Abstract
Fusion protein constructs of the 56 amino acid globular protein GB-1 with various peptide sequences, coupled with the incorporation of a histidine tag for affinity purification, have generated high-yield fusion protein constructs. Methionine residues were inserted into the constructs to generate pure peptides following CNBr cleavage, yielding a system that is efficient and cost effective for isotopic labeling of peptides for NMR studies and other disciplines such as mass spectroscopy. Six peptides of varying sequences and hydrophobicities were expressed using this GB-1 fusion protein technique and produced soluble fusion protein constructs in all cases. The ability to easily express and purify recombinant peptides in high yields is applicable for biomedical research and has medicinal and pharmaceutical applications.
Keywords: NMR, GB-1, isotopic labeling, CNBr cleavage, affinity purification
Polypeptides represent a significant group of biomolecules, which can play an important role in various cellular functions. Included in this group of biomolecules is the 51-residue polypeptide insulin, which initiates a hormonal cellular signaling cascade in response to blood glucose concentrations (Tyrberg and Levine 2001). Indications that polypeptides behave as independent domains are exemplified by the 26-residue α-helical lytic bee venom polypeptide melittin (Hristova et al. 2001), and an effector of intracellular Ca2+-signaling, phospholamban, an integral membrane 52-residue polypeptide composed of two α-helical regions connected by a β-turn (Hutter et al. 2002; Pollesello and Annila 2002). Polypeptides attributed to diseased states such as the 37-residue islet amyloid polypeptide (hIAPP), which has been found to accumulate as amyloid fibrils in the pancreas of individuals with type II diabetes (Azriel and Gazit 2001; Mazor et al. 2002), as well as polypeptides such as LL-37, have demonstrated strong antimicrobial properties, leading to a novel class of treatment remedies of antibiotic-resistant pathogens (Gudmundsson et al. 1996; Saiman et al. 2001). Polypeptide regions can be generated from larger multidomain proteins using various proteolytic procedures, with generated regions retaining full biological activity of the parent domains. Biomedical advancements have exploited this ability of peptides to retain activity to allow them to be used as tools in research, such as for potential therapeutic agents against disease (Jacobsen 2002; Nathisuwan and Talbert 2002) and as peptide vaccines (Celis 2002).
The current peptide production technique commonly used is solid-state synthesis; yet this can be hindered by high cost and relatively low yields, as well as the inability to accurately produce peptides of lengths over ~50 residues with acceptable yields. Furthermore, the ability to introduce isotopic labels (13C, 15N, 2H) within polypeptides by solid-state methodology for NMR studies is not a viable option, because the costs involved quickly escalate. The ability to recombinantly produce peptides is an alternative to solid-state synthesis, as cell culture expression can yield high production at relatively low cost, with easy incorporation of isotopic labels. Unfortunately, expression of peptides in vivo has met with limited success because peptides are relatively poorly expressed in cell culture, attributed to an unstructured state in solution that is susceptible to cellular proteases, as well as to solubility concerns of the expressed peptide. Peptide production may also produce highly toxic effects on cells during expression (Majerle et al. 2000).
One solution to high-yield recombinant peptide production is the use of a fusion protein construct, which helps in peptide stability and solubility following expression. Fusion proteins offer protection of expressed peptides from cellular proteases, incorporation of various affinity tags for ease in fusion protein purification, and introduction of proteolytic and/or chemical cleavage sites for generation of subsequent peptide fragments. Several groups have reported success with various fusion protein constructs, with many using a hydrophobic fusion protein construct, which produces a construct that is sequestered within inclusion bodies (Jones et al. 2000; Majerle et al. 2000; Sharon et al. 2002). However, reconstitution of inclusion body proteins provides additional work and use of chemical denaturants for subsequent peptide production.
In this study, we report the production of six different fusion protein constructs using the B1 immunoglobulin binding domain of streptococcal protein G (Gronenborn et al. 1991), termed GEV-1 vector (Gronenborn and Clore 1996; Huth et al. 1997; Lindhout et al. 2002), a 56-amino acid soluble globular domain combined with a poly histidine tag affinity purification protocol. The advantage of the GB-1 fusion approach over other previous techniques is the production of a soluble fusion protein construct, with high expression yields and ease of purification. The ability of GB-1 to express multiple differing peptide fragments of various composition and length is demonstrated in this study. We have cloned, expressed, and purified six different peptides. These include cIp-RR-20, a 20-residue peptide of human cardiac troponin I (cTnI128–147) that plays a role in regulation of heart muscle contractility (Campbell et al. 1992; Tripet et al. 1997; Li et al. 2000), as well as two mutants of cIp-RR-20 (cIp-RG-20 and cIp-GR-20) that yield altered activities, with cIp-GR-20 being associated with the heart disease familial hypertrophic cardiomyopathy (Redwood et al. 1999; Hernandez et al. 2001). We have also produced and purified TRTK-12, a 12-residue homology peptide of the actin capping protein (CapZα1265–276), which has been shown to interact with the calcium-binding protein S-100 (Ivanenkov et al. 1995); FLQS-26, a 26-residue peptide from human sodium proton exchanger isoform 1 (NHE1155–179), which constitutes a predicted hydrophobic transmembrane helix (Counillon et al. 1997; Wakabayashi et al. 2000); and EDQL-26, a 26-residue peptide of cardiac troponin T (cTnT226–251), which makes a helical coiled-coil interaction with cardiac troponin I (Pearlstone and Smillie 1985). Using synthetic DNA oligonucleotides to code for the peptides of interest, as well as the introduction of methionine residues for subsequent CNBr cleavage following fusion protein purification, we have demonstrated a system that can be implemented, cloned, expressed, and purified in as little as 2 wk with high yields and minimal effort. Moreover, we demonstrate the ability of this system to produce isotopic labeling of peptides for NMR studies and to provide a strong prospect for future production of peptides for various industrial, pharmaceutical, and general research applications.
Results and Discussion
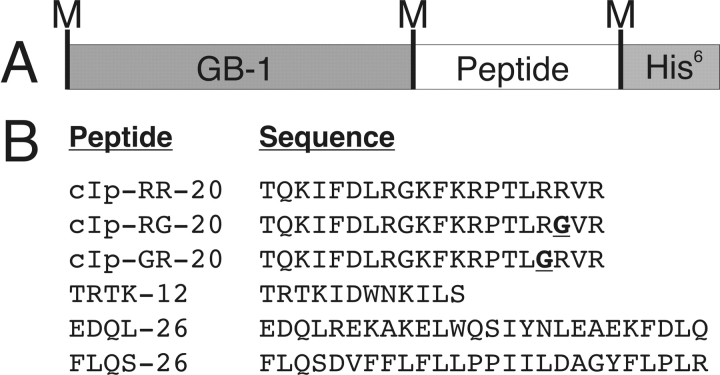
Figure 1 ▶ shows the primary amino acid sequence of each peptide cloned into a GB-1 fusion protein, ranging from 12 to 26 residues in length. In this study, we have used peptides of various lengths, hydrophobicities, and predicted secondary structure to evaluate the efficiency of peptide production. The cloning site for the inserted synthetic peptide oligonucleotides is C terminal to the GB-1 domain and N terminal to a poly-6-his tag. Three methionine residues are present in each fusion construct; one on the N terminus to initiate transcription (start codon), which is not posttranslationally cleaved, and two met residues immediately N and C terminal to the peptide of interest, which were coded for within the synthetic DNA oligonucleotides.
Figure 1.
(A) GB-1 fusion protein construction, with location of methionine residues denoted by M. (B) Peptide name and amino acid sequence of expressed peptides.
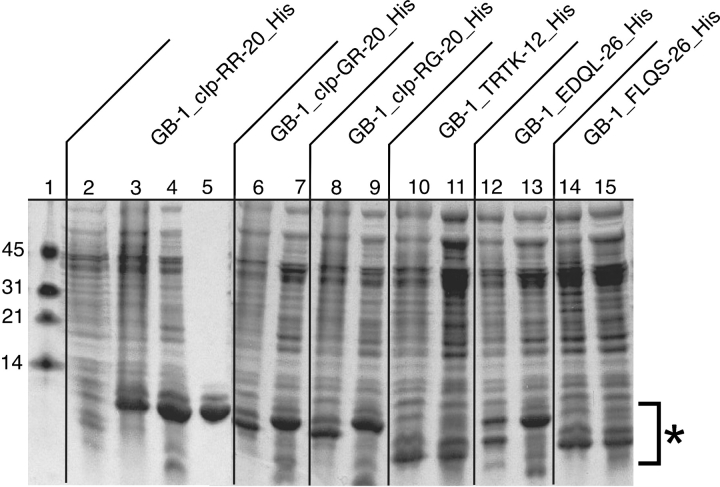
Figure 2 ▶ shows the SDS-PAGE gel of the expression of six fusion protein constructs. Lane 1 contains a low MW marker (Biorad), with MW standards as indicated. For simplicity, we have chosen to show the uninduced and induced lanes (lanes 2, 3) for the GB-1_cIp-RR-20_His construct, followed by French press lysate supernatant following centrifugation (lane 4) and GB-1_cIp-RR-20_His following purification on a Ni2+ affinity column (lane 5). As can be observed, the induced GB-1_cIp-RR-20_His expressed very well, with an average yield of 80 mg/L from expression in 2xYT. We have also included the induced cells and French press lysis supernatant for each of the remaining five fusion constructs to compare expression yields and overall solubility of expressed proteins (lanes 6–15). All six fusion constructs expressed to approximately equivalent yields, with uninduced lanes being identical (data not shown). As can be seen from the gel, all six fusion protein constructs produce a soluble fusion protein following French pressure lysis, regardless of amino acid composition of the desired peptide. This is of particular importance regarding construct GB-1_FLQS-26_His, as the peptide insert of FLQS-26 is a predicted transmembrane helix with a majority of hydrophobic residues. Thus, the ability of the GB-1 domain to solubilize a fusion protein–peptide construct is demonstrated and all six constructs are well behaved on a Ni2+ purification column producing pure fusion protein constructs. The construct GB-1_FLQS-26_His unexpectedly runs at a lower MW (lanes 14–15) than would be expected when compared with fusion construct GB-1_EDQL-26_His (lanes 12–13), which shares an approximate equivalent MW. This is attributed to the difference in amino acid sequence with an increase in hydrophobic amino acids for the GB-1_FLQS-26_His construct.
Figure 2.
An 8%–22% gradient SDS-PAGE gel of expressed fusion proteins. MW markers (lane 1), uninduced cells (lane 2), induced cells (lane 3), French press lysate supernatant (lane 4), and purified GB-1_cIp-RR-20_His (lane 5). Lanes 6–15 contain induced cells and the French press lysate supernatant of each indicated fusion protein construct, with regions of fusion protein constructs denoted by an asterisk.
For production of isotopic labeling of GB-1 fusion protein constructs, expression in M9 minimal media with only 15N-(NH4)2SO4 as the sole nitrogen source (for 15N-labeling) or 15N-(NH4)2SO4/13C-glucose as the only nitrogen/carbon source produced very limited yield of fusion proteins. The addition of 50 mL of 15N and/or 15N/13C enriched media (Cambridge Isotope Laboratories) to each liter of minimal media restored expression levels to ~75% that of the 2xYT media, giving high yields with relatively low cost associated with expression of labeled fusion constructs. The addition of enriched media to the minimal media is predicted to supply minerals and nutrients essential to expression of fusion protein constructs.
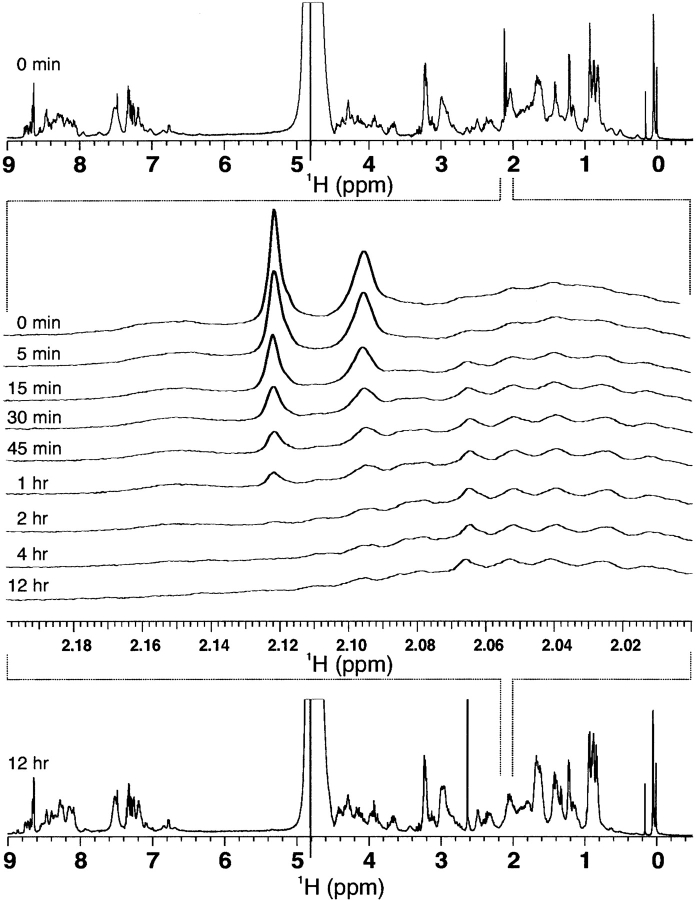
Cleavage of the GB-1 fusion constructs was performed by CNBr cleavage, which under acidic conditions (0.1 N HCl) will cleave C terminal to all methionine residues, chemically modifying all methionine residues into homoserine residues. Monitoring of ɛ-CH3 groups of the fusion protein GB-1_RR-20_His by 1H NMR spectroscopy (Fig. 3 ▶) indicates that cleavage is quantitative after 12 h at room temperature as peak intensity of the ɛ-CH3 groups diminishes over time. ɛ-CH3 groups of methionine residues will resonant as a sharp singlet within the 1H-NMR spectrum around 2.0 ppm, as no J couplings of the methyl group protons to neighboring side-chain CH2 groups are observed, because of a separation by a sulfur atom within the side chain. Three singlets are expected for GB-1_RR-20_His because three methionine residues are present within the construct (Fig. 1A ▶), yet only two peaks are observed because there is peak overlap of two ɛ-CH3 groups at 2.095 ppm. The amide bond linkage between Met-Thr residues has been previously reported to be inefficient under standard CNBr cleavage conditions, yet we have proven >99% cleavage for fusion constructs GB-1_cIp-RR-20_His, GB-1_cIp-RG-20_His, GB-1_cIp-GR-20_His, and GB-1_TRTK-12_His, all of which contain a Met–Thr cleavage site. Following cleavage by CNBr, HPLC purification and verification by MALDI-TOF mass spectroscopy of all eluted peaks indicated that pure peptide was obtained with a C-terminal homoserine residue present for all constructs as a consequence of CNBr cleavage. For production of peptides containing methionine residues, CNBr cleavage is not an option and thus the cleavage sites must be altered to proteolytic cleavage sites (i.e., thrombin) or other chemical cleavage sites for yield of peptide constructs. For this study, CNBr was chosen because of its speed and reproducible results for yielding pure peptide constructs. Expression of GB-1 fusion constructs resulted in ~20 mg/15 mg of pure peptide per liter of 2xYT/minimal media.
Figure 3.
CNBr cleavage of GB-1_cIp-RR-20_His fusion protein as monitored by 1H-NMR. Reduction of peak intensity of methionine ɛ-CH3 side chains as a function of time indicates efficient cleavage of the peptide amide bond.
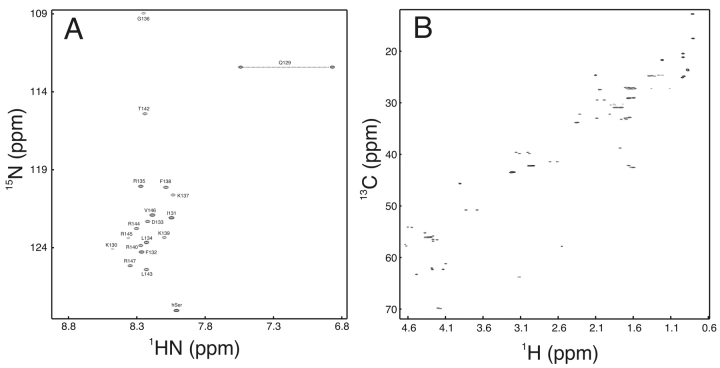
The efficiency of labeling of cIp-RR-20 is exhibited in Figure 4 ▶, with {1H, 15N}-HSQC of 15N-cIp-RR-20 and {1H, 13C}-HSQC of 15N/13C-cIp-RR-20 being displayed. The complete backbone amide assignment of cIp-RR-20 is shown in Figure 4A ▶, with residues T128 and Q129 not observed because of exchange with water, and one homoserine resonance being observed. cIp-RR-20 is unstructured in solution, as evidenced by the limited spectral dispersion in the {1H, 15N}-HSQC NMR spectrum.
Figure 4.
(A) {1H, 15N-HSQC} and (B) {1H, 13C-HSQC} NMR spectra of 15N- and 15N/13C-cIp-RR-20. Backbone amide assignments for {1H, 15N-HSQC} are as indicated.
We have demonstrated the feasibility of a GB-1 fusion protein approach for use in recombinant peptide labeling and expression, with high purity of product and high product yield. The applications for labeled peptides for NMR studies is large, yielding the ability to measure relaxation rates of bound peptides to a target protein, as well as giving the ability to perform multidimensional heteronuclear NMR experiments for solution structure determination. As peptide interactions with targeted domains become more documented in medicine and cellular functions, leading toward an increased demand, the ability to exploit a well-behaved, well-characterized, and time-efficient system for expressing and purifying recombinant peptides becomes a major beneficial goal of molecular biology.
Materials and methods
Preparation of cIp-RR-20, cIp-RG-20, cIp-GR-20, TRTK-12, EDQL-26, and FLQS-26 expression vectors
Sense and antisense DNA oligonucleotide primers were synthesized using optimized codons for Escherichia coli (K12) for the primary amino acid sequence for each of the six peptides, as indicated in Figure 1 ▶. In addition to coding for amino acid sequences, we introduced a methionine codon on either side of the peptide sequence, a SmaI cut site on the 3′ end of the sense strand, and NheI /XhoI cut sites for cloning into GEV-1 vector. Sense and antisense oligonucleotides were annealed together at 100°C for 1 h, followed by ethanol precipitation. GEV-1 vector was simultaneously digested with NheI/XhoI, agarose gel purified, and ligated to individual annealed oligonucleotides. Following transformation into E. coli XL1 Blue, positive colonies were selected using SmaI restriction digests and respective vectors were transformed into E. coli BL21 DE3 (pLysS) for subsequent expression.
Expression of GB-1 fusion proteins
BL21 DE3 (pLysS) cells containing each respective GB-1 fusion vector were grown in 50 mL 2xYT containing 100 μg/mL ampicillin overnight at 37°C with shaking. One-liter cultures of 2xYT containing 100 μg/mL ampicillin were inoculated with 5 mL of overnight culture of each fusion protein construct, grown to OD600 0.8 at 37°C with shaking, and induced with 1 mM IPTG for 4 h. To determine the extent of soluble expression, we pelleted each fusion protein construct, broke it in a French pressure cell, cleared it by centrifugation, and analyzed the supernatant on an 8%–22% gradient SDS-PAGE gel (Biorad).
Labeling of GB-1 fusion protein cIp-RR-20
For both 15N and 13C labeling of fusion proteins, starter cultures were grown in 50 mL 2xYT containing 100 μg/mL ampicillin at 37°C until OD600 0.8 was reached. For 15N labeling of fusion proteins, 1-L cultures of M9 minimal media containing 50 mL of 15N-enriched media (Cambridge Isotope Laboratories) and 1.1 g 15N-(NH4)2SO4 as the sole nitrogen sources, and 10 g of glucose, were inoculated with 5 mL of starter culture, grown to OD600 0.8 at 37°C, inoculated with 1 mM IPTG, and allowed to induce for 4 h prior to cell harvesting. For 13C/15N labeling of fusion proteins, 1-L cultures of M9 minimal media were used, containing 50 mL of 13C/15N-enriched media (Cambridge Isotope Laboratories), 1.1 g 15N-(NH4)2SO4, and 2.5 g of 13C-glucose.
Purification of GB-1 fusion proteins
BL21 DE3 (pLysS) cells that expressed respective fusion proteins were pelleted, resuspended in 50 mM Tris buffer (pH 8.0), broken in a French pressure cell, centrifuged, and passed through a 0.22-μm filter to remove all insoluble cellular debris. Supernatant was loaded onto a 25-mL column of fast-flow chelating Sepharose, previously charged with 50 mM NiSO4, and equilibrated in 5 mM imidazole, 500 mM NaCl, and 20 mM TrisHCl (pH 7.9). The column was then washed with 40 mM imidazole, 500 mM NaCl, and 10 mM TrisHCl (pH 7.9), followed by fusion protein elution with 100 mM EDTA, 500 mM NaCl, and 20 mM TrisHCl (pH 7.9). Fusion protein elutant was lyophilized to dryness, desalted on a G25 Sephadex column (10mM NH4HCO3 at pH 8.0), and lyophilized to dryness.
CNBr cleavage of GB-1 fusion proteins and purification of peptides
GB-1 fusion proteins were dissolved in 0.1 N HCl (5 mg/mL), to which solid CNBr was added to a final molar concentration of 100:1, and left in the dark at room temperature for 24 h. Solutions were then diluted 10-fold with distilled deionized H2O (ddH2O) and lyophilized to dryness. Samples were subjected to a reverse-phase HPLC column, and individual peaks were collected with MWs verified by MALDI-TOF mass spectroscopy. Peaks containing pure peptide were pooled and twice lyophilized to dryness to remove all organic solvents.
NMR spectroscopy
NMR experiments were performed on a Varian INOVA 500-MHz NMR spectrometer equipped with a triple resonance probe and Z-axis pulsed field gradients. All recorded spectra were referenced to an external DSS standard. Five-point-three mg of 1H-GB-1_cIp-RR-20_His was dissolved in 525 μL of 0.1 N HCl with 25 μL of 2H2O, to which a 100-fold excess of CNBr was added directly to the NMR tube. 1-dimensional 1H scans were taken at 15-min intervals at 25°C for 24 h. All one-dimensional spectra were processed and analyzed using vnmr (Varian associates). Two and a half mg of 15N-cIp-RR-20 and 3.1 mg of 15N/13C-cIp-RR-20 were dissolved separately into 500 μL NMR buffer (100 mM KCl, 10 mM imidazole, 0.015% NaN3 at pH 6.7), with all two-dimensional spectra processed with nmrPipe (Delaglio et al. 1995) and analyzed with the program nmrView (Johnson and Blevins 1994). Both {1H, 15N}-HSQC spectra (Kay et al. 1992) and {1H, 13C}-HSQC spectra (Neri et al. 1989) were acquired with 16 transients and 128 increments, zero filled, and the number of points doubled with linear prediction during spectral processing.
Acknowledgments
We gratefully acknowledge Dr. G. Marius Clore and coworkers (NIH, Bethesda, MD) for the kind gift of the GEV-1 vector and for helpful discussions. We thank Mr. Paul Semchuck and Mr. Lorne Burke for assistance with HPLC purification and MALDI mass spectroscopy measurements, as well as Mr. Gerry McQuaid for spectrometer maintenance and Mr. David Corson for helpful discussions.
Supported by the Canadian Institutes of Health Research (CIHR) and the Heart and Stroke Foundation of Canada. D.A.L. is supported by an Alberta Heritage Foundation for Medical Research Studentship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
NMR, nuclear magnetic resonance
GB-1, B1 immunoglobulin domain of streptococcal protein G
cTnI, human cardiac troponin I
cTnT, human cardiac troponin T
CNBr, cyanogen bromide
His, 6-poly histidine
IPTG, isopropyl β-D-thiogalactopyranoside
MW, molecular weight
NHE1, human sodium proton exchanger isoform 1
CapZα1, actin capping protein α
2xYT, 2X-yeast-tryptone media
HPLC, high performance liquid chromatography
MALDI-TOF, matrix-assisted laser desorption/ionization-time of flight
HSQC, heteronuclear single quantum coherence
OD, optical density
EDTA, ethylenediaminetetraacetic acid
DSS, sodium 2,2-dimethyl-,2-silapantane-5-sulfonate
Article and publication are at http://www.genome.org/cgi/doi/10.1110/ps.0376003.
References
- Azriel, R. and Gazit, E. 2001. Analysis of the minimal amyloid-forming fragment of the islet amyloid polypeptide. An experimental support for the key role of the phenylalanine residue in amyloid formation. J. Biol. Chem. 276 34156–34161. [DOI] [PubMed] [Google Scholar]
- Campbell, A.P., Van Eyk, J.E., Hodges, R.S., and Sykes, B.D. 1992. Interaction of troponin I and troponin C: Use of the two-dimensional transferred nuclear Overhauser effect to determine the structure of a Gly-110 inhibitory troponin I peptide analog when bound to cardiac troponin C. Biochim. Biophys. Acta 1160 35–54. [DOI] [PubMed] [Google Scholar]
- Celis, E. 2002. Getting peptide vaccines to work: Just a matter of quality control? J. Clin. Invest. 110 1765–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counillon, L., Noel, J., Reithmeier, R.A., and Pouyssegur, J. 1997. Random mutagenesis reveals a novel site involved in inhibitor interaction within the fourth transmembrane segment of the Na+/H+ exchanger-1. Biochemistry 36 2951–2959. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Gronenborn, A.M. and Clore, G.M. 1996. Rapid screening for structural integrity of expressed proteins by heteronuclear NMR spectroscopy. Protein Sci. 5 174–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronenborn, A.M., Filpula, D.R., Essig, N.Z., Achari, A., Whitlow, M., Wingfield, P.T., and Clore, G.M. 1991. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science 253 657–661. [DOI] [PubMed] [Google Scholar]
- Gudmundsson, G.H., Agerberth, B., Odeberg, J., Bergman, T., Olsson, B., and Salcedo, R. 1996. The human gene FALL39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes. Eur. J. Biochem. 238 325–332. [DOI] [PubMed] [Google Scholar]
- Hernandez, O.M., Housmans, P.R., and Potter, J.D. 2001. Invited review: Pathophysiology of cardiac muscle contraction and relaxation as a result of alterations in thin filament regulation. J. Appl. Physiol. 90 1125–1136. [DOI] [PubMed] [Google Scholar]
- Hristova, K., Dempsey, C.E., and White, S.H. 2001. Structure, location, and lipid perturbations of melittin at the membrane interface. Biophys. J. 80 801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huth, J.R., Bewley, C.A., Jackson, B.M., Hinnebusch, A.G., Clore, G.M., and Gronenborn, A.M. 1997. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 6 2359–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter, M.C., Krebs, J., Meiler, J., Griesinger, C., Carafoli, E., and Helms, V. 2002. A structural model of the complex formed by phospholamban and the calcium pump of sarcoplasmic reticulum obtained by molecular mechanics. Chembiochem 3 1200–1208. [DOI] [PubMed] [Google Scholar]
- Ivanenkov, V.V., Jamieson Jr., G.A., Gruenstein, E., and Dimlich, R.V. 1995. Characterization of S-100b binding epitopes. Identification of a novel target, the actin capping protein, CapZ. J. Biol. Chem. 270 14651–14658. [DOI] [PubMed] [Google Scholar]
- Jacobsen, J.S. 2002. Alzheimer’s disease: An overview of current and emerging therapeutic strategies. Curr. Top. Med. Chem. 2 343–352. [DOI] [PubMed] [Google Scholar]
- Johnson, A.J. and Blevins, R.A. 1994. NMRView: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4 603–614. [DOI] [PubMed] [Google Scholar]
- Jones, D.H., Ball, E.H., Sharpe, S., Barber, K.R., and Grant, C.W. 2000. Expression and membrane assembly of a transmembrane region from Neu. Biochemistry 39 1870–1878. [DOI] [PubMed] [Google Scholar]
- Kay, L.E., Keifer, P., and Saarinen, T. 1992. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114 10663–10665. [Google Scholar]
- Li, M.X., Spyracopoulos, L., Beier, N., Putkey, J.A., and Sykes, B.D. 2000. Interaction of cardiac troponin C with Ca2+ sensitizer EMD 57033 and cardiac troponin I inhibitory peptide. Biochemistry 39 8782–8790. [DOI] [PubMed] [Google Scholar]
- Lindhout, D.A., Li, M.X., Schieve, D., and Sykes, B.D. 2002. Effects of T142 phosphorylation and mutation R145G on the interaction of the inhibitory region of human cardiac troponin I with the C-domain of human cardiac troponin C. Biochemistry 41 7267–7274. [DOI] [PubMed] [Google Scholar]
- Majerle, A., Kidric, J., and Jerala, R. 2000. Production of stable isotope enriched antimicrobial peptides in Escherichia coli: An application to the production of a 15N-enriched fragment of lactoferrin. J. Biomol. NMR 18 145–151. [DOI] [PubMed] [Google Scholar]
- Mazor, Y., Gilead, S., Benhar, I., and Gazit, E. 2002. Identification and characterization of a novel molecular-recognition and self-assembly domain within the islet amyloid polypeptide. J. Mol. Biol. 322 1013–1024. [DOI] [PubMed] [Google Scholar]
- Nathisuwan, S. and Talbert, R.L. 2002. A review of vasopeptidase inhibitors: A new modality in the treatment of hypertension and chronic heart failure. Pharmacotherapy 22 27–42. [DOI] [PubMed] [Google Scholar]
- Neri, D., Szyperski, T., Ottig, G., Senn, H., and Wuthrich, K. 1989. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28 7510–7516. [DOI] [PubMed] [Google Scholar]
- Pearlstone, J.R. and Smillie, L.B. 1985. The interaction of rabbit skeletal muscle troponin-T fragments with troponin-I. Can. J. Biochem. Cell Biol. 63 212–218. [DOI] [PubMed] [Google Scholar]
- Pollesello, P. and Annila, A. 2002. Structure of the 1–36 N-terminal fragment of human phospholamban phosphorylated at Ser-16 and Thr-17. Biophys. J. 83 484–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redwood, C.S., Moolman-Smook, J.C., and Watkins, H. 1999. Properties of mutant contractile proteins that cause hypertrophic cardiomyopathy. Cardiovasc. Res. 44 20–36. [DOI] [PubMed] [Google Scholar]
- Saiman, L., Tabibi, S., Starner, T.D., San Gabriel, P., Winokur, P.L., Jia, H.P., McCray Jr., P.B., and Tack, B.F. 2001. Cathelicidin peptides inhibit multiply antibiotic-resistant pathogens from patients with cystic fibrosis. Antimicrob. Agents Chemother. 45 2838–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet, B., Van Eyk, J.E., and Hodges, R.S. 1997. Mapping of a second actin-tropomyosin and a second troponin C binding site within the C terminus of troponin I, and their importance in the Ca2+-dependent regulation of muscle contraction. J. Mol. Biol. 271 728–750. [DOI] [PubMed] [Google Scholar]
- Tyrberg, B. and Levine, F. 2001. Current and future treatment strategies for type 2 diabetes: The β-cell as a therapeutic target. Curr. Opin. Investig. Drugs 2 1568–1574. [PubMed] [Google Scholar]
- Wakabayashi, S., Pang, T., Su, X., and Shigekawa, M. 2000. A novel topology model of the human Na(+)/H(+) exchanger isoform 1. J. Biol. Chem. 275 7942–7949. [DOI] [PubMed] [Google Scholar]