rhodopsin kinase. Biochemistry 38 12893–11898. [DOI] [PubMed] [Google Scholar]
- Dunfield, L.G., Burgess, A.W., and Scheraga, H.A. 1978. Energy parameters in polypeptides. 8. Empirical potential energy algorithm for the conformational analysis of large molecules. J. Phys. Chem. 82 2609–2616. [Google Scholar]
- Farrens, D.L., Altenbach, C., Yang, K., Hubbell, W.L., and Khorana, H.G. 1996. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science 274 768–770. [DOI] [PubMed] [Google Scholar]
- Feig, M., Karanicolas, J., and Brooks, C.L.I. 2001. MMTSB Tool Set. MMTSB NIH Research Resource, The Scripps Research Institute, La Jolla, CA.
- Galasco, A., Crouch, R.K., and Knapp, D.R. 2000. Intrahelic arrangement in the integral membrane protein rhodopsin investigated by site-specific chemical cleavage and mass spectrometry. Biochemistry 39 4907–4914. [DOI] [PubMed] [Google Scholar]
- Godzik, A., Kolinski, A., and Skolnick, J. 1992. Topology fingerprint approach to the inverse folding problem. J. Mol. Biol 227 227–238. [DOI] [PubMed] [Google Scholar]
- Gromiha, M.M. 1999. A simple method for predicting transmembrane α-helices with better accuracy. Protein Eng. 12 557–561. [DOI] [PubMed] [Google Scholar]
- Hirokawa, T., Boon-Chieng, S., and Mitaku, S. 1998. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 14 378–379. [DOI] [PubMed] [Google Scholar]
- Nikiforovich, G.V., Galaktionov, S., Balodis, J., and Marshall, G.R. 2001. Novel approach to computer modeling of seven-helical transmembrane proteins: Current progress in the test case of bacteriorhodopsin. Acta Biochim. Pol. 48 53–64. [PubMed] [Google Scholar]
- Rabenstein, M.D. and Shin, Y.K. 1995. Determination of the distance between two spin labels attached to a macromolecules. Proc. Natl. Acad. Sci. 92 8239–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale, K.L., Sar, C., Sharp, K.A., Hideg, K., and Fajer, P. 2002. Structural determination of spin label immobilization and orientation: A Monte Carlo minimization approach. J. Magn. Reson. 156 104–112. [DOI] [PubMed] [Google Scholar]
- Sheikh, S.P., Zvyaga, T.A., Lichtarge, O., Sakmar, T.P., and Bourne, H.R. 1996. Rhodopsin activation blocked by metal-ion-binding sites linking transmembrane helices C and F. Nature 383 347–350. [DOI] [PubMed] [Google Scholar]
- Unger, V.M. and Schertler, G.F. 1995. Low resolution structure of bovine rhodopsin determined by electron cryo-microscopy. Biophys. J. 68 1776–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidehi, N., Floriano, W.B., Trabanino, R., Hall, S.E., Freddolino, P., Choi, E.J., Zamanakos, G., and Goddard III, W.A. 2002. Prediction of structure and function of G protein-couple receptors. Proc. Natl. Acad. Sci. 99 12622–12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend, G. 1990. WHAT IF: A molecular modeling and drug design program. J. Mol. Graphics 8 52–56. [DOI] [PubMed] [Google Scholar]
- Xiang, Z., Soto, C., and Honig, B. 2002. Evaluating conformational free energies: The colony energy and its application to the problem of loop prediction. Proc. Natl. Acad. Sci. 99 7432–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, K., Farrens, D.L., Hubbell, W.L., and Khorana, H.G. 1996. Structure and function in rhodopsin. Single cysteine substitution mutants in the cytoplasmic interhelical E-F loop region show position-specific effects in transducin activation. Biochemistry 35 14040–14046. [DOI] [PubMed] [Google Scholar]
- Yeagle, P.L., Choi, G., and Albert, A.D. 2001. Studies on the structure of the G-protein-coupled receptor rhodopsin including the putative G-protein binding site in unactivated and activated forms. Biochemistry 40 11932–11937. [DOI] [PubMed] [Google Scholar]
- Young, M.M., Tang, N., Hempel, J.C., Oshiro, C.M., Taylor, E.W., Kuntz, I.D., Gibson, B.W., and Dollinger, G. 2000. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc. Natl. Acad. Sci. 97 5802–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H., Kono, M., McKee, T.D., and Oprian, D.D. 1995. A general method for mapping tertiary contacts between amino acid residues in membrane embedded proteins. Biochemistry 34 14963–14969. [DOI] [PubMed] [Google Scholar]
- Yu, H., Kono, M., and Oprian, D.D. 1999. State-dependent disulfide cross-linking in rhodopsin. Biochemistry 38 12028–12032. [DOI] [PubMed] [Google Scholar]