Abstract
Recombinant collagens are attractive proteins for a number of biomedical applications. To date, significant progress was made in the large-scale production of nonmodified recombinant collagens; however, engineering of novel collagen-like proteins according to customized specifications has not been addressed. Herein we investigated the possibility of rational engineering of collagen-like proteins with specifically assigned characteristics. We have genetically engineered two DNA constructs encoding multi-D4 collagens defined as collagen-like proteins, consisting primarily of a tandem of the collagen II D4 periods that correspond to the biologically active region. We have also attempted to decrease enzymatic degradation of novel collagen by mutating a matrix metalloproteinase 1 cleavage site present in the D4 period. We demonstrated that the recombinant collagen α-chains consisting predominantly of the D4 period but lacking most of the other D periods found in native collagen fold into a typical collagen triple helix, and the novel procollagens are correctly processed by procollagen N-proteinase and procollagen C-proteinase. The nonmutated multi-D4 collagen had a normal melting point of 41°C and a similar carbohydrate content as that of control. In contrast, the mutant multi-D4 collagen had a markedly lower thermostability of 36°C and a significantly higher carbohydrate content. Both collagens were cleaved at multiple sites by matrix metalloproteinase 1, but the rate of hydrolysis of the mutant multi-D4 collagen was lower. These results provide a basis for the rational engineering of collagenous proteins and identifying any undesirable consequences of altering the collagenous amino acid sequences.
Keywords: Recombinant collagen, protein engineering, extracellular matrix
Collagens are crucial components of the extracellular matrix supporting and holding together multicellular organisms. In a family of collagenous proteins, fibrillar collagens play a central role (Piez 1984; Prockop and Kivirikko 1995). Not only are they the most abundant among all proteins but also they possess the ability to self-assemble into highly organized fibrils that strengthen connective tissues and to interact with other structural macromolecules (Eyre et al. 1987; Weber et al. 1996), growth factors (Suzawa et al. 1999), and cells (Loeser 1993; Svoboda 1998).
The structure of fibrillar collagens makes these proteins unique in their mechanical properties, resistance to some proteolytic enzymes, and ability to interact with other macromolecules and cells. Collagens are composed of three collagen α-chains that fold into a compact triple helix. Individual chains consist of ∼1000 amino acids organized as a continuous sequence of -Gly-X-Y- repeats, in which proline frequently occupies the -X- position and hydroxyproline is frequently present in the -Y- position (Piez 1984). Hydroxyprolines are particularly important because they take part in stabilizing the structure of collagen triple helix (Sakakibara et al. 1973; Bella et al. 1995), and their position in the collagen α-chains defines collagen-stabilizing domains (Sieron et al. 1993; Zafarullah et al. 1997; Arnold et al. 1998). It has been suggested that those domains vary in thermostability and undergo micro-unfolding in the physiological range of temperatures. It has also been demonstrated that the thermostability of those regions can be altered by mutations (Royce and Steinmann 2002).
Because of their mechanical and biological characteristics, collagens have numerous biomedical applications. In tissue engineering, for example, collagens are used as material for matrices that promote tissue repair by supporting cell growth (Stone et al. 1990, 1992; Nehrer et al. 1997) and by facilitating delivery of growth factors to a site of injury (Pandit et al. 1999). Almost all of the collagen available on the market today is obtained from bovine hide and bones. These sources allow for the extraction of large quantities of collagenous proteins in a cost-effective manner, but at the same time, they do not permit molecular-level modifications that could enhance biological or mechanical characteristics of the product. Expressing recombinant collagens circumvents this problem and offers a significant potential for the rational engineering of collagenous proteins. As suggested by John et al. (1999), such rational engineering of collagens with enhanced biological properties would generate products for use in tissue engineering, fibrosis, wound healing, and drug and cell delivery. Successes in expressing recombinant fibrillar collagens in mammalian cells (Geddis and Prockop 1993; Fertala et al. 1994) prompted the development of market-oriented, large-scale production technologies, and at present, recombinant collagens are expressed in bacteria, (Buechter et al. 2002), yeast (Myllyharju 2000; Toman et al. 2000), insect cells (Myllyharju 2000), transgenic animals (John et al. 1999; Toman et al. 1999), and transgenic plants (Ruggiero et al. 2000; Merle et al. 2002). All these technologies, however, focus on expressing the recombinant collagens identical to those extracted from animal tissues, and to date, rational engineering of collagens with specifically assigned biological characteristics has not been addressed.
Because most biological processes in which fibrillar collagens take part depend on site-specific interactions (Tuckwell et al. 1994; Knight et al. 1998, 2000; Keene et al. 2000; Fertala et al. 2001b; Di Lullo et al. 2002; Sieron et al. 2002), the concept of engineering collagen-like proteins that consist of repeats of a collagen domain critical for particular site-specific function is very important. Such rational engineering would create a custom-tailored "super-collagen" with enhanced specific biological properties and characterized by a high density of the active sites. Moreover, by adding or removing specific domains, it will be possible to modulate collagen-dependent signaling and to influence stability and self-assembly of collagen molecules. Such collagen-like proteins have the potential to be used in "smart-scaffolds" that encode specific commands for controlling tissue formation. It is not known, however, whether engineered collagen α-chains consisting predominantly of multiplied specific domain but lacking most of other domains found in native collagen would fold into a thermostable collagen triple helix, a structure critical for biological and mechanical functions.
Earlier studies with the use of collagen II variants that lack specific D-periods defined as consecutive fragments of 234 amino acids each demonstrated that the C-terminal region corresponding to the D4 period is essential for collagen–cell interaction (Fertala et al. 2001a). In addition, this region is important because it contains a site located between residues 775 and 776 that is recognized and cleaved by matrix metalloproteinase 1 (MMP-1), an enzyme that in vivo takes part in degradation of collagen. It has also been suggested that the corresponding region of collagen I is critical for interaction with numerous extracellular macromolecules and integrins (Di Lullo et al. 2002).
In this present study, we explored the possibility of engineering collagen-like proteins for novel scaffolding materials containing a high density of sites critical for interaction with cells. These proteins were engineered to contain a tandem of the D4 periods of collagen II. In addition, we attempted to engineer a collagen-like variant characterized by higher resistance to MMP-1. We show that the engineered multi-D4 procollagen-like proteins fold into a triple helix, and their morphology and structure are similar to that of native procollagen molecules. We demonstrated that single amino acid substitution in the D4 period intended to alter the MMP-1 cleavage site changed the thermostability of the collagen-like protein. The results provide a basis for the rational engineering of novel collagen-like proteins customized for specific biomedical applications and identifying the region of collagen II sensitive to changes in the amino acid sequence.
Results
Expression of recombinant multi-D4 procollagens
By using a DNA cassette system (for details, see Arnold et al. 1997, 1998), we engineered DNA constructs encoding multi-D4 procollagens consisting predominantly of the D4-period of collagen II (mD4; Figs. 1, 2 ▶ ▶). Even though the role of the procollagen N-terminal propeptide that, in native fibrillar procollagens, folds back on the region located in the D1 period (Holmes et al. 1991) is not fully understood (Hu et al. 1995; Bornstein 2002; Bornstein et al. 2002), we decided to include this domain as part of the recombinant proteins. Hence, the 5′ end of each construct was flanked with a sequence encoding the N-terminal propeptide, telopeptide, and the D1 period (Arnold et al. 1998). To facilitate correct folding of the protein and its secretion from cells (Bulleid et al. 1997; Lees et al. 1997; McLaughlin and Bulleid 1998), the 3′ end of each construct was flanked with sequences encoding a C-terminal propeptide, telopeptide, and the D0.4 period (Figs. 1, 2 ▶ ▶). To test the possibility of creating the MMP-1–resistant multi-D4 collagen (mD4M), the -CTG- codon for leucine present at position 776 has been changed to the -TTC- codon for phenylalanine. The decision to introduce this particular substitution was based on previous observations that the presence of phenylalanine at P1′ lowers the rate of hydrolysis of a synthetic linear peptide by MMP-1 (Netzel-Arnett et al. 1991).
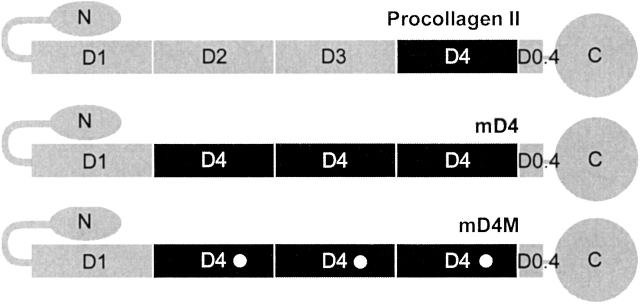
Figure 1.
Schematic of the recombinant procollagen-like proteins consisting predominantly of the D4 period of collagen II. To facilitate correct folding of the collagen triple helix, the multi-D4 procollagen includes procollagen C-propeptide and the D0.4 period. Because the correct folding of the N-propeptide depends on its interaction with D1 period, the multi-D4 procollagen includes this domain. Procollagen II indicates normal recombinant procollagen II; mD4, engineered procollagen-like protein consisting of a tandem D4 periods; and mD4M, engineered procollagen-like protein consisting of tandem D4 periods, in which leucine 776 was purposely substituted with phenylalanine. The white dot indicates the approximate position of mutation. N indicates procollagen N-terminal propeptide and telopeptide; D1, D2, D3, and D4, consecutive fragments of 234 amino acids corresponding to the D-periods seen in the banded collagen fibrils; D0.4, C-terminal D period consisting of 78 amino acid; and C, procollagen C-terminal propeptide and telopeptide.
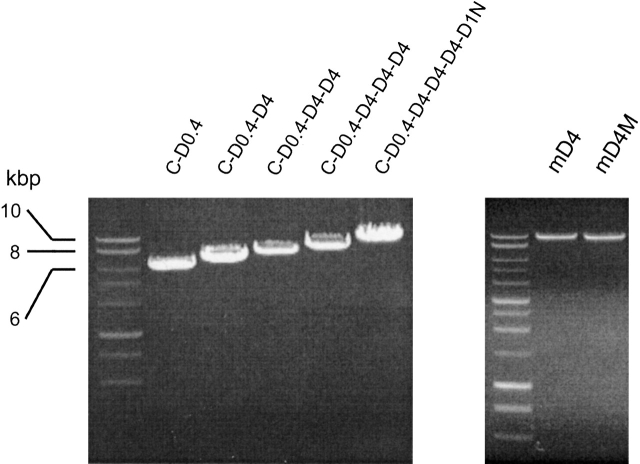
Figure 2.
Engineering of a DNA construct encoding the mD4 and mD4M procollagen-like proteins. (Left) The constructs were assembled from the 3′ end toward the 5′ end from the DNA cassettes that encode specific regions of human procollagen II. N indicates DNA cassette that encodes procollagen N-terminal propeptide and telopeptide; D1 and D4, DNA cassettes that encode consecutive fragments of 234 amino acids corresponding to the D1 and D4 periods of collagen II; D0.4, DNA cassette that encodes C-terminal D period consisting of 78 amino acids; and C, DNA cassette that encodes procollagen C-terminal propeptide and telopeptide. In the mD4M, the D4 cassette was mutated prior to the assembly of the full-length construct. (Right) Full-length mD4 and mD4M DNA constructs cloned into pcDNA2.1 vector. The constructs were linearized with the HindIII restriction enzyme.
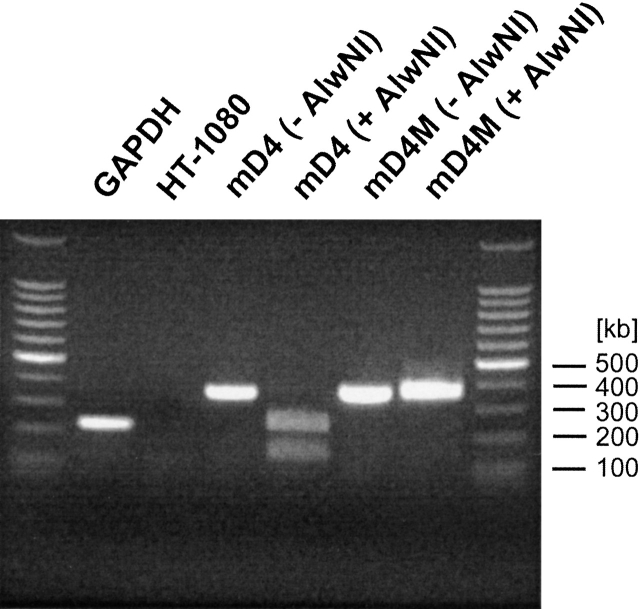
The fidelity of all DNA constructs used for transfection of HT-1080 cells was confirmed by DNA sequencing. To validate HT-1080 clones secreting recombinant collagens, the expression of the DNA constructs was analyzed by RT-PCR of total RNA extracted from clonal lines. The forward primer recognized a unique sequence of the junction between sequences encoding consecutive D4 periods of the multi-D4 collagens, and the reverse primer recognized an internal sequence present in the fragment encoding a D4 period. Correct PCR products of 350 bp derived from positive clones are depicted in Figure 3 ▶. Because mutating the -CTG- codon for leucine to the -TTC- codon for phenylalanine alters the -CAGNNN/CTG- site recognized by the restriction enzyme AlwNI, we were able to confirm expression of the mutated DNA construct as evident by resistance of the PCR product derived from cells expressing the mD4M collagen to the cleavage by AlwNI (Fig. 3 ▶). Specificity of RT-PCR was confirmed by use of nontransfected HT-1080 cells. For RT-PCR, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a positive internal standard (Fig. 3 ▶).
Figure 3.
RT-PCR analysis of mD4 and mD4M constructs from HT-1080 cells expressing recombinant collagens. GAPDH indicates internal positive control; HT-1080, nontransfected cells used as a negative control; mD4, RT-PCR product derived from cells transfected with the mD4 construct; mD4M, RT-PCR product derived from cells transfected wit the mD4M construct; and +AlwNI, RT-PCR products tested for the AlwNI restriction site that is present in the mD4 but not in the mD4M construct.
The multi-D4 procollagens were secreted from transfected HT-1080 cells as intact proteins, which was evidenced by the presence of single-protein bands. The mD4 migrated as a band of ∼140 kD. The mD4M migrated as a band of ∼160 kD, and its migration corresponded to that of recombinant procollagen II (Fig. 4 ▶).
Figure 4.
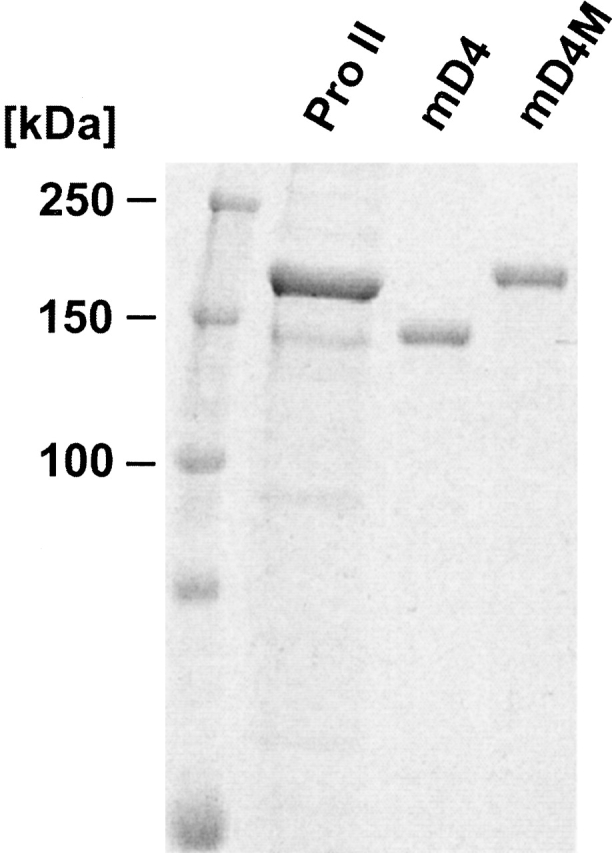
Analysis of purified mD4 and mD4M procollagens. Purified proteins were separated in 7.5% polyacrylamide-SDS gel in reduced conditions. The apparent difference in migration between normal procollagen II and mD4 procollagen is most likely a result of a significant dissimilarity in amino acid composition. Slower migration of the mD4M in comparison with the mD4 is most likely a result of an overmodification.
Analysis of amino acid composition and carbohydrate content
To characterize the recombinant mD4 and mD4M collagens, their amino acid composition was analyzed. The observed amino acid composition was consistent with composition predicted from the cDNA sequence (data not shown). In another series of experiments, the carbohydrate content of the recombinant procollagens was determined with the use of a carbohydrate analyzer. The result showed that the carbohydrate content of the mD4 procollagen was 12% w/w and was similar to that of normal recombinant procollagen II (Fertala et al. 1994). In contrast, the level of glycosylation of the mD4M variant was determined to be 27% w/w of the protein, which was significantly higher than that of control.
Morphometry of procollagen molecules
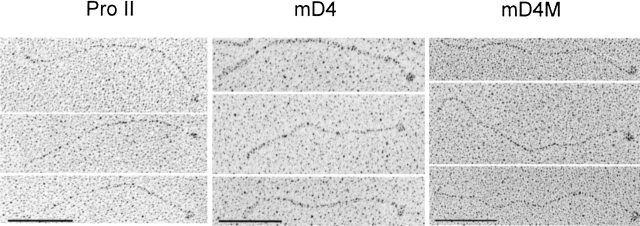
Electron micrographs of rotary-shadowed mD4 and mD4M molecules revealed the extended structure of these proteins (Fig. 5 ▶). At the C terminus, each procollagen molecule was flanked with a globular domain of 10 to 12 nm in diameter. As determined with the use of an image analysis program, the mean length of the mD4 procollagen was 290.3 nm (±4.04 SEM, n = 33), the mean length of the mD4M was 336 nm (±3.37 SEM, n = 15), and the mean length of normal procollagen II was 302 nm (±4.31 SEM, n = 16).
Figure 5.
Electron microscopy of the rotary shadowed procollagen molecules. All procollagens had an extended structure. Globular C-propeptide is apparent in all procollagen variants. Pro II indicates procollagen II; mD4, mD4 procollagen; and mD4M, mD4M procollagen. The mean length of the mD4 procollagen was 290.3 nm (±4.04 SEM, n = 33), the mean length of the mD4M was 336 nm (±3.37 SEM, n = 15), and the mean length of normal procollagen II was 302 nm (±4.31 SEM, n = 16). Bar, 100 nm.
Thermostability of multi-D4 collagen
Collagen α-chains correctly folded into a collagen triple helix are resistant to digestion with trypsin and chymotrypsin unless the triple helix unfolds to expose single α-chains. To test whether the recombinant multi-D4 collagens were able to fold into the thermostable triple helices, the proteins were exposed to digestion with a mixture of trypsin and chymotrypsin at temperatures ranging from 25°C to 42°C. The melting temperature of the mD4 triple helix, defined as the temperature in which 50% of the α-chains remain resistant to digestion with chymotrypsin-trypsin was 41°C; a value closely corresponding to the melting temperature of nonmodified recombinant collagen II (Fig. 6 ▶). In contrast, the thermostability assays demonstrated that the midpoint for unfolding of the recombinant mD4M variant was ∼6°C lower. Moreover, protease digestion of the mD4M at 25°C generated an intermediate product represented by a band of faster mobility in an electrical field.
Figure 6.
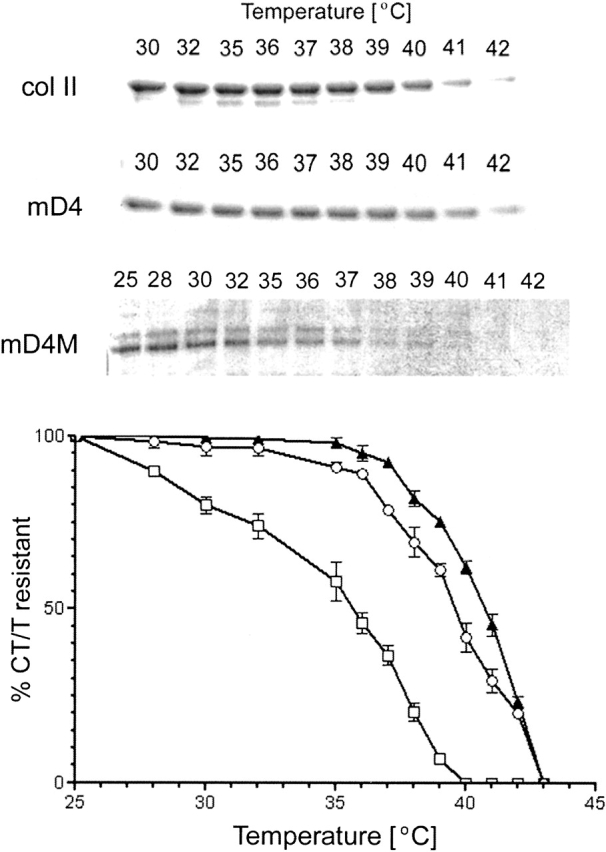
Analysis of the thermostability of novel collagens. Structural integrity of procollagen II, mD4, and mD4M was assayed by brief digestion with chymotrypsin and trypsin (CT/T) at the indicated temperatures. (Top) Products of enzymatic digestion were separated in a 7.5% polyacrylamide gel, visualized by staining with Coomassie blue, and then quantified by densitometry. (Bottom) Results from three independent experiments were plotted as function of temperature. The melting temperature was defined as the temperature at which 50% of the α-chains remain resistant to digestion with chymotrypsin and trypsin. The thermostability of the mD4 triple helix was 41°C; a value closely corresponding to the melting temperature of the normal collagen II. The thermostability of the mD4M was ∼36°C. Circles indicate normal collagen II; triangles, mD4; and squares, mD4M. Bars, SEM.
Cleavage of mD4 and mD4M procollagens with MMP-1
Engineered procollagens were tested as substrates for MMP-1. Proteins were digested with MMP-1 at 30°C, and then, products of digestion were reduced and separated in 12% polyacrylamide gel in the presence of SDS. Because of comigration of bands derived from MMP-1 with products of digestion of multi-D4 procollagens, proteins were transferred from a gel onto a nitrocellulose membrane. Bands derived from digestion of procollagens with MMP-1 were detected with the use of the anticollagen II polyclonal antibodies that recognize all collagen II D-fragments (Sieron et al. 2002). Because of three MMP-1 cleavage sites present in the multi-D4 procollagen, the predicted molecular masses of the products of a complete cleavage of a substrate are ∼40, 21, 21, and 51 kD. The 51-kD fragment is identical to the C-terminal B fragment generated after cleavage of native procollagen II (Fig. 7 ▶). As predicted, the mD4 procollagen was cleaved by MMP-1 at multiple sites. After 1 h of digestion, intermediate products of ∼90 and 51 kD were apparent. After 6 h of digestion, intermediates of ∼51, 40, and ∼20 kD were present. The 20-kD products appeared as weak bands, most likely because of the low number of epitopes recognized by polyclonal antibodies. Because of the possibility that the short products of digestion are not stable and lack triple helical conformation, the digestion was not carried out for >6 h. Mutating the MMP-1 cleavage site did not prevent digestion of the mD4M. The mD4M, however, was processed significantly slower, as evident by the presence of intact substrate and the number of partially processed intermediates still present at the sixth hour of digestion (Fig. 7 ▶).
Figure 7.
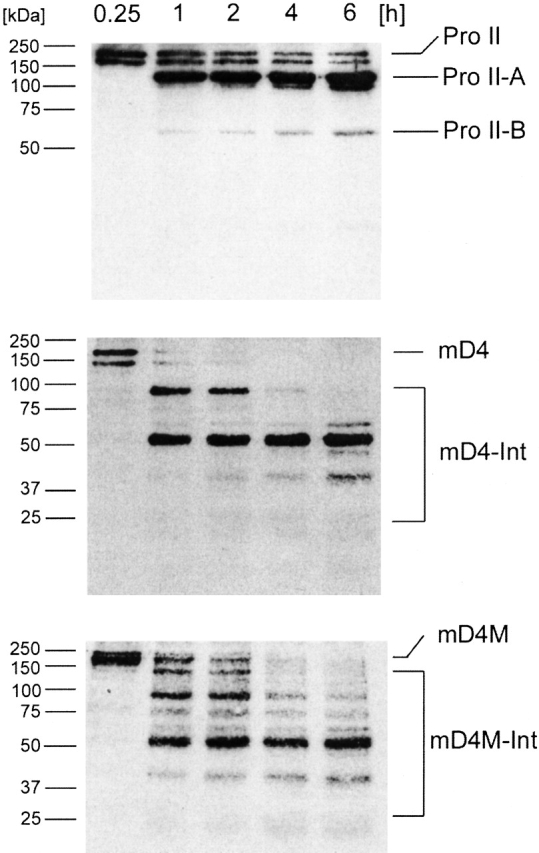
Enzymatic cleavage of mD4 and mD4M procollagens with MMP-1. Recombinant multi-D4 procollagens were tested for their sensitivity to MMP-1. Products of enzymatic digestion were separated in a 12% polyacrylamide gel, transferred onto a nitrocellulose membrane, and then detected with the use of collagen II–specific polyclonal antibodies. As expected, the substrates were cleaved in multiple sites present in three D4 periods. As evident by the presence of intact substrate and partially processed intermediates after 6 h of digestion, the cleavage of the mD4M collagen containing mutation in the MMP-1 cleavage site was slower than the cleavage of the mD4. Pro II indicates procollagen II; Pro II-A, larger of two procollagen II fragments derived from cleavage by MMP-1; Pro II-B, smaller of two procollagen II fragments derived from cleavage by MMP-1; mD4, recombinant mD4 procollagen; mD4-Int, intermediates generated by cleavage of the mD4 procollagen by MMP-1; mD4M, recombinant mD4M procollagen; and mD4M-Int, intermediates generated by cleavage of the mD4M procollagen by MMP-1. Time of digestion with MMP-1 and molecular masses of protein markers are also indicated.
Cleavage of mD4 and mD4M procollagens with procollagen N-proteinase and procollagen C-proteinase
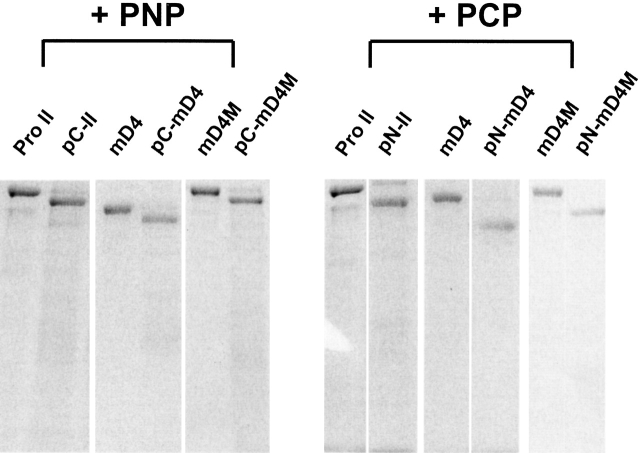
Folding of individual pro α-chains into procollagen begins at the C-terminal propeptide and is initiated by association of the individual collagen chains (Bulleid et al. 1997). Assembly of the N-propeptide completes the folding of a procollagen molecule. Correct folding of the procollagen N-propeptide and, to some extent, the procollagen C-propeptide is required for specific processing by the procollagen N-proteinase (PNP) and procollagen C-proteinase (PCP), respectively (Arnold et al. 1998). Therefore, cleavage of a procollagen with PNP and PCP is an important assay for correctly folded termini of procollagens. To test whether the propeptides of the mD4 and mD4M were correctly folded, the proteins were used as a substrate for PNP and PCP. Electrophoresis of the products of the enzymatic cleavage of the mD4 and mD4M procollagens demonstrated that the sizes of the fragments generated by digestion with PNP or PCP were consistent with the prediction that the cleavage occurs only at a single PNP or PCP cleavage site, respectively (Fig. 8 ▶).
Figure 8.
Enzymatic cleavage of mD4 and mD4M procollagens with procollagen N-proteinase (PNP) and procollagen C-proteinase (PCP). To test the folding of procollagen propeptides, purified procollagens were treated with PNP or PCP. The size of the fragments generated by digestion with PNP or PCP was consistent with the prediction that the cleavage occurs only at the single PNP or PCP cleavage site, respectively. Pro II indicates procollagen II; mD4, mD4 procollagen; mD4M, mD4M procollagen; pC, procollagens in which N-propeptides were enzymatically removed; and pN, procollagens in which C-propeptides were enzymatically removed.
Discussion
Recombinant collagens are proteins with enormous potential for use in tissue engineering, wound healing, cosmetics, and delivery of cells and growth factors. Rational design of gene engineered collagen-like proteins holds the promise of inventing novel molecules characterized by enhanced biological properties. Successful engineering of the collagen-like proteins must meet two fundamental requirements: (1) Novel proteins have to be triple helical, and (2) they must be thermostable. Triple helicity is not only important for the stability of collagens, but it also plays an important biological role. For instance, the integrin-mediated binding of cells to collagens depends on the correct structure of a triple helix, and thermal denaturation of the triple helix alters the cell-collagen interaction (Cardarelli et al. 1992; Tuckwell et al. 1994). Because most functions of fibrillar collagens depend on the site-specific interactions between distinctive regions of these proteins and domains of other macromolecules, engineering of the collagen-like proteins according to the required specifications and consisting of repeats of a specific amino acid sequence will be critical for creation of advanced biomaterials with a high density of biologically active sites. Another approach to producing collagenous molecules containing regions important for biomedical applications is the synthesis of short collagen-like peptides that correspond to the specific regions of the collagen molecule. Such short peptides, however, are characterized by low thermostability, and to remain triple helical, they must be stabilized by an addition of collagenous Gly-Pro-Hyp repeats or by flanking with noncollagenous stabilizing domains (Shah et al. 1996; Frank et al. 2001). Such bulky noncollagenous fragments, however, could adversely affect crucial biological properties of purposely engineered collagenous fragments. Therefore, engineering collagen-like proteins consisting predominantly of a multiplied specific domain and having the same length as native collagen circumvents this problem.
Among the regions of fibrillar collagens, the region that corresponds to the D4-period is of special importance; it participates in a number of interactions that include binding of collagen telopeptides (Prockop and Fertala 1998), integrins (for review, see Di Lullo et al. 2002), bone morphogenetic protein-2 (Sieron et al. 2002), fibronectin (Kleinman and McGoodwin 1976), collagen IX (Fertala et al. 2001b), and cartilage oligomeric matrix protein (Rosenberg et al. 1998).
In the present study, we engineered collagen-like proteins consisting primarily of the repeats of the region that extends between residues 703 and 936 of the collagen II triple helix. Because of the importance of the D1 period for the folding and enzymatic cleavage of the N-terminal propeptide (Arnold et al. 1998) and the D0.4 period for the folding and stability of entire procollagen molecule (Arnold et al. 1998), both periods were included in the engineered procollagens. Even though it has already been demonstrated that truncated collagens lacking short fragments of the triple helix fold into stable triple helices (Sieron et al. 1993; Zafarullah et al. 1997; Arnold et al. 1998), here we demonstrate for the first time that the recombinant, full-length collagen-like proteins composed mainly of multiplied D4 period but lacking most of other D-periods fold into collagen triple helix and are correctly processed by PNP and PCP.
The mD4 collagen had normal thermostability and was correctly glycosylated. Because hydroxylation of prolines and posttranslational glycosylation of the α-chains stabilize the triple helix in native collagens and contribute to its resistance to proteolysis (Bann et al. 2000), correct posttranslational modification of the mD4 procollagen ensures its procollagen-like characteristics. Even though the size of DNA constructs encoding recombinant mD4 and mD4M is the same as the size of DNA constructs encoding recombinant procollagen II, and the predicted molecular mass of these proteins is similar, the electrophoretic mobility of the mD4 differed from the electrophoretic mobility of recombinant procollagen II. Migration of fibrillar collagens, however, frequently does not follow rules that apply to globular proteins. Perhaps the best example illustrating this problem is the migration of the α2-chain of collagen I with a similar predicted molecular mass to the α1-chain of collagen I still migrating significantly faster than the α1-chain. Slower mobility of the mD4M collagen in comparison with the mD4 is most likely a result of its higher glycosylation, which affects both the molecular mass and the charge of this protein.
The D4 period is unique in that it contains the MMP-1 cleavage site. It is not clear, however, why only one site per α-chain is cleaved, whereas many other potential sites pres-ent in another D periods are not. It has been suggested that the MMP-1 cleavage site is preceded by a region of tightly-coiled collagen triple helix followed by a longer micro-unfolded region, and that this conformation is critical for the MMP-1 cleavage (Fields 1991). In an attempt to engineer an MMP-1–resistant multi-D4 collagen, we converted a codon for leucine located at position 776 to a codon for phenylalanine. As reported by Netzel-Arnet et al. (1991), such a mutation decreased the rate of hydrolysis of a synthetic non–triple helical peptide used as a substrate for MMP-1 to ∼5% in comparison to control. Even though cleavage of the mD4M by MMP-1 was slower than mD4, it is apparent that it was still >5% reported by Netzel-Arnet et al. Because denatured collagen is processed by MMP-1 ∼60 times slower than the nondenatured triple helical substrate (Knauper et al. 1996), it is possible that the previously reported slow cleavage of a mutated linear peptide was in part due to a lack of conformation optimal for processing by MMP-1 (Netzel-Arnett et al. 1991).
We found that converting a codon for leucine located at position 776 to a codon for phenylalanine caused a significant decrease in the thermostability of the mD4M collagen. This observation is consistent with previous reports that mutations in the region surrounding the MMP-1 cleavage site of collagen I cause a significant decrease in thermostability of collagen from a patient with osteogenesis imperfecta (Fertala et al. 1993). Moreover, this observation supports a report that the presence of phenylalanine in the -X- position of the -Gly-X-Hyp- triplet present in a synthetic triple helical peptide, markedly decreases its thermostability (Persikov et al. 2000).
One of the theories explaining this decrease in thermostability in mutated collagens is destabilization of cooperative blocks predicted to function in the collagen triple helix (for review, see Privalov 1982). Because protease digestion of normal collagen II and mD4 did not create stable intermediates, it may indicate that the unfolding of these proteins is a highly cooperative process. Because the mD4M was cleaved to stable intermediates at 25°C, the results indicate that the mutation destabilizes a cooperative block, which in the mD4M is triplicated, so the protein is cleaved by proteases at a significantly lower temperature than is the nonmutated mD4. Such destabilization of cooperative blocks was previously suggested by Westerhausen et al. (1990) to explain low thermostability of collagen I with serine for glycine α1–598 substitution found in a patient with osteogenesis imperfecta.
Lower thermostability of the mD4M collagen was most likely the reason for its higher glycosylation. Similar overmodification of collagens with low thermostability was found in a number of collagen I mutants (for review, see Royce and Steinmann 2002).
There is a general agreement that the contour length of triple helical region of the fibrillar collagens determined by electron microscopy is 300 nm; however, the reported values vary. More recently, by using optical tweezers and using two different techniques to capture collagen, Sun et al. (2002) reported a contour length of 315 nm (±44) and 300 nm (±35), respectively. It is not clear whether the apparent difference in the contour lengths of mD4 and mD4M is due to an electron microscopy artifact caused by variations in dehydration of samples or their different interaction with the surface onto which the collagen molecules adhered, or whether it results from structural differences in these two proteins.
Overall, our results determined for the first time that full-length multi-D collagen-like proteins lacking specific domains otherwise present in native fibrillar collagens can be successfully expressed as triple helical and thermostable recombinant proteins. With an increasing number of collagen domains with specific biological functions that have been defined and with advances in the production of large quantities of recombinant collagens, our results provide a basis for the rational design of gene-engineered collagen-like proteins for use in a vast area of biotechnology and medicine.
Materials and methods
Engineering of mD4 and mD4M procollagen DNA constructs
A DNA construct encoding the mD4 procollagen was assembled from DNA cassettes as described by Arnold et al. (1997). The following DNA cassettes were used: (1) Nt-D1 cassette, encoding a signal peptide, procollagen II N-propeptide, N-telopeptide, and the D1-period that corresponds to the fragment of collagen II triple helix located between residues 1 and 234; (2) three D4 cassettes, each encoding the D4-period that corresponds to the fragment of procollagen II triple helix located between residues 703 and 936; and (3) D0.4-Ct-UTR cassette, encoding the fragment of collagen II triple helix located between residues 937 and 1014, C-propeptide, C-telopeptide, and a 2-kb PpuMI-PvuII fragment of the α1(II) 3′ UTR (Baldwin et al. 1989). The cassettes were assembled according to the procedure described by Arnold et al. (1997) into the complete construct (described as Nt-D1-D4-D4-D4-D0.4-Ct-UTR) and cloned into the pcDNA2.1 vector (Invitrogen Corp.). The structures of all junctions between D cassettes were verified by DNA sequencing.
For the expression in HT-1080 cells, the DNA construct was cloned into the pcDNA3.1 mammalian expression vector (Invitrogen Corp.) containing a cytomegalovirus promoter and a gene encoding neomycin resistance. The mD4M collagen variant was engineered in the same way except that prior to the assembly of the construct, two point mutations were introduced into the D4 DNA cassette. The mutations were generated by the use of the QuickChange site-directed mutagenesis kit (Stratagene). To change the -CTG- codon for the leucine 776 to the -TTC- codon for phenylalanine, we used 5′-CCAGGTCCCCAGGGTTTCGCTGGT CAGAGAGGCATCG-3′ and 5′-CGATGCCTCTCTGACCAGC GAAACCCTGGGGACC TGG-3′ primers. The presence of mutations was confirmed by sequencing of DNA used to transfect HT-1080 cells.
Expression of the mD4 and mD4M in HT-1080 cells
HT-1080 (ACTT, CCL-121) cells were transfected with the mD4 and mD4M DNA construct with the use of the calcium phosphate precipitation method (Stratagene). By using anticollagen II polyclonal antibodies (Biosciences Inc.) the G418-resistant clones were analyzed for production of recombinant proteins, as described previously (Sieron et al. 1993). In addition, expression of the DNA constructs in the G418-resistant clones that secreted recombinant collagens was analyzed by RT-PCR. Isolation of total RNA and RT-PCR was carried out with the use of the OneStep RT-PCR kit according to manufacturer’s protocol (QIAGEN Inc.). The forward primer (5′-GCTTCACGGGGCGTGTTGGAC-3′) recognized a unique sequence of the junction between sequences encoding consecutive D4 periods of the multi-D4 collagens, and the reverse primer (5′-GACCTCTGTCTCCAGATGCTCCAGGAGC-3′) recognized an internal sequence present in the fragment encoding a D4 period. Because mutating the -CTG- codon for leucine to the -TTC- codon for phenylalanine alters the -CAGNNN/CTG- site recognized by the restriction enzyme AlwNI, the PCR products derived from cells expressing multi-D4 collagens were subjected to cleavage by AlwNI (New England BioLabs Inc.). Specificity of RT-PCR was confirmed by use of nontransfected HT-1080 cells. GAPDH was used as a positive internal standard.
Purification of recombinant procollagens
The mD4 and mD4M procollagens were purified from cell culture media according to the method described by Fertala et al. (1994). Briefly, ∼4 L of media harvested from each 24-h period was filtered through a 1.6-μm glass-fiber filter (Millipore) and supplemented with the following reagents at the indicated concentrations: 0.1 M Tris-HCl buffer, 0.4 M NaCl, 25 mM EDTA, and 0.02% NaN3 adjusted to pH 7.4. High-molecular-weight proteins in the medium were concentrated ∼10-fold at 4°C by the use of cartridges with a 100-kD molecular-weight cut-off (Prep/Scale-TFF filter; Millipore). Proteins in the concentrated media were precipitated overnight at 4°C with 175 mg/mL of ammonium sulfate and collected by centrifugation at 15,000g for 1 h at 4°C. Recombinant proteins were purified by using three-step ion exchange chromatography, as described by Fertala et al. (1994).
Analysis of amino acid composition and sugar content
The amino acid composition was determined by the AAA Laboratory (Mercer Island, WA). The carbohydrate content was determined by chromatographic methods developed for analysis of acidic and neutral oligosaccharides (Anumula and Taylor 1991). The oligosaccharides were released from the recombinant procollagens with peptide N-glycosidase F, endo-β-acetylglucosaminidase F, and endo-β-N-acetylglucosaminidase H and were then analyzed with a carbohydrate analyzer (Dionex BioLC).
Electron microscopy of procollagen molecules
For rotary shadowing, the mD4, mD4M, and recombinant procollagen II were dissolved in 0.5 M acetic acid at a concentration of 50 μg/mL. Equal volumes of procollagen samples and 65% glycerol were mixed according to the procedure of Tyler and Branton (1980). The samples were nebulized onto freshly cleaved mica surface, and then the mica plates were placed on a rotating stage of a JEOL JFD9000 freeze-fracture apparatus (JEOL Co.). The specimens were vacuum-dried at 5 × 10−6 torr for 20 to 30 min at room temperature. The dried specimens were set to rotate at ∼60 rpm and then shadowed with platinum-carbon at a low angle of 6°. The replicas were immediately reinforced with an additional carbon layer at an angle of 90° and then floated on distilled water and picked up on 150-mesh copper grids. Replicas were examined with a transmission electron microscope Hitachi H-8100 (Hitachi Co.), operating at 100 kV. Photographs were taken at 54,000× magnification. Measurements of the length of procollagen molecules were performed by using the SigmaScan Pro image analysis program (SPSS Inc.), and data were analyzed by use of a statistical program (GraphPad Prizm program, GraphPad Software Inc.).
Analysis of thermostability
The samples dissolved in 0.4 M NaCl and 0.1 M Tris-HCl buffer (pH 7.4) were preincubated at the indicated temperatures for 6 min and then digested for 2 min with 100 μg/mL trypsin and 250 μg/mL chymotrypsin (Sigma Aldrich Corp.) as described earlier (Sieron et al. 1993). Enzymatic digestion was terminated by the addition of 0.1 volume of 5 mg/mL soya-bean trypsin inhibitor (Sigma Aldrich Corp.). The samples were assayed by electrophoresis in a 7.5% polyacrylamide-SDS gel. Proteins were visualized by staining with Coomassie G250 stain (Bio-Rad) and then quantified by densitometry. Recombinant human collagen II was used as a control (Fertala et al. 1994). Mean values of the percentage of the triple helix resistant to proteases digestion as derived from three independent measurements were plotted against temperature.
Cleavage of mD4 and mD4M procollagens with MMP-1
For digestion with MMP-1 (Calbiochem) the enzyme was first activated at 37°C for 4 h in the presence of p-aminophenylmercuric acetate. Subsequently, 2 μg of procollagen II, mD4 procollagen, or mD4M procollagen was digested with MMP-1 (0.5 μg of enzyme/μg of procollagen) for 6 h at 30°C. The reaction buffer was 50 mM Tris-HCl (pH 7.0), 300 mM NaCl, 5 mM CaCl2, 1 μM ZnCl2, 0.05% Brij 35, and 0.02% NaN3. The reaction was terminated by an addition of EDTA to a final concentration of 10 mM. Products of enzymatic digestion were reduced and then separated in a 12% polyacrylamide gel in the presence of SDS. Because of comigration of bands derived from MMP-1 with products of digestion of recombinant procollagens, instead of being stained, the proteins were transferred onto a nitrocellulose membrane, and then procollagen-derived bands were specifically detected with the use of the anticollagen II polyclonal antibodies (Biodesign International) that recognize all collagen II D-periods (Sieron et al. 2002). Next, the procollagen-derived bands were visualized by chemiluminescence with the use of goat antirabbit antibodies conjugated to horseradish peroxidase (Sigma-Aldrich).
Cleavage of mD4 and mD4M procollagens with procollagen N- and C-proteinases
The N-terminal propeptide and the C-terminal propeptide were cleaved by procollagen N-proteinase (Hojima et al. 1989) and procollagen C-proteinase (Hojima et al. 1985), respectively. Enzymatic cleavage was carried out in 25 mM Tris-HCl buffer (pH 7.5) containing 7 mM CaCl2, 0.1 M NaCl, 0.015% Brij, and 0.02% NaN3. The reaction mixture contained ∼10 μg of procollagen and 5 U N-proteinase or 10 U C-proteinase. The reaction was carried out for 4 h at 35°C. The enzymes were inactivated by an addition of EDTA to a final concentration of 10 mM.
Acknowledgments
This work was supported in part by grants to A.F. from the National Aeronautics and Space Administration (NAG9-1342) and the NIH (AR48544 and AR49537) and by a grant to E.A. from the Academic Frontier Project by the Ministry of Education, Science, Sports and Culture of Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0385103.
References
- Anumula, K.R. and Taylor, P.B. 1991. Rapid characterization of asparagine-linked oligosaccharides isolated from glycoproteins using a carbohydrate analyzer. Eur. J. Biochem. 195 269–280. [DOI] [PubMed] [Google Scholar]
- Arnold, W.V., Sieron, A.L., Fertala, A., Bachinger, H.P., Mechling, D., and Prockop, D.J. 1997. A cDNA cassette system for the synthesis of recombinant procollagens: Variants of procollagen II lacking a D-period are secreted as triple-helical monomers. Matrix Biol. 16 105–116. [DOI] [PubMed] [Google Scholar]
- Arnold, W.V., Fertala, A., Sieron, A.L., Hattori, H., Mechling, D., Bachinger, H.P., and Prockop, D.J. 1998. Recombinant procollagen II: Deletion of D period segments identifies sequences that are required for helix stabilization and generates a temperature-sensitive N-proteinase cleavage site. J. Biol. Chem. 273 31822–31828. [DOI] [PubMed] [Google Scholar]
- Baldwin, C.T., Reginato, A.M., Smith, C., Jimenez, S.A., and Prockop, D.J. 1989. Structure of cDNA clones coding for human type II procollagen: The α 1(II) chain is more similar to the α1 (I) chain than two other α chains of fibrillar collagens. Biochem. J. 262 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bann, J.G., Peyton, D.H., and Bachinger, H.P. 2000. Sweet is stable: Glycosylation stabilizes collagen. FEBS Lett. 473 237–240. [DOI] [PubMed] [Google Scholar]
- Bella, J., Brodsky, B., and Berman, H.M. 1995. Hydration structure of a collagen peptide. Structure 3 893–906. [DOI] [PubMed] [Google Scholar]
- Bornstein, P. 2002. The NH2-terminal propeptides of fibrillar collagens: Highly conserved domains with poorly understood functions. Matrix Biol. 21 217–226. [DOI] [PubMed] [Google Scholar]
- Bornstein, P., Walsh, V., Tullis, J., Stainbrook, E., Bateman, J.F., and Hormuzdi, S.G. 2002. The globular domain of the pro-α1(I) N-propeptide is not required for secretion, processing by procollagen N-proteinase, or fibrillogenesis of type I collagen in mice. J. Biol. Chem. 277 2605–2613. [DOI] [PubMed] [Google Scholar]
- Buechter, D.D., Paolella, D.N., Leslie, B.S., Brown, M.S., Mehos, K.A., and Gruskin, E.A. 2002. Co-translational incorporation of trans-4-hydroxyproline into recombinant proteins in bacteria. J. Biol. Chem. 278 645–650. [DOI] [PubMed] [Google Scholar]
- Bulleid, N.J., Dalley, J.A., and Lees, J.F. 1997. The C-propeptide domain of procollagen can be replaced with a transmembrane domain without affecting trimer formation or collagen triple helix folding during biosynthesis. EMBO J. 16 6694–6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardarelli, P.M., Yamagata, S., Taguchi, I., Gorcsan, F., Chiang, S.L., and Lobl, T. 1992. The collagen receptor α2 β1, from MG-63 and HT1080 cells, interacts with a cyclic RGD peptide. J. Biol. Chem. 267 23159–23164. [PubMed] [Google Scholar]
- Di Lullo, G.A., Sweeney, S.M., Korkko, J., Ala-Kokko, L., and San Antonio, J.D. 2002. Mapping the ligand-binding sites and disease-associated mutations on the most abundant protein in the human, type I collagen. J. Biol. Chem. 277 4223–4231. [DOI] [PubMed] [Google Scholar]
- Eyre, D.R., Wu, J.J., and Apone, S. 1987. A growing family of collagens in articular cartilage: Identification of five genetically distinct types. J. Rheumatol. 14: Spec No: 25–27. [PubMed] [Google Scholar]
- Fertala, A., Westerhausen, A., Morris, G., Rooney, J.E., and Prockop, D.J. 1993. Two cysteine substitutions in procollagen I: A glycine replacement near the N-terminus of α1 (I) chain causes lethal osteogenesis imperfecta and a glycine replacement in the α2 (I) chain markedly destabilizes the triple helix. Biochem. J. 289 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fertala, A., Sieron, A.L., Ganguly, A., Li, S.W., Ala-Kokko, L., Anumula, K.R., and Prockop, D.J. 1994. Synthesis of recombinant human procollagen II in a stably transfected tumour cell line (HT1080). Biochem. J. 298 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fertala, A., Han, W.B., and Ko, F.K. 2001a. Mapping critical sites in collagen II for rational design of gene-engineered proteins for cell-supporting materials. J. Biomed. Mater. Res. 57 48–58. [DOI] [PubMed] [Google Scholar]
- Fertala, A., Sieron, A.L., Adachi, E., and Jimenez, S.A. 2001b. Collagen II containing a Cys substitution for Arg-α1–519: Abnormal interactions of the mutated molecules with collagen IX. Biochemistry 40 14422–14428. [DOI] [PubMed] [Google Scholar]
- Fields, G.B. 1991. A model for interstitial collagen catabolism by mammalian collagenases. J. Theor. Biol. 153 585–602. [DOI] [PubMed] [Google Scholar]
- Frank, S., Kammerer, R.A., Mechling, D., Schulthess, T., Landwehr, R., Bann, J., Guo, Y., Lustig, A., Bachinger, H.P., and Engel, J. 2001. Stabilization of short collagen-like triple helices by protein engineering. J. Mol. Biol. 308 1081–1089. [DOI] [PubMed] [Google Scholar]
- Geddis, A.E. and Prockop, D.J. 1993. Expression of human COL1A1 gene in stably transfected HT1080 cells: The production of a thermostable homotrimer of type I collagen in a recombinant system. Matrix 13 399–405. [DOI] [PubMed] [Google Scholar]
- Hojima, Y., van der Rest, M., and Prockop, D.J. 1985. Type I procollagen carboxyl-terminal proteinase from chick embryo tendons: Purification and characterization. J. Biol. Chem. 260 15996–16003. [PubMed] [Google Scholar]
- Hojima, Y., McKenzie, J.A., van der Rest, M., and Prockop, D.J. 1989. Type I procollagen N-proteinase from chick embryo tendons: Purification of a new 500-kD form of the enzyme and identification of the catalytically active polypeptides. J. Biol. Chem. 264 11336–11345. [PubMed] [Google Scholar]
- Holmes, D.F., Mould, A.P., and Chapman, J.A. 1991. Morphology of sheet-like assemblies of pN-collagen, pC-collagen and procollagen studied by scanning transmission electron microscopy mass measurements. J. Mol. Biol. 220 111–123. [DOI] [PubMed] [Google Scholar]
- Hu, G., Gura, T., Sabsay, B., Sauk, J., Dixit, S.N., and Veis, A. 1995. Endoplasmic reticulum protein Hsp47 binds specifically to the N-terminal globular domain of the amino-propeptide of the procollagen I α1 (I)-chain. J. Cell Biochem. 59 350–367. [DOI] [PubMed] [Google Scholar]
- John, D.C., Watson, R., Kind, A.J., Scott, A.R., Kadler, K.E., and Bulleid, N.J. 1999. Expression of an engineered form of recombinant procollagen in mouse milk. Nat. Biotechnol. 17 385–389. [DOI] [PubMed] [Google Scholar]
- Keene, D.R., San Antonio, J.D., Mayne, R., McQuillan, D.J., Sarris, G., Santoro, S.A., and Iozzo, R.V. 2000. Decorin binds near the C terminus of type I collagen. J. Biol. Chem. 275 21801–21804. [DOI] [PubMed] [Google Scholar]
- Kleinman, H.K. and McGoodwin, E.B. 1976. Localization of the cell attachment region in types I and II collagens. Biochem. Biophys. Res. Commun. 72 426–432. [DOI] [PubMed] [Google Scholar]
- Knauper, V., Lopez-Otin, C., Smith, B., Knight, G., and Murphy, G. 1996. Biochemical characterization of human collagenase-3. J. Biol. Chem. 271 1544–1550. [DOI] [PubMed] [Google Scholar]
- Knight, C.G., Morton, L.F., Onley, D.J., Peachey, A.R., Messent, A.J., Smethurst, P.A., Tuckwell, D.S., Farndale, R.W., and Barnes, M.J. 1998. Identification in collagen type I of an integrin α2 β1-binding site containing an essential GER sequence. J. Biol. Chem. 273 33287–33294. [DOI] [PubMed] [Google Scholar]
- Knight, C.G., Morton, L.F., Peachey, A.R., Tuckwell, D.S., Farndale, R.W., and Barnes, M.J. 2000. The collagen-binding A-domains of integrins α1β1 and α2β1 recognize the same specific amino acid sequence, GFOGER, in native (triple-helical) collagens. J. Biol. Chem. 275 35–40. [DOI] [PubMed] [Google Scholar]
- Lees, J.F., Tasab, M., and Bulleid, N.J. 1997. Identification of the molecular recognition sequence which determines the type-specific assembly of procollagen. EMBO J. 16 908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeser, R.F. 1993. Integrin-mediated attachment of articular chondrocytes to extracellular matrix proteins. Arthritis Rheum. 36 1103–1110. [DOI] [PubMed] [Google Scholar]
- McLaughlin, S.H. and Bulleid, N.J. 1998. Molecular recognition in procollagen chain assembly. Matrix Biol. 16 369–377. [DOI] [PubMed] [Google Scholar]
- Merle, C., Perret, S., Lacour, T., Jonval, V., Hudaverdian, S., Garrone, R., Ruggiero, F., and Theisen, M. 2002. Hydroxylated human homotrimeric collagen I in Agrobacterium tumefaciens–mediated transient expression and in transgenic tobacco plant. FEBS Lett. 515 114–118. [DOI] [PubMed] [Google Scholar]
- Myllyharju, J. 2000. Recombinant collagen trimers from insect cells and yeast. Methods Mol. Biol. 139 39–48. [DOI] [PubMed] [Google Scholar]
- Nehrer, S., Breinan, H.A., Ramappa, A., Shortkroff, S., Young, G., Minas, T., Sledge, C.B., Yannas, I.V., and Spector, M. 1997. Canine chondrocytes seeded in type I and type II collagen implants investigated in vitro. J. Biomed. Mater. Res. 38 95–104. [DOI] [PubMed] [Google Scholar]
- Netzel-Arnett, S., Fields, G.B., Birkedal-Hansen, H., Van Wart, H.E., and Fields, G. 1991. Sequence specificities of human fibroblast and neutrophil collagenases. J. Biol. Chem. 266 6747–6755. [PubMed] [Google Scholar]
- Pandit, A., Ashar, R., and Feldman, D. 1999. The effect of TGF-β delivered through a collagen scaffold on wound healing. J. Invest. Surg. 12 89–100. [DOI] [PubMed] [Google Scholar]
- Persikov, A.V., Ramshaw, J.A., Kirkpatrick, A., and Brodsky, B. 2000. Amino acid propensities for the collagen triple-helix. Biochemistry 39 14960–14967. [DOI] [PubMed] [Google Scholar]
- Piez, K.A. 1984. Extracellular matrix biochemistry, pp. 1–40. Elsevier, New York.
- Privalov, P.L. 1982. Stability of proteins: Proteins which do not present a single cooperative system. Adv. Protein Chem. 35 1–104. [PubMed] [Google Scholar]
- Prockop, D.J. and Fertala, A. 1998. Inhibition of the self-assembly of collagen I into fibrils with synthetic peptides: Demonstration that assembly is driven by specific binding sites on the monomers. J. Biol. Chem. 273 15598–15604. [DOI] [PubMed] [Google Scholar]
- Prockop, D.J. and Kivirikko, K.I. 1995. Collagens: Molecular biology, diseases, and potentials for therapy. Annu. Rev. Biochem. 64 403–434. [DOI] [PubMed] [Google Scholar]
- Rosenberg, K., Olsson, H., Morgelin, M., and Heinegard, D. 1998. Cartilage oligomeric matrix protein shows high affinity zinc-dependent interaction with triple helical collagen. J. Biol. Chem. 273 20397–20403. [DOI] [PubMed] [Google Scholar]
- Royce, P.M. and Steinmann, B. 2002. Connective tissue and its heritable disorders: Molecular, genetic, and medical aspects, 2nd ed. John Wiley & Sons, New York.
- Ruggiero, F., Exposito, J.Y., Bournat, P., Gruber, V., Perret, S., Comte, J., Olagnier, B., Garrone, R., and Theisen, M. 2000. Triple helix assembly and processing of human collagen produced in transgenic tobacco plants. FEBS Lett. 469 132–136. [DOI] [PubMed] [Google Scholar]
- Sakakibara, S., Inouye, K., Shudo, K., Kishida, Y., Kobayashi, Y., and Prockop, D.J. 1973. Synthesis of (Pro-Hyp-Gly) n of defined molecular weights: Evidence for the stabilization of collagen triple helix by hydroxypyroline. Biochim. Biophys. Acta 303 198–202. [DOI] [PubMed] [Google Scholar]
- Shah, N.K., Ramshaw, J.A., Kirkpatrick, A., Shah, C., and Brodsky, B. 1996. A host-guest set of triple-helical peptides: Stability of Gly-X-Y triplets containing common nonpolar residues. Biochemistry 35 10262–10268. [DOI] [PubMed] [Google Scholar]
- Sieron, A.L., Fertala, A., Ala-Kokko, L., and Prockop, D.J. 1993. Deletion of a large domain in recombinant human procollagen II does not alter the thermal stability of the triple helix. J. Biol. Chem. 268 21232–21237. [PubMed] [Google Scholar]
- Sieron, A.L., Louneva, N., and Fertala, A. 2002. Site-specific interaction of bone morphogenetic protein 2 with procollagen ii. Cytokine 18 214–221. [DOI] [PubMed] [Google Scholar]
- Stone, K.R., Rodkey, W.G., Webber, R.J., McKinney, L., and Steadman, J.R. 1990. Future directions: Collagen-based prostheses for meniscal regeneration. Clin. Orthop. 252 129–135. [PubMed] [Google Scholar]
- ———. 1992. Meniscal regeneration with copolymeric collagen scaffolds: In vitro and in vivo studies evaluated clinically, histologically, and biochemically. Am. J. Sports Med. 20 104–111. [DOI] [PubMed] [Google Scholar]
- Sun, Y.L., Luo, Z.P., Fertala, A., and An, K.N. 2002. Direct quantification of the flexibility of type I collagen monomer. Biochem. Biophys. Res. Commun. 295 382–386. [DOI] [PubMed] [Google Scholar]
- Suzawa, M., Takeuchi, Y., Fukumoto, S., Kato, S., Ueno, N., Miyazono, K., Matsumoto, T., and Fujita, T. 1999. Extracellular matrix-associated bone morphogenetic proteins are essential for differentiation of murine osteoblastic cells in vitro. Endocrinology 140 2125–2133. [DOI] [PubMed] [Google Scholar]
- Svoboda, K.K. 1998. Chondrocyte-matrix attachment complexes mediate survival and differentiation. Microsc. Res. Tech. 43 111–122. [DOI] [PubMed] [Google Scholar]
- Toman, P.D., Pieper, F., Sakai, N., Karatzas, C., Platenburg, E., de Wit, I., Samuel, C., Dekker, A., Daniels, G.A., Berg, R.A., et al. 1999. Production of recombinant human type I procollagen homotrimer in the mammary gland of transgenic mice. Transgenic Res. 8 415–427. [DOI] [PubMed] [Google Scholar]
- Toman, P.D., Chisholm, G., McMullin, H., Giere, L.M., Olsen, D.R., Kovach, R.J., Leigh, S.D., Fong, B.E., Chang, R., Daniels, G.A., et al. 2000. Production of recombinant human type I procollagen trimers using a four-gene expression system in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 275 23303–23309. [DOI] [PubMed] [Google Scholar]
- Tuckwell, D.S., Ayad, S., Grant, M.E., Takigawa, M., and Humphries, M.J. 1994. Conformation dependence of integrin-type II collagen binding: Inability of collagen peptides to support α 2 β 1 binding, and mediation of adhesion to denatured collagen by a novel α 5 β1-fibronectin bridge. J. Cell. Sci. 107 993–1005. [DOI] [PubMed] [Google Scholar]
- Tyler, J.M. and Branton, D. 1980. Rotary shadowing of extended molecules dried from glycerol. J. Ultrastruct. Res. 71 95–102. [DOI] [PubMed] [Google Scholar]
- Weber, I.T., Harrison, R.W., and Iozzo, R.V. 1996. Model structure of decorin and implications for collagen fibrillogenesis. J. Biol. Chem. 271 31767–31770. [DOI] [PubMed] [Google Scholar]
- Westerhausen, A., Kishi, J., and Prockop, D.J. 1990. Mutations that substitute serine for glycine α 1–598 and glycine α 1–631 in type I procollagen: The effects on thermal unfolding of the triple helix are position-specific and demonstrate that the protein unfolds through a series of cooperative blocks. J. Biol. Chem. 265 13995–14000. [PubMed] [Google Scholar]
- Zafarullah, K., Sieron, A.L., Fertala, A., Tromp, G., Kuivaniemi, H., and Prockop, D.J. 1997. A recombinant homotrimer of type I procollagen that lacks the central two D-periods: The thermal stability of the triple helix is decreased by 2°C to 4°C. Matrix Biol. 16 245–253. [DOI] [PubMed] [Google Scholar]