Abstract
The reconstituted pea chloroplastic outer envelope protein of 16 kDa (OEP16) forms a slightly cation-selective, high-conductance channel with a conductance of Λ = 1,2 nS (in 1 M KCl). The open probability of OEP16 channel is highest at 0 mV (Popen = 0.8), decreasing exponentially with higher potentials. Transport studies using reconstituted recombinant OEP16 protein show that the OEP16 channel is selective for amino acids but excludes triosephosphates or uncharged sugars. Crosslinking indicates that OEP16 forms a homodimer in the membrane. According to its primary sequence and predicted secondary structure, OEP16 shows neither sequence nor structural homologies to classical porins. The results indicate that the intermembrane space between the two envelope membranes might not be as freely accessible as previously thought.
Plant cells house an additional organelle type, the plastid family. Most prominent are chloroplasts, which carry out oxygenic photosynthesis in the green parts of plants. All types of plastids are surrounded by a pair of double membranes called the outer and inner envelope membrane. Both membranes contain distinct protein translocation machineries (1–3). The envelope membranes have essential capacities in the biosynthesis of lipids, isoprenoids, and prenylquinones (4). Most important, however, is the flux of photosynthesis-related low molecular weight substances into and out of chloroplasts. Furthermore, plastids are the sole site of nitrite reduction that yields ammonia, which is provided for the rest of the cell as amino acid-bound nitrogen (5). Concomitantly chloroplasts take up inorganic cations (K+, Na+, Mg2+, Ca2+, and others), anions (NO2−, SO42−, PO43−), and a variety of organic biosynthetic pathway intermediates (e.g., sugars, amino acids, or acetate for fatty acid biosynthesis). The inner envelope membrane forms a barrier for small molecular weight solutes, which are thought to pass freely across the chloroplastic outer envelope membrane (4, 6, 7). Whereas specific carrier proteins located in the inner envelope membrane have been identified on a molecular level for a variety of substrates, the porin or permease-like proteins in the outer envelope membrane remain unidentified.
A number of electrophysiological studies using either giant chloroplasts from Nitellopsis (8, 9) or isolated envelope membranes from spinach (6, 7) described the presence of different voltage-dependent, high-conductance channels in the inner and outer membranes of Nitellopsis or spinach, respectively. A pore channel with an unusually large exclusion limit of about 10 kDa has been described in reconstituted spinach envelope membranes (6). So far, attempts have failed to detect any polypeptides in the chloroplastic outer envelope that show homology to bacterial or mitochondrial porins. Only recently (10) a protein from pea proplastids was described that showed significant homology on a primary sequence basis to bacterial and mitochondrial porins (11–13). However, the proplastid-localized porin failed to import into isolated chloroplasts. Further biochemical data suggested that this protein was present only in proplastids but not in chloroplasts.
In this work we describe the molecular identification and functional reconstitution of the chloroplastic outer envelope protein of 16 kDa (OEP16). Reconstituted OEP16 forms a voltage-regulated, high-conductance channel that revealed a selective permeability for amino acids.
MATERIALS AND METHODS
A random primed digoxigenin-labeled probe was synthesized from an EcoRI/XhoI fragment of the Arabidopsis EST cDNA clone 107L13T7 (GenBank accession no. T22850) by PCR according to the manufacturer’s recommendation (Boehringer, Mannheim) and used to screen a pea cDNA expression library (14) (Uni Zap XR, Stratagene), resulting in the isolation of peacOEP16 coding for OEP16. Both DNA strands were sequenced (ref. 15; accession no. Z73553). An NdeI site was introduced at the first methionine of the coding region of peacOEP16 by PCR using the forward primer 5′-GGGGGGCATATGCCTCGTAGCAGTTTTTCAGG3-′ and universal primer. The resulting PCR product was blunt-ended by treatment with Klenow-fragment and ligated into the vector pBSC SK(+) (Stratagene) after linearization with SmaI, resulting in peacOEP16pbsc. A NdeI/XhoI fragment from peacOEP16pbsc was subcloned into the vector pET21b (Novagen) resulting in peacOEP16pet. Overexpression of OEP16 was done after transforming the vector into E. coli BL21(DE3) cells (Novagen).
Overexpressed OEP16 was recovered from insoluble inclusion bodies (16) and purified by ion exchange chromatography using an anion exchange resin (Source Q, Pharmacia) as a first step in the purification protocol. OEP16 was recovered in the flow-through and passed over a MonoS cation exchanger (6 M urea/50 mM Hepes/KOH, pH 7.6/1 mM EDTA/10 mM 2-mercaptoethanol) from which it was eluted at 210 mM NaCl. The protein was dialyzed against water, freeze-dried, and used for the reconstitution experiments.
Immunological Methods.
An antiserum was raised in a rabbit against OEP16 that was purified by SDS/PAGE from isolated chloroplastic outer envelope membranes. The antiserum was affinity-purified by passing over an OEP16 Sepharose column (17).
Liposomes.
Purified OEP16 was resuspended in 50 mg/ml l-α-phosphatidylcholine (from soybean type IV-S, Sigma) in 80 mM MEGA-9/20 mM KCl/10 mM Tris⋅HCl, pH 7.0. After 1 h of incubation at room temperature the suspension was subjected to dialysis at 4°C for 12 h and used for bilayer measurements.
CD Spectroscopy.
CD spectra were measured using a Jasco-J-5000A spectropolarimeter (Easton, MD) after calibration with (+)-10-camphorsulfonic acid. The spectra were recorded at 20°C in a quartz-cell with 0.5-cm optical path length. Scans were performed at a rate of 20 nm/min with a sampling interval of 0.2 nm and averaged (n = 60) to improve the signal/noise ratio. Both samples (OEP16 in 8 M urea and OEP16 liposomes) were adjusted to the same protein concentration: 50 ± 20 μg protein (18).
Optical Measurements.
Time-scan (duration 2 min) absorbance measurements were performed in an Aminco DW200 UV-Vis spectrophotometer at 400 nm. Liposomes with an optical density of 0.15–0.2 were equilibrated in 1 ml of 10 mM tetraethylammonium (TEA)/Hepes, pH 7.0/10 mM KCl. Absorbance at 400 nm was monitored for 15 s and the osmotically active solute was added in a total volume of 200 μl after stirring for 10 s. The change of absorbance was continuously monitored up to 2 min. The pH of the stock solutions was titrated either by addition of TEA/OH or solid Hepes when required.
Electrophysiological Measurements.
Planar lipid bilayers were produced by the painting technique (19, 20). A solution of 50 mg/ml l-α-phosphatidylcholine in n-decane was applied to a hole (100–200 μm diameter) in a Teflon septum, separating the two bath chambers (total volume, ≈3 ml). The resulting bilayers had a typical capacitance of ≈0.5 μF/cm2 and a resistance of >100 GΩ. The noise was 1 pA (rms) at 5 kHz bandwidth. After a stable bilayer was formed in symmetrical solutions of 20 mM KCl/10 mM Hepes/Tris, pH 7.0, the solutions were changed to asymmetric concentrations (cis-chamber: 250 mM KCl/10 mM CaCl2). The liposomes were added to the cis compartment so that the liposomes slowly flowed directly across the bilayer (21). When necessary the solution in the cis-chamber was stirred to promote fusion. After fusion the electrolytes were changed to the final composition by perfusion. The Ag/AgCl electrodes were connected to the chambers through 1 M KCl-agar bridges. The electrode of the trans compartment was directly mounted to the headstage of a current amplifier (EPC 7, List Medical, Darmstadt, Germany). Reported membrane potentials are referred to the trans compartment. The amplified currents were recorded on a modified DAT recorder (SV 3700, Panasonic, Secaucus, NJ). For analysis (22), current recordings were low-pass filtered with an eight-pole bessel filter, typically at 1–2 kHz, digitized at a sampling interval of 0.2 ms, and fed into an Axolab 1100 A/D converter (Axon Instruments, Foster City, CA).
Electrophysiologically Monitored Osmotic Permeation Assay.
Bilayers were produced as described above. Then, the solution in the cis-chamber was changed to the final composition by perfusion with a 750 mM solution of the solute to be tested and addition of concentrated CaCl2 to reach a final concentration of 20 mM. Liposomes with reconstituted OEP16 were applied as described above. A membrane potential of 100 mV was applied (trans-chamber positive). The pH of the solute stock solutions was adjusted by addition of either TEA/OH (20% wt/wt solution in H2O) or solid Hepes when required. The inability of both ions to promote fusion was checked.
RESULTS
Attempts to identify possible “candidates” for a solute pore in the pea chloroplastic outer envelope were based on the following observations. Membrane vesicles were treated with the protease thermolysin to distinguish between protease-sensitive and protease-resistant proteins (refs. 23–25; also see below). The rationale behind this approach was (i) that a major solute channel of the chloroplastic outer envelope should be an abundant protein as the central protein translocation channel OEP75 (26, 27), and (ii) that it should be fairly resistant to protease treatment. Several abundant outer envelope proteins have been identified before as constituents of the protein import machinery (e.g., OEP86, OEP75, and OEP34; refs. 27–30) or as functionally not assigned components of the outer envelope (e.g., OEP14; ref. 31). Furthermore, the pea chloroplastic outer envelope lacks abundant proteins in the range between 25 and 33 kDa, the average size of classical porins. OEP16 was a polypeptide that met the criteria mentioned above (i.e., abundance and low protease accessibility; see below). Moreover, OEP16 behaves as an integral membrane protein in that it is resistant to extraction by 1 M NaCl or pH 11 (32) (not shown). We therefore decided to further analyze the structure and function of OEP16.
Primary Structure and Tissue Distribution of OEP16.
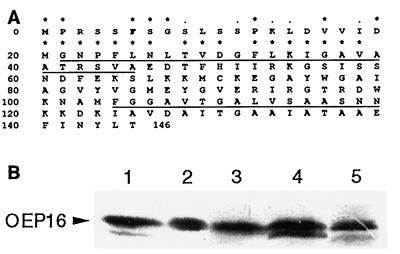
Internal amino acid sequence information was obtained after CNBr cleavage of SDS/PAGE-purified OEP16. A database search (May 23, 1996) gave significant homologies with cDNA clones from Arabidopsis. The Arabidopsis cDNA (T22850) was used to isolate the corresponding cDNA clone from a pea cDNA expression library. A cDNA clone was isolated that contained an open reading frame coding for a protein of 146 amino acids with a calculated molecular mass of 15.5 kDa (Fig. 1A, accession no. Z73553). The peptide sequences obtained after CNBr cleavage of OEP16 were all present in the deduced amino acid sequence (Fig. 1A). Computer alignments revealed the presence of OEP16 in the dicotyledonous plant Arabidopsis thaliana [expressed sequence tags (ESTs) T33953, T45509, T46371, T88007, T21986, T21804, T88008, T73663, T22880, T21031, H76061, T20890, R65250] and the monocotyledonous plants Zea mays (EST T25210) and Oryza sativa (rice, ESTs D49286, D40577). A sequence comparison of the N-terminal 40 amino acids of these proteins shows 62% identical amino acids and an overall similarity of 75% (Fig. 1A). Further analysis was not possible because pea OEP16 is the only full-length sequence described. It is obvious that OEP16 is highly conserved and present in a wide variety of plants. OEP16 is present in plastids of roots, shoots, and leaves of pea plants grown either in the dark or in the light, demonstrating its presence in different plastid types (i.e., etioplasts, proplastids, amyloplasts, and chloroplasts; Fig. 1B).
Figure 1.
Deduced amino acid sequence and tissue distribution of pea OEP16. (A) The deduced amino acid sequence of the pea cDNA clone for OEP16 is shown (accession no. Z73553). The peptide sequences obtained after CNBr cleavage are underlined. N-terminal 40 amino acids of the OEP16 are compared with homologues from arabidopsis, maize, and rice. An asterisk indicates identities, and dots indicate homologous exchanges in all four species, respectively. (B) Immunodetection of OEP16 in chloroplasts from leaves (lane 2), shoots (lane 3), and root-plastids (lane 4) of light-grown pea plants or in leaves from etiolated plants (lane 5). Total plastid protein (100 μg) was used. Lane 1, presence of OEP16 in purified chloroplastic outer envelopes (1 μg protein). Organelles were isolated and subfractionated as described (33–35).
Functional Characterization of OEP16.
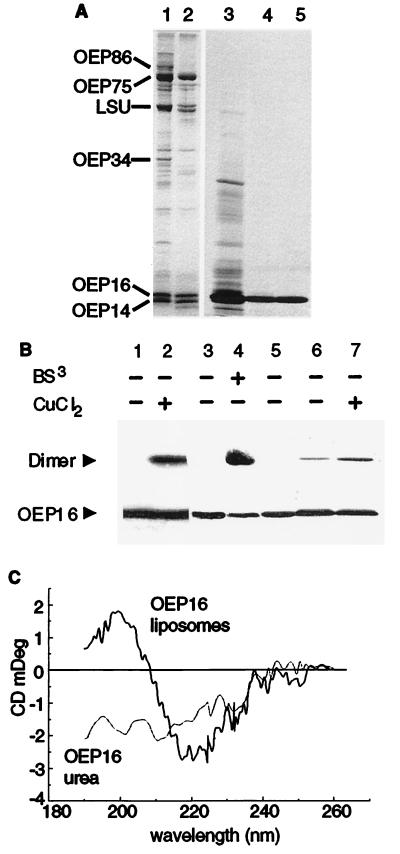
OEP16 was synthesized in E. coli cells, recovered from insoluble inclusion bodies, and purified to homogeneity by ion exchange chromatography (Fig. 2A). Recombinant OEP16 was solubilized in MEGA-9 and reconstituted into lipid vesicles (OEP16 liposomes). Two criteria were used to ensure that OEP16 attained a native-like structure in OEP16 liposomes: (i) crosslinking studies of OEP16 in isolated envelope vesicles and in liposomes and (ii) CD spectra of the unfolded and reconstituted OEP16.
Figure 2.
Characterization of pea OEP16. (A) Chloroplastic outer membrane polypeptides were analyzed by SDS/PAGE either before (lane 1) or after (lane 2) treatment with the protease thermolysin (24, 25). The position of known polypeptides is indicated. Silver-stained polypeptide composition of heterologuously expressed OEP16 recovered in inclusion bodies (lane 3) after anion- (lane 4) and cation-exchange chromatography (lane 5). (B) OEP16 forms a homodimer. Purified outer envelope membranes (10 μg protein) were either not treated or treated with the crosslinker BS3 (500 μM, 10 min on ice) or with 1 mM CuCl2 (lanes 1–4). E. coli-expressed OEP16 (lane 5) was reconstituted into liposomes and incubated in the absence (lane 6) or presence (lane 7) of 1 mM CuCl2. Products were separated by SDS/PAGE (36) in the absence of reducing agents and analyzed by immunoblotting using αOEP16. (C) CD spectra of OEP16 either denatured in 8 M urea or reconstituted into liposomes.
First, purified outer envelope vesicles were treated with the homobifunctional crosslinker bis(sulfosuccinimidyl)suberate (BS3). In the presence of BS3 a product of 30-kDa molecular size was formed, which could represent a homodimer of OEP16 (Fig. 2B, lanes 3 and 4). The putative OEP16 dimer was also formed after treating purified outer envelope membrane vesicles with the thiol-oxidant CuCl2 (Fig. 2B, lanes 1 and 2; refs. 37, 38). The CuCl2-formed dimer could be dissociated into monomers in the presence of excess DTT or mercaptoethanol (not shown), clearly demonstrating that a disulfide bridge was responsible for forming the OEP16 homodimer. Because CuCl2 functions as a thiol crosslinker without an additional spacer arm, the thiolgroups must be in very close physical proximity to yield a reversible covalent linkage between polypeptide chains. The dimer was also formed when OEP16 liposomes were treated with CuCl2 (Fig. 2B, lanes 5–7), indicating that OEP16 adapts a conformation that brings the only cysteine present in OEP16 in such close physical proximity that a homodimer can be formed. BS3 has a spacer arm of 11.4 Å and should therefore be able to yield crosslink products also with other proteins in the vicinity of OEP16; however, no further OEP16-containing crosslink products were observed (not shown).
Second, CD-spectra (Fig. 2C) obtained from OEP16 liposomes indicate the presence of β-sheet and helical secondary structure motifs as deduced from the ellipticities between 200 and 230 nm. In contrast to this, the spectrum of urea-denatured OEP16 shows that the protein adopts a largely extended conformation (39).
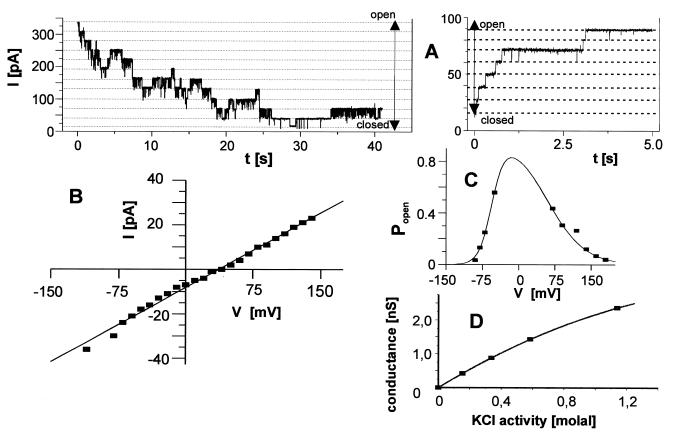
After fusion of OEP16 liposomes with planar bilayers, voltage-dependent single-channel currents could be observed (Fig. 3A). Multiple channel copies were detectable in the bilayer, which opened or closed successively in a voltage-dependent manner (Fig. 3A). The channels exhibited mainly one conductance state of Λ = 330 pS (in symmetrical 250 mM KCl), concomitant with rarely occurring openings reaching 70% of the main amplitude. In asymmetric KCl solutions (250:20 mM KCl) a reversal potential of Vrev = 32 mV was obtained (VK+ = 60 mV) (Fig. 3B). This shows that the OEP16 channel is slightly cation selective (PK+/PCl− = 6.5). The open probability of the channel was strongly dependent on the applied membrane potential (Fig. 3C). Around Vm = 0 mV the channel was almost completely open (Popen = 0.8), whereas Popen decreased steeply with increasing positive and negative potentials. Measurements of the relative selectivity of OEP16 channels yielded the following sequence: Cs+ = Rb+(1.5) > K+ = Na+(1) > Li+ (0.78) > Tris+ (0.57). This sequence roughly reflects the mobility of these ions in water, indicating that the channel has a wide pore and that its interaction with these ions is weak (40). Tris+-cations were conducted nearly as efficiently as the smaller alkali metal ions. In contrast to this, TEA+-cations, though not blocking the pore, were very poorly conducted. From this observation a pore diameter of 0.8–1.0 nm (the rough cross-section of the Tris+-cations) can be estimated. This is consistent with the channel diameter calculated from the conductivity (Λ = 330 pS in 250 mM KCl) assuming a simple water-filled pore of 5 nm length (40). Conductance measurements in symmetrical solutions (0.2 M, 0.4 M, 0.6 M, and 2 M KCl) on both sides of the membrane resulted in an almost linearly increasing concentration–conductivity relationship (Fig. 3D). A fit of the flux-concentration data with a saturation function (Michaelis–Menten type) yielded a saturation conductivity (Λmax) of 7 nS with a half-maximal conductance at 4 molal KCl.
Figure 3.
Reconstituted OEP16 constitutes a voltage-sensitive, cation-selective pore. (A) Liposomes containing E. coli-overexpressed OEP16 were fused to black lipid membranes. The cis chamber contained 250 mM KCl and the trans chamber, 20 mM KCl. A current trace from a bilayer containing 11 active pores is shown directly after a voltage jump from 0 to 150 mV (Left). The channels, which were open at 0 mV, close after the increase to 150 mV. When the identical experiment (Left) was continued, opening of several channels was measured after lowering the potential in one step to 80 mV (Right). (B) Current–voltage relationship from the data presented in A. The slope of the linear regression is 330 pS and the zero current potential is 32 mV. (C) Open probabilities of the OEP16 pore. Conditions were as in A, except that 250 mM KCl was used on both sides of the bilayer. Voltages were applied for 10 min, but only the mean current of the last minute was taken as the steady-state current. The quotient of this current and the total current for 24 open channels (estimated number of channels present in this experiment) are plotted. All potentials are referred to the trans compartment. (D) The conductivity of OEP16 depends nearly linear on solution activity of KCl. All measurements were performed on the same bilayer. Solutions were exchanged by perfusion, and current traces were recorded for at least eight different voltages.
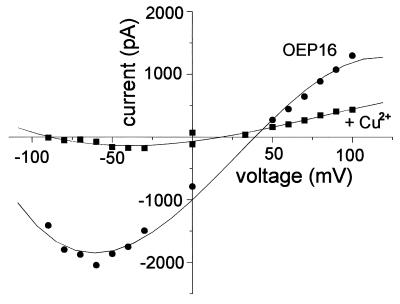
When CuCl2 was added to either cis or trans or to both bilayer compartments to induce disulfide bridge formation between two OEP16 monomers, a massive decrease of macroscopic current was observed in the bilayer that contained multiple OEP16 copies (Fig. 4). The observed inhibition was between 40 and 75%. After removal of CuCl2 by perfusion and reduction of the disulfide bridges by DTT, OEP16 channel activity could be recovered. (Fifty to 80% of the controls, not shown). Recombinant OEP16, which contains a cysteine-to-serine mutation, still forms a solute channel but cannot further be inhibited by CuCl2 (K.P. and J.S., unpublished data), clearly showing that channel-forming activity observed in proteoliposomes did not derive from E. coli BL21(DE3) cells (not shown).
Figure 4.
Current–voltage relationship of bilayers containing multiple copies of OEP16 channels in the absence and presence of 10 mM CuCl2 on the cis side (cis compartment 250 mM KCl, trans 20 mM KCl). Mean values were calculated from 5-s current-recording intervals at the given voltages. Considering the single-channel conductance of OEP16 (330 pS in 250 mM KCl), the bilayer contained about 200 active channels.
Light-scattering properties of OEP16 liposomes were used to investigate the permeability of OEP16 to different solutes. Turbidity changes of liposome suspensions are mainly related to volume changes. After a fast increase of the osmolarity of the surrounding medium, the following is to be expected. If the liposome membrane is impermeable to the osmoticum the liposomes will shrink, whereas no, or only very small, volume changes are expected when the membrane is permeable to the solute. Depending on the distribution and average size of the vesicles (41), shrinking of the liposomes may induce a positive or negative change of the turbidity. The size distribution can be measured qualitatively by the wavelength dependence of the turbidity, which can be approximated to λ−x in the region from 350 to 800 nm (42). The wider the size distribution becomes, the smaller is the value of x. For example, a suspension of small liposomes with a broad size distribution has a value of x ≈ 0.8, and E. coli suspensions approach a value of x ≈ 2.3 (42). We measured a value of x = 0.8 for control liposomes and a value of x = 1.3 for the OEP16 liposomes, revealing both a larger and narrower size distribution of the latter (42).
Addition of sucrose and KCl to OEP16 liposomes produced a different response of the suspension turbidity. Whereas (ΔOsm)KCL = 200 mOsm produced a turbidity change of (ΔA/A)400 nm = +0.05, the addition of sucrose (ΔOsm)sucrose = 200 mOsm produced a turbidity change of (ΔA/A)400 nm = −0.3 in the opposite direction. Control liposome suspensions containing no protein showed identical turbidity changes ((ΔA/A)400 nm ≈ +0.35) for KCl and sucrose upon changes in the osmolarity from 10 to 200 mOsm. We therefore used this “calibration” to further investigate the permeability of OEP16 to different solutes. Because the OEP16 channel has its highest open probability at Vm = 0 mV, this method is ideally suited for testing permeation of even uncharged substances through the open pore. By using the same proteoliposome preparation it was possible to obtain relative permeability measurements for a variety of compounds (Table 1). Surprisingly OEP16 was permeable to amino acids (e.g., glycine, valine, arginine, lysine, glutamic acid, or glutamine) (Table 1), whereas phosphoglyceric acid could not pass the OEP16 channel. Uncharged sugar molecules like fructose, glucose, sucrose, sorbitol, or even the small molecule dihydroxy acetone were not permeated through OEP16 (Table 1). Cadaverine (2,5-diaminopentane), a polyamine known to block E. coli type porins (46) is taken up rapidly into OEP16 liposomes, further supporting the differences in solute selectivity between OEP16 and general diffusion pores. The timecourse of the turbidity recordings indicated moreover that KCl and glycine were taken up more rapidly into OEP16 liposomes than the larger amino acids.
Table 1.
Selectivity of reconstituted OEP16 channels
As a second method to assess the selectivity of OEP16 we used the osmotically induced fusions of OEP16 liposomes with the planar bilayer (43, 44). Osmotic pressure was induced by raising the concentration of the osmotically active molecules on the cis side of the bilayer to hyperosmolar concentrations with respect to the trans side, the difference being typically ≥500 mOsm. Provided the liposome membrane is not permeable to the solute, no swelling of the bilayer bound liposomes is expected unless an open channel permeable to the osmotically active solute is present in the liposomes (45). Because the OEP16 channel has its highest open probability at Vm = 0 mV, permeation of even uncharged substances through the open pore and subsequent fusion of liposomes with the bilayer can be directly monitored electrically by the measurement of the bilayer resistance. The electrophysiological data obtained by this method gave identical solute selectivities for OEP16 as obtained with the first approach (Table 1). The OEP16 channel obviously does not select solely on the basis of size and charge but seems to be rather selective for amino acids and compounds containing primary NH2 groups.
DISCUSSION
Channel Properties of OEP16.
The electrophysiological data show that the reconstituted OEP16 constitutes an approximately 1-nm-wide voltage-gated pore that is impermeable to the large TEA+-cation with a unit conductance of Λ = 1,2 nS in 1 M KCl. Although the voltage dependence of the OEP16 channel gating remains elusive in vivo, its similarity to the mitochondrial outer membrane voltage-dependent anion channels and porins from the outer membrane of Gram-negative bacteria is striking (13, 47). Voltage gating as the sole regulatory mechanism seems unlikely because, similar to the outer mitochondrial and outer membrane of Gram-negative bacteria, no significant steady-state membrane potential for the chloroplast outer envelope has been described so far. Preliminary results indicate that OEP16 is phosphorylated within the N terminus (K.P. and J.S., unpublished data), which could influence the open probability of OEP16. The substrate specificities of OEP16 (i.e., impermeable to sugars but permeable to cadaverine) is in striking contrast to mitochondrial voltage-dependent anion channels and the porins from the outer membrane of Gram-negative bacteria, which behave exactly in the opposite manner.
Putative Structure of the OEP16 Channel.
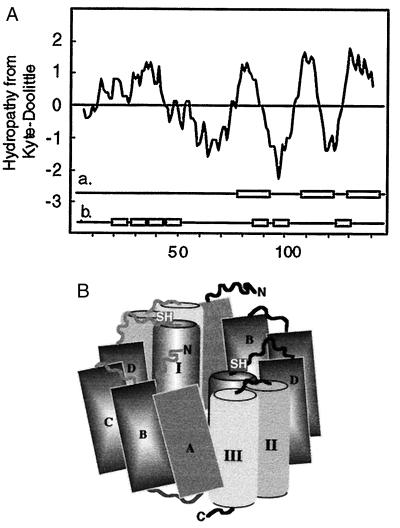
Hydropathy analysis (48) and secondary structure prediction algorithms (49, 50) indicate that OEP16 traverses the chloroplastic outer envelope with four β-strands and three α-helices (Fig. 5A). The CD spectra obtained from reconstituted OEP16 (Fig. 2C) support this theoretical prediction. The putative structure would be distinct from the classical porin structure, which is made up of amphiphilic β-strands (51). Crosslinking approaches using thiol- or amine-reactive crosslinkers indicated that OEP16 forms a homodimer in situ. A hypothetical model of the OEP16 dimer is proposed in Fig. 5B. This model is corroborated by two findings, first by the sequence similarities (52) we observed between OEP16 helix 1 and a sodium/amino acid cotransporter from pig (accession no. P31636) and a branched chain amino acid permease from E. coli (accession no. P08340). These similarities range between 50 and 70% on the primary amino acid sequence basis. Helix 1 shows the potential to be amphiphilic and could constitute parts of the channel-forming unit of the OEP16 dimer. Second, oxidation of Cys-71 by CuCl2 reduces the transport capacities of OEP16 liposomes dramatically, probably because the disulfide bridge brings the two helices very close together and functions in vitro as a floodgate that closes the channel. The calculated pore diameter of 1 nm seems unusually large for a dimer; however, this could be accomplished by the hypothetical pore structure of OEP16 as proposed in Fig. 5B.
Figure 5.
Protein structure prediction of OEP16. (A) A hydropathy analysis was carried out according to the Kyte–Doolittle algorithm using a window of 11 amino acids. The prediction for the presence of hydrophobic α-helices (a) was done according to ref. 50. The possible presence of β-strands in OEP16 (b) was predicted according to ref. 49. (B) Hypothetic model for the membrane arrangement of the pea OEP16 dimer. Putative β-strands A–D are shown as rectangles; the putative helices are shown as columns. The position of cysteine-71 is indicated by an SH. The model is not drawn to scale.
Possible Physiological Role of OEP16.
Plants use NO3− as their major nitrogen source. Nitrate is converted to NO2− in the cytoplasm, transported into chloroplasts by a poorly characterized mechanism (53), and converted to NH4+ in a light-dependent reaction. NH4+ is immediately incorporated into glutamic acid to yield glutamine. Chloroplasts support the surrounding cell compartments with essential NH4+ for different biosynthetic processes via the export of amino acids (5). OEP16 shows a broad specificity for amino acids; it allows the transport of glutamine and glutamic acids, two amino acids that might be important in the export of NH4+ from the chloroplasts (5). Positively charged lysine or arginine is transported with similar rates as negatively charged glutamic acid or branched chain valine. Furthermore, chloroplasts synthesize most proteinogenic amino acids themselves, including the essential aromatic and branched chain amino acids. On the other hand, during chloroplast differentiation, maturation, and senescence, a number of biosynthetic capacities are lost from the organelle and it becomes more dependent on cooperation with other compartments (54, 55). Chloroplasts therefore require amino acid transport facilities in both directions. The OEP16 channel excludes uncharged C5 and C6 sugars as well as phosphoglyceric acid or dihydroxyacetone, though the pore size (1 nm) of OEP16 should be large enough to allow passage of most of the above-listed compounds. Substrate specificity is most likely composed by the amino acid backbone that lines the aqueous channel formed by OEP16.
It seems to be generally accepted that the outer membranes of mitochondria and plastids do not represent a permeability barrier for low molecular weight solutes because of the presence of nonselective channel proteins. At least the chloroplastic outer envelope membrane from pea seems to deviate from this general assumption because it houses an amino acid-selective channel, namely OEP16, and maybe other yet to be identified channels selective for different solute classes such as triosephosphates, organic acids, or uncharged sugars.
Acknowledgments
We thank Dr. L. Heins for helpful discussions. This work was supported by grants from the Deutsche Forschungsgemeinschaft to J.S. and R.W., the Fonds der Chemischen Industrie (J.S.) and Stiftung, Volkswagenwerk (R.W.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: OEP, outer envelope protein; EST, expressed sequence tag; BS3, bis(sulfosuccinimidyl)suberate.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. 273553).
References
- 1.Gray J C, Row P E. Trends Cell Biol. 1995;5:243–251. doi: 10.1016/s0962-8924(00)89018-2. [DOI] [PubMed] [Google Scholar]
- 2.Schnell D J. Cell. 1995;83:521–524. doi: 10.1016/0092-8674(95)90090-x. [DOI] [PubMed] [Google Scholar]
- 3.Soll J. Bot Acta. 1995;108:277–282. [Google Scholar]
- 4.Joyard J, Block M A, Douce R. Eur J Biochem. 1991;199:489–509. doi: 10.1111/j.1432-1033.1991.tb16148.x. [DOI] [PubMed] [Google Scholar]
- 5.Lam H-M, Coschigano K T, Oliveira I C, Melo-Oliveira R, Coruzzi G M. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:569–593. doi: 10.1146/annurev.arplant.47.1.569. [DOI] [PubMed] [Google Scholar]
- 6.Flügge U-I, Benz R. FEBS Lett. 1984;169:85–89. [Google Scholar]
- 7.Heiber T, Steinkamp T, Hinnah S, Schwarz M, Flügge U-I, Weber A, Wagner R. Biochemistry. 1995;34:15906–15917. doi: 10.1021/bi00049a005. [DOI] [PubMed] [Google Scholar]
- 8.Pottosin I I. FEBS Lett. 1992;308:87–90. doi: 10.1016/0014-5793(92)81057-s. [DOI] [PubMed] [Google Scholar]
- 9.Pottosin I I. FEBS Lett. 1993;330:211–214. doi: 10.1016/0014-5793(93)80275-y. [DOI] [PubMed] [Google Scholar]
- 10.Fischer K, Weber A, Brink S, Arbinger B, Schünemann D, Borchert S, Heldt H W, Popp B, Benz R, Link T A, Eckerskorn C, Flügge U-I. J Biol Chem. 1994;269:25754–25760. [PubMed] [Google Scholar]
- 11.Heins L, Mentzel H, Schmid A, Benz R, Schmitz U K. J Biol Chem. 1994;269:26402–26410. [PubMed] [Google Scholar]
- 12.Benz R. Biochim Biophys Acta. 1994a;1197:167–196. doi: 10.1016/0304-4157(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 13.Benz R. Am Rev Microbiol. 1994b;42:359–393. doi: 10.1146/annurev.mi.42.100188.002043. [DOI] [PubMed] [Google Scholar]
- 14.Lübeck J, Soll J, Akita M, Nielsen E, Keegstra K. EMBO J. 1996;15:4230–4238. [PMC free article] [PubMed] [Google Scholar]
- 15.Sanger F, Nickler S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waegemann K, Soll J. Methods Cell Biol. 1995;50:255–267. doi: 10.1016/s0091-679x(08)61035-3. [DOI] [PubMed] [Google Scholar]
- 17.Harlow E, Lane D. Antibodies. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 18.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 19.Mueller P, Rudin D O, Tien H, Wescott W C. Nature (London) 1992;194:979–980. doi: 10.1038/194979a0. [DOI] [PubMed] [Google Scholar]
- 20.White S H. In: Ion Channel Reconstitution. Miller C, editor. New York and London: Plenum; 1984. pp. 3–36. [Google Scholar]
- 21.Cohen F S. In: Ion Channel Reconstitution. Miller C, editor. New York and London: Plenum; 1984. pp. 131–140. [Google Scholar]
- 22.Schwarz M, Gross A, Steinkamp T, Flügge U-I, Wagner R. J Biol Chem. 1994;269:29481–29489. [PubMed] [Google Scholar]
- 23.Waegemann K, Eichacker S, Soll J. Planta. 1992;187:89–94. doi: 10.1007/BF00201628. [DOI] [PubMed] [Google Scholar]
- 24.Joyard J, Billecocq A, Bartlett S G, Block M A, Chua N H, Douce R. J Biol Chem. 1983;258:10000–10006. [PubMed] [Google Scholar]
- 25.Cline K, Werner-Washburne M, Andrews J, Keegstra K. Plant Physiol. 1984;75:675–678. doi: 10.1104/pp.75.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnell D J, Kessler F, Blobel G. Science. 1994;266:1007–1012. doi: 10.1126/science.7973649. [DOI] [PubMed] [Google Scholar]
- 27.Hirsch S, Muckel E, Heemeyer F, von Heijne G, Soll J. Science. 1994;266:1989–1992. doi: 10.1126/science.7801125. [DOI] [PubMed] [Google Scholar]
- 28.Kessler F, Blobel G, Patel H A, Schnell D J. Science. 1994;266:1035–1039. doi: 10.1126/science.7973656. [DOI] [PubMed] [Google Scholar]
- 29.Tranel J T, Froehlich J, Goyal A, Keegstra K. EMBO J. 1995;14:2436–2446. doi: 10.1002/j.1460-2075.1995.tb07241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seedorf M, Waegemann K, Soll J. Plant J. 1995;7:401–411. doi: 10.1046/j.1365-313x.1995.7030401.x. [DOI] [PubMed] [Google Scholar]
- 31.Li H-M, Moore T, Keegstra K. Plant Cell. 1991;3:709–717. doi: 10.1105/tpc.3.7.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujiki Y, Hubbard A L, Fowler S, Lazarow P B. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waegemann K, Soll J. Plant J. 1991;1:149–158. [Google Scholar]
- 34.Soll J, Wanner G, Henkelmann G, Röper M, Schulze M. Plant Biol. 1986;2:229–234. [Google Scholar]
- 35.Keegstra K, Youssif A E. Methods Enzymol. 1986;118:316–325. [Google Scholar]
- 36.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Kobashi K. Biochim Biophys Acta. 1968;158:239–245. doi: 10.1016/0304-4165(68)90136-0. [DOI] [PubMed] [Google Scholar]
- 38.Seedorf M, Soll J. FEBS Lett. 1995;367:19–22. doi: 10.1016/0014-5793(95)00529-i. [DOI] [PubMed] [Google Scholar]
- 39.Venyaminov S Y, Yang J T. In: Circular Dichroism and the Conformational Analysis of Biomolecules. Fasman G D, editor. New York: Plenum; 1996. [Google Scholar]
- 40.Hille B. Ionic Channels of Excitable Membranes. Sunderland, MA: Sinauer; 1992. [Google Scholar]
- 41.Viera L I, Senistra G A, Disalvo E A. Chem Phys Lipids. 1996;81:45–54. doi: 10.1016/0009-3084(96)02532-7. [DOI] [PubMed] [Google Scholar]
- 42.Yoshikawa W, Akutsu H, Kyogoku Y. Biochim Biophys Acta. 1983;735:397–406. [Google Scholar]
- 43.Miller C, Arvan P, Telford J N, Racker E. J Membr Biol. 1976;30:271–282. doi: 10.1007/BF01869672. [DOI] [PubMed] [Google Scholar]
- 44.Woodbury D J, Miller C. Biophys J. 1990;58:833–839. doi: 10.1016/S0006-3495(90)82429-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woodbury D J, Hall J E. Biophys J. 1988;54:1053–1063. doi: 10.1016/S0006-3495(88)83042-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeLaVega A L, Delcour A H. J Bacteriol. 1996;178:3715–3720. doi: 10.1128/jb.178.13.3715-3721.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manella C A, Forte M, Colombini M. J Bioenerg Biomembr. 1992;24:7–19. doi: 10.1007/BF00769525. [DOI] [PubMed] [Google Scholar]
- 48.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 49.Eisenberg D, Weiss R M, Terwilliger T C. Proc Natl Acad Sci USA. 1984;81:140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Claros M G, von Heijne G. Cabios. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 51.Covan S W, Rosenberg J P. Science. 1994;264:914–916. [Google Scholar]
- 52.Worley K C, Wiese B A, Smith R F. Genome Res. 1995;5:173–184. doi: 10.1101/gr.5.2.173. [DOI] [PubMed] [Google Scholar]
- 53.Shingles R, Roh M H, McCarty R E. Plant J. 1996;112:1375–1381. doi: 10.1104/pp.112.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoppe P, Heintze A, Riedel A, Creuzer C, Schultz G. Planta. 1993;190:253–262. [Google Scholar]
- 55.Preiss M, Bustanur R, Hoppe P, Schultz G. J Plant Physiol. 1993;142:525–530. [Google Scholar]