Abstract
Low in vivo solubility of recombinant proteins expressed in Escherichia coli can seriously hinder the purification of structural samples for large-scale proteomic NMR and X-ray crystallography studies. Previous results from our laboratory have shown that up to one half of all bacterial and archaeal proteins are insoluble when overexpressed in E. coli. Although a number of strategies may be used to increase in vivo protein solubility, there are no generally applicable methods, and the expression of each insoluble recombinant protein must be individually optimized. For this reason, we have tested a generic denaturation/refolding protein purification procedure to assess the number of structural samples that could be generated by using this methodology. Our results show that a denaturation/refolding protocol is appropriate for many small proteins (≤18 kD) that are normally soluble in vivo. In addition, refolding the purified proteins by using dialysis against a single buffer allowed us to obtain soluble protein samples of 58% of small proteins that were found in the insoluble fraction in vivo, and 10% of the initial number of proteins provided good heteronuclear single quantum coherence (HSQC) NMR spectra. We conclude that a denaturation/refolding protocol is an efficient way to generate structural samples for high-throughput studies of small proteins.
Keywords: Protein structure/folding, protein solubility, protein purification, NMR spectroscopy, circular dichroism spectroscopy, structural proteomics
Structure determination using NMR spectroscopy and X-ray crystallography requires the generation of large amounts of soluble recombinant protein. Most often, recombinant proteins for structural biology studies are produced in Escherichia coli because the cells grow rapidly and to high density in inexpensive medium, the expression system is well characterized, and a large number of expression vector systems and mutant host strains are available. In addition, under appropriate conditions, the recombinant protein can comprise >50% of the total cellular protein. However, the use of the E. coli expression system is limited because the target proteins often segregate partially or completely into the insoluble fraction of the cell. Recent studies of protein expression on a large number of prokaryotic and eukaryotic proteins indicate that >50% of recombinant proteins may be found in the insoluble fraction of bacterial cell lysates (Christendat et al. 2000a,b; Yee et al. 2002).
Several strategies have been used to increase the probability of producing soluble recombinant proteins in bacterial cells, including co-expressing protein folding modulators, manipulating the temperature of growth and induction, and producing the protein as a fusion with another soluble protein. However, not one of these techniques has been uniformly successful. Recently, the effect of altering fusion protein partners was evaluated systematically. Two groups monitored the solubility of different proteins expressed as fusions with six (Hammarstrom et al. 2002) or eight (Shih et al. 2002) different soluble proteins or affinity tags. By testing a variety of constructs, the percentage of recombinant proteins that could be detected in the soluble fraction of the cell increased from ∼50% to 80% to 85% overall.
An alternative approach to generate recombinant proteins for functional and structural studies is to purify proteins from inclusion bodies. In this approach, the insoluble proteins must first be solubilized with a denaturant and then refolded into a soluble native conformation to be useful for structural studies. Currently, there is no generally applicable method to refold insoluble proteins. One of the emerging trends is to use an array of refolding conditions to screen for a single condition that is compatible with a given protein. However, there is no body of evidence that describes the success rates of these procedures. The challenges associated with developing generic refolding procedures, as well as the perception that refolding methods, are usually unsuccessful and have limited the widespread use of refolding as a first-tier protein purification strategy. If such approaches could be developed, the use of insoluble protein as a starting material has several advantages. First, it would not be necessary to use widespread screens of different protein constructs and expression vectors in an attempt to produce protein that is soluble in vivo. Second, the use of chaperones to assist protein folding in vivo would not be required. Third, insoluble proteins are less susceptible to proteolytic degradation compared with soluble proteins. Finally, it would be possible to express toxic proteins, which inhibit cell growth if they are expressed in the soluble fraction.
The refolding of proteins from the insoluble cellular fraction is commonly accomplished by solubilizing the protein in a chaotropic agent, such as guanidine hydrochloride (GuHCl) or urea, and then removing the denaturant by dialysis or rapid dilution. The efficiency of protein renaturation depends on the competition between correct folding and aggregation, and there is evidence that the presence of contaminants in the refolding buffer can significantly decrease the yield of refolded protein (Maachupalli-Reddy et al. 1997). In a study of lysozyme refolding, aggregation increased when plasmid DNA, lipopolysaccharides, or proteins that aggregate upon refolding were added to the renaturation mixture. Other studies clearly show that the removal of contaminants before preparative refolding increases the yield (Babbitt et al. 1990; Wong et al. 1996; Tran-Moseman et al. 1999).
Although refolding strategies have been successful on a case-by-case basis, the proportion of proteins that can be denatured and refolded is unknown, and the success rate of refolding methods can only be inferred from anecdotal information. We set out to estimate the success rate of refolding methods by performing denaturation/refolding experiments on a large number of proteins chosen from four test organisms under study in our laboratory. Proteins from E. coli, a Gram-negative bacterium (Blattner et al. 1997); Thermotoga maritima, a thermophilic eubacterium with an optimal growth temperature of 80°C (Nelson et al. 1999); Methanobacterium thermoautotrophicum, a lithoautotrophic thermophilic archaeon that grows optimally at 65°C (Smith et al. 1997); and the unicellular eukaryotic budding yeast Saccharomyces cerivisiae (Goffeau et al. 1996) were cloned into overexpression vectors and purified from E. coli. Our aim was to determine if the success rate for refolding methods was high enough to warrant their inclusion in the standard protein purification arsenal, particularly in a structural proteomics setting in which a large number of proteins are expressed and purified at once.
Results
Target selection
Two sets of protein targets were selected for denaturation/refolding analysis. The first set comprised 70 proteins, the "insoluble proteins", which were known to partition to >90% in the insoluble fraction of an E. coli cell lysate. These 70 proteins, which were chosen from three organisms (S. cerivisiae, T. maritima, and M. thermoautotrophicum) did not have predicted transmembrane regions or known structural homologs. The second set comprised 25 proteins from four organisms (E. coli, S. cerivisiae, T. maritima, and M. thermoautotrophicum) that were produced in the soluble fraction of an E. coli cell lysate, and had been previously characterized by using NMR spectroscopy. This set was included in our analysis to compare the suitability of denaturation/refolding strategies for proteins that could otherwise be produced in soluble form using a native protein purification strategy.
All proteins in both sets ranged between 6 and 18 kD, so that the quality of the refolded sample could be analyzed by NMR spectroscopy. Each of the 25 soluble proteins (labeled with 15N) had previously been purified from the soluble fraction and analyzed by two-dimensional 15N-edited HSQC. On the basis of the HSQC spectrum, which provides a "signature" pattern of amide 1H-15N resonances, the proteins were classified as good, promising, dilute, or poor candidates for NMR structure determination. The good spectra showed well-dispersed peaks of approximately equal intensity and of the number expected for the sequence of the protein. Promising spectra showed well-dispersed peaks that were either too few or too many, or of unequal intensities, indicating conformational heterogeneity or the presence of dynamic processes on an intermediate timescale that broaden or obscure NMR signals. The quality of these protein samples may be improved by changing the solution conditions or the size of the protein construct. Dilute protein samples were those that precipitated out of solution and, thus, gave extremely weak or no NMR signal. The classification of poor spectra was used for both unfolded and aggregated protein samples. Unfolded proteins show many sharp intense peaks with chemical shifts consistent with random coil conformation. Aggregated proteins are characterized by too few peaks, which are broadened and clustered in the center of the spectrum. Proteins that form large stable oligomers will generate spectra that will be classified as poor. Most of the 25 proteins generated either good or promising HSQC spectra or produced crystals after native protein purification. The classifications of the NMR spectra of these 25 soluble proteins were as follows: 17 good, 4 promising, 2 dilute, and 2 poor (Table 2).
Table 2.
Comparison of the HSQC results for the protein samples that were expressed in the soluble fraction in vivo
| gi#a | No. of amino acids | % Refolded | Denaturing HSQC | Native HSQC |
| Escherichia coli | ||||
| 1789047 | 61 | 100 | good | good |
| 1790579 | 112 | 80 | poor | promising |
| 1786899 | 148 | 100 | good | promising |
| 1788196 | 167 | 0 | good | |
| 1786341 | 179 | 50 | good | good |
| 1787927 | 82 | 80 | good | good |
| 1786351 | 114 | 50 | good | good |
| 1789689 | 127 | 100 | good | dilute |
| Thermotoga maritima | ||||
| 4980762 | 83 | 100 | good | good |
| 4981224 | 151 | 50 | good | good |
| 4981243 | 152 | 0 | dilute | |
| 4981256 | 149 | 80 | good | poor |
| 4981409 | 158 | 95 | good | good |
| 4981518 | 87 | 80 | good | good |
| 4981520 | 118 | 5 | good | |
| 4981522 | 79 | 100 | good | good |
| 4981537 | 135 | 50 | promising | good |
| 4981552 | 158 | 95 | good | good |
| 4981598 | 101 | 100 | good | promising |
| 4981628 | 124 | 100 | good | good |
| 4981728 | 137 | 100 | good | poor |
| 4982075 | 150 | 100 | good | good |
| Saccharomyces cerivisiae | ||||
| 6322902 | 133 | 80 | good | good |
| Methanobacterium thermoautotrophicum | ||||
| 2621511 | 145 | 90 | good | promising |
| 2621893 | 62 | 100 | good | good |
aNational Center for Biotechnology Information (NCBI) protein identification number (PID).
Purification and refolding of well-expressed insoluble proteins from E. coli
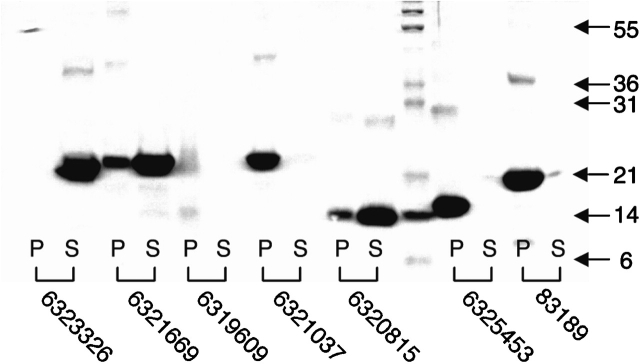
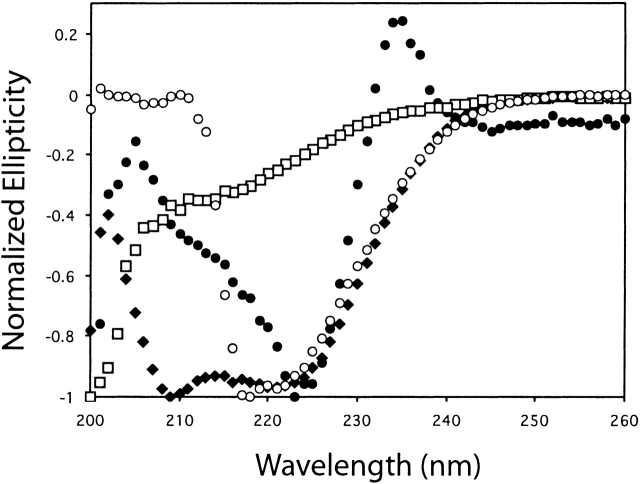
The 70 insoluble proteins were expressed in E. coli, and the whole-cell pellets were solubilized in a solution containing 6.9 M GuHCl. Purification of the His-tagged protein was performed in the same solution by using Ni-affinity chromatography, and the samples were refolded by dialysis against buffer containing 25 mM phosphate (pH 6.8) and 250 mM NaCl. A total of 41 of the 70 (58%) samples remained soluble after dialysis with at least 50% recovery as estimated by SDS-PAGE (Fig. 1 ▶, Table 1). The three-dimensional conformation of each of the 41 soluble protein samples was probed by using far-UV circular dichroism (CD) spectroscopy. In the far-UV region (<250 nm), the spectral characteristics of a protein are primarily determined by the conformation of its polypeptide backbone, especially its secondary structure. Each protein was classified according to its spectrum into one of four groups: α-helical, β-sheet, unusual, or random coil (Table 1). Representative CD spectra are shown in Figure 2 ▶. Of the 41 protein samples that remained soluble after dialysis, 21 showed significant α-helical content, 5 showed β-strand character, 5 were unusual, and 7 showed spectra consistent with random coil. Three samples could not be concentrated to yield a sufficient CD signal, due to precipitation. In summary, by using a simple refolding strategy, 31 of the 70 insoluble proteins could be purified and refolded, resulting in a protein sample that demonstrated some secondary structure and remained in solution at low concentration.
Figure 1.
SDS-polyacrylamide gels showing the fractionation of seven refolded yeast proteins into the pellet (P) or supernatant (S) after centrifugation of the sample at 15,000 rpm for 15 min. Protein marker molecular weights (kD) are indicated at right.
Table 1.
Refolding and biophysical analysis of the 70 protein samples that are insoluble in vivo
| gi#a | No. of Residues | % Refolded | CD result | HSQC |
| Saccharomyces cerivisiae | ||||
| 6322622 | 116 | 100 | α-helical | poor |
| 6325453 | 109 | 100 | β-sheet | poor |
| 6320900 | 107 | 100 | β-sheet | unfolded |
| 6324270 | 110 | 0 | ||
| 6322568 | 123 | 0 | ||
| 6325316 | 128 | 0 | ||
| 6321730 | 73 | 0 | ||
| 6319465 | 145 | 0 | ||
| 6321928 | 148 | 50 | α-helical | poor |
| 6321669 | 137 | 100 | random coil | unfolded |
| 14318509 | 148 | 0 | ||
| 6324454 | 102 | 100 | dilute | poor |
| 6321245 | 103 | 100 | dilute | poor |
| 6319472 | 104 | 80 | α-helical | poor |
| 10383788 | 136 | 100 | random coil | unfolded |
| 6321445 | 84 | 100 | unusual | poor |
| 6322784 | 147 | 0 | ||
| 6320815 | 87 | 100 | α-helical | poor |
| 14318500 | 117 | 0 | ||
| 6321306 | 111 | 100 | α-helical | poor |
| 6319745 | 105 | 80 | random coil | unfolded |
| 6323397 | 110 | 60 | random coil | poor |
| 6321317 | 126 | 0 | ||
| 6322326 | 105 | 100 | no signal | poor |
| 6321850 | 130 | 100 | unusual | poor |
| 6323850 | 74 | 0 | ||
| 6321037 | 122 | 100 | unusual | poor |
| 6323326 | 124 | 100 | random coil | unfolded |
| 6324523 | 106 | 80 | α-helical | dilute |
| 6324941 | 117 | 0 | ||
| 83189 | 146 | 5 | ||
| 6324001 | 146 | 100 | α-helical | poor |
| 6325210 | 99 | 50 | α-helical | unfolded |
| 6322681 | 137 | 80 | uunusual | good |
| 6320760 | 122 | 5 | ||
| 6323190 | 114 | 10 | ||
| 6320458 | 149 | 5 | ||
| 6319609 | 133 | 100 | α-helical | poor |
| 6325115 | 105 | 50 | α-helical | good |
| 10383803 | 146 | 50 | β-sheet | dilute |
| 7839146 | 126 | 0 | ||
| 6322262 | 108 | 5 | ||
| Thermotoga maritima | ||||
| 4981121 | 151 | 100 | α-helical | promising |
| 4981364 | 101 | 0 | ||
| 4981437 | 101 | 100 | unusual | poor |
| 4981475 | 118 | 100 | α-helical | 2.8-Åb |
| 4981497 | 135 | 50 | α-helical | poor |
| 4981533 | 101 | 0 | ||
| 4981568 | 111 | 0 | ||
| 4981614 | 114 | 0 | ||
| 4981889 | 111 | 100 | random coil | unfolded |
| 4981890 | 144 | 0 | ||
| 4982100 | 92 | 80 | β-sheet | good |
| 4982192 | 121 | 0 | ||
| 4982245 | 117 | 0 | ||
| Methanobacterium thermoautotrophicum | ||||
| 2621100 | 114 | 80 | α-helical | dilute |
| 2621252 | 127 | 100 | random coil | unfolded |
| 2621282 | 125 | 5 | ||
| 2621294 | 138 | 30 | ||
| 2621458 | 126 | 50 | α-helical | promising |
| 2621459 | 148 | 0 | ||
| 2622053 | 106 | 50 | α-helical | poor |
| 2622351 | 148 | 100 | β-sheet | poor |
| 2622414 | 63 | 50 | α-helical | poor |
| 2622420 | 136 | 60 | α-helical | poor |
| 2622488 | 132 | 100 | α-helical | good |
| 2622555 | 169 | 90 | α-helical | dilute |
| 2622607 | 148 | 0 | ||
| 2622899 | 135 | 100 | α-helical | good |
| 2621045 | 50 | 100 | random coil | unfolded |
aNational Center for Biotechnology Information (NCBI) protein identification number (PID).
bThis sample produced a poor HSQC, but formed native crystals that diffract to 2.8 Å.
Figure 2.
Representative circular dichroism spectra used for protein secondary structure classification. Proteins were classified as either α-helical (gi 2621893; diamonds), β-sheet (gi 4981224; empty circles), unusual (gi 4981537; solid circles), or unfolded (gi 10383788; squares).
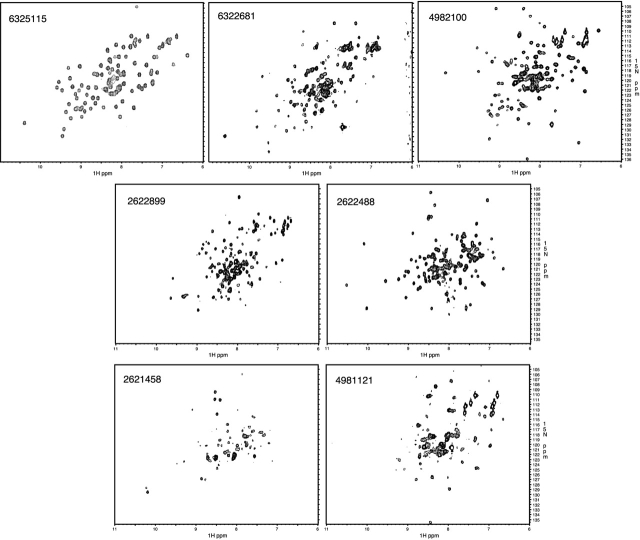
To assess the suitability of proteins for NMR spectroscopy or for crystal trials, it is necessary to concentrate the proteins to >0.3 mM. Of the 31 proteins that could be solubilized and refolded, 24 could be concentrated at >0.3 mM. These samples were uniformly labeled with 15N and concentrated by ultrafiltration for NMR data collection and for crystal trials. Seven of the 24 samples yielded 15N-HSQC spectra that could be considered good or promising (Table 1, Fig. 3 ▶), and one sample formed a crystal that diffracted to 2.8 Å. Therefore, by using a simple refolding strategy, we were able to rapidly generate purified protein for 58% of the 70 small insoluble proteins and to generate structural samples for ∼10%.
Figure 3.
15N-HSQC spectra of the seven proteins expressed in the insoluble fraction of E. coli that provided good or promising structural samples.
Comparison of native and denaturing protein purification protocols
We performed denaturation/refolding studies on 25 well-characterized proteins that are soluble in vivo for two reasons. First, we wanted to compare the success rates of the native and denaturing protein purification methods on a common set of proteins. Second, if some soluble proteins could also be purified by using the denaturation/refolding approach, we wanted to ensure that the renatured proteins adopted the same three-dimensional conformation as those purified from the soluble fraction by using a native purification procedure.
The set of 25 well-expressed soluble proteins was purified by using both methods. Of the 25 samples, 22 (88%) refolded. The 88% refolding rate for this set of proteins is significantly higher than the rate of 55% achieved with the insoluble proteins, indicating that proteins that are soluble in vivo will be better behaved in vitro. Two of the three proteins that were unable to be refolded had been classified as good, with the remaining third as dilute when purified by using the native purification protocol. However, although two good samples were unable to be purified by denaturation/refolding, the renaturation approach actually improved the behavior of four other proteins. Two proteins classified as poor, one classified as dilute, and one classified as promising when purified by using the native protocol were classified as good when purified using the denaturation/refolding protocol. Altogether, there were 21 samples classified as good or promising for each of the denaturing and native protein purification strategies. Figure 4 ▶ compares the HSQC spectra obtained for six samples using both denaturing and native purification protocols. The nearly identical spectra obtained by using the two purification protocols illustrates that the renatured proteins are adopting the same three-dimensional conformations as the proteins purified under native conditions.
Figure 4.
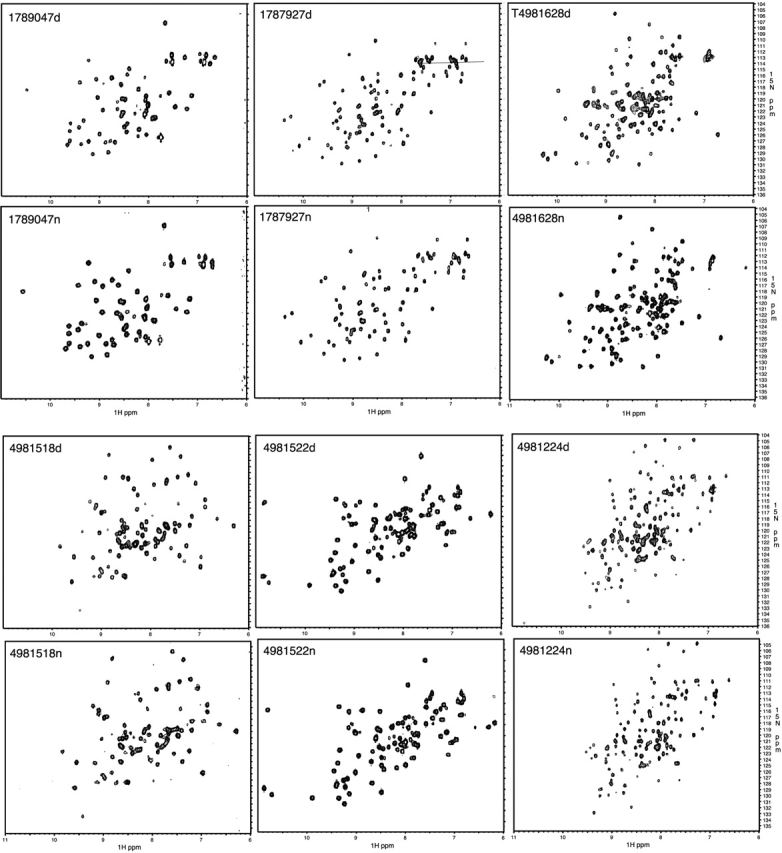
Representative 15N-HSQC spectra of 6 of the 14 proteins that produced good structural samples when prepared by both native and denaturing protocols. Spectra for the proteins purified by both denaturing (d, upper panels) and native (n, lower panels) methods are shown.
Discussion
Up to 50% of cytosolic bacterial and archaeal proteins are sequestered to the insoluble fraction of the cell when overexpressed in E. coli. High-throughput structural proteomic projects will face an increasingly difficult task as the three-dimensional structures of the soluble proteins are solved, and new approaches to deal with nonideal protein samples will need to be developed. The development of a simple and efficient renaturation procedure that can be applied to the insoluble proteins provides the most straightforward strategy to produce large amounts of recombinant protein for structural and functional studies.
We have examined the refolding behavior of a group of 95 small proteins. A simple denaturing/refolding protocol provided a source of soluble folded protein for 58% of proteins that were insoluble in vivo. Slightly >10% of the proteins that were purified from the insoluble fraction generated good samples for NMR spectroscopy. In previous studies, we have shown that 33% of the small proteins expressed in the soluble fraction of E. coli provide good HSQC spectra (Yee et al. 2002). The success rate with which we recovered good samples from the insoluble fraction is significantly lower, although only one set of buffer refolding conditions was examined. A larger percentage of structural samples might be recovered from the insoluble fraction by exploring a wider array of refolding protocols, such as rapid dilution or refolding while immobilized on a column, which may help prevent aggregation of folding intermediates.
It may also be possible to increase the fraction of refolded proteins by exploring a number of different solution conditions, especially for those proteins that contain common metal ions and/or cofactors. There are commercially available kits that can be used for this purpose. Rapid dilution or column-based refolding could be easily automated by using 96-well plates. The extent of refolding in this format could be monitored by using UV spectrophotometry, NMR, or CD spectroscopy. There are several advantages to using CD spectroscopy as a monitor for protein structure, including the speed of data collection, the relatively simple interpretation of the spectra, the ability to collect spectra under a wide variety of conditions, the small amount of sample required, and the ability to recover the sample. The major disadvantage is that the technique gives only a global "average" view of the protein. Although deconvolution of the spectra can approximate the amount of secondary structure present, it cannot be related to the exact structure of the protein. This is not a concern for screening buffer conditions, as the samples could be evaluated according to presence of identifiable secondary structure and absence of light scattering, which indicates a soluble folded protein.
Finally, the addition of a crystal screening step could also potentially increase the number of structures determined for proteins recovered from the insoluble fraction of the cell. We set up crystal trials with 12 samples that gave poor HSQC spectra and found that one of them formed a crystal that diffracted to 2.8 Å. A previous study of 46 small proteins comparing the effectiveness of NMR and crystallography in generating structural samples found that three proteins that exhibited poor HSQCs could be solved to high resolution by using X-ray crystallography (Savchenko et al. 2003). Other unpublished results from our laboratory that support this finding include the high-resolution crystal structures of three proteins from a group of 55 small proteins that exhibited poor HSQCs (A. Yee, D. Christendat, A.M. Edwards, and C.H. Arrowsmith, pers. comm.). These results indicate that we could produce good structural samples for another 5% of proteins that are insoluble in vitro by combining NMR and crystallographic studies.
Denaturing purifications may be desirable not only for totally insoluble proteins but also for proteins that are not expressed to high levels in E. coli and for those that are partially sequestered to inclusion bodies. By denaturing the total cellular protein, it would be possible to recover a larger fraction of the recombinant protein, not just the amount present in the soluble fraction. This could translate to significant cost savings when labeling proteins with 13C and 15N for NMR studies, or with selenomethionine for crystallographic studies. There are also a number of technical advantages to using the denaturing protein purification protocol. For example, many of the strategies used to increase protein solubility in vivo function simply by decreasing the rate of protein expression. These methods include performing the induction step at low temperature, inducing with a nonmetabolizable carbon source such as desoxyglucose, or inducing with limited amounts of inducer added to the culture. If the requirement to express the protein in the soluble fraction of E. coli was obviated, then induction of the recombinant protein could be carried out under conditions that optimize expression levels, but not solubility (i.e., induce for a few hours at 37°C).
It is important to assess the conformational differences between proteins purified by native and denaturing methods, particularly when studying proteins for which an activity assay is unavailable. No studies have systematically compared the three-dimensional structures of proteins purified by using both purification methods. In this study, we examined the 15N-HSQC NMR spectra of 22 proteins that were purified by both native and denaturing protocols, and we found that the spectra look very similar in each case. The minor differences observed between the pairs of proteins are likely due to small amounts of degradation or slight differences in the sample buffer. These results give us confidence that the three-dimensional structures of the proteins that were refolded from denaturant are the same as those purified by using a native purification protocol.
Conclusion
The recovery of recombinant protein from the insoluble fraction of E. coli cell lysates was thought to require technically diverse and often complex refolding procedures. We purified 25 proteins by using native and renaturation approaches. Each of the procedures generated 21 good or promising structural samples. A denaturation/refolding protocol is therefore appropriate for most small proteins that would normally be found in the soluble fraction. By using a simple denaturing protein purification strategy, we were able to obtain soluble protein samples for 58% of small proteins that were insoluble in vivo, with 10% providing good HSQC NMR spectra. We conclude that, particularly for high-throughput studies of small proteins, it is more efficient to perform a denaturing/refolding protocol than a native purification.
Materials and methods
Expression and solubility tests
Target proteins were PCR-amplified from genomic DNA and cloned into the expression vector pET15b (Novagen) as a fusion with an N-terminal 6-His affinity tag and a thrombin cleavage site, or a modified pET15b vector with a TEV protease cleavage site. The fusion proteins were overexpressed in the E. coli strain BL21 STAR (Novagen). Initial trials to determine protein solubility were performed by using a culture volume of 3 mL of Luria broth (LB) in 24-well polypropylene microtiter plates. Three to five colonies were picked from fresh transformations and used to inoculate the LB cultures, which were grown at 37°C to an A600 of ∼0.6. Protein expression was induced by the addition of 175 μg/mL of IPTG, followed by incubation overnight at room temperature. The cells were harvested and lysed in BugBuster (Novagen), to release any soluble proteins. The insoluble proteins and cell debris were then removed by centrifugation, and the soluble and insoluble cell fractions were analyzed by SDS-PAGE followed by Coomassie staining.
For large-scale production of the protein samples, cells were grown at 37°C in M9 minimal medium enriched with 0.7 g/L of 15N-NH4Cl to an A600 of 1.0. Protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (final concentration, 175 μg/mL), followed by incubation for 4 h at 37°C. The cells were harvested by centrifugation and lysed in 6 M GuHCl, 100 mM NaH2PO4, 10 mM Tris-HCl, and 10 mM imidazole (pH 8.0) and were purified in the same buffer via a batch method by using nickel-nitrilotriacetic acid-agarose resin (Qiagen). The pure proteins were eluted with 6 M GuHCl and 0.2 M acetic acid and were refolded by dialysis into 25 mM phosphate (pH 6.8), 250 mM NaCl, and 2 mM DTT. After dialysis, any protein precipitate was collected by centrifugation at 15,000 rpm for 15 min, and was resuspended in 8 M urea, 100 mM NaH2PO4, and 10 mM Tris-HCl (pH 8.0). Equal amounts of the soluble fraction of the sample and the solubilized precipitate were analyzed by SDS-PAGE followed by Coomassie staining. The percentage of soluble protein present in solution versus the insoluble precipitate was then estimated.
Biophysical analysis
CD wavelength scan experiments were performed in an Aviv 62A DS CD spectrometer. The soluble fractions of the refolded protein solutions were analyzed immediately after removal from dialysis. The protein concentrations ranged from 20 to 50 μM. Data was collected at 25°C from 260 to 200 nm (1-nm increments), with a 2-s averaging time.
All 1H-15N HSQC spectra were acquired at 25°C in a Varian INOVA 500- or 600-MHz spectrometer equipped with a pulse-field gradient unit and actively shielded z-gradient triple resonance probes. The total number of t1 increments was 64, with the number of scans per increment ranging from eight to 64, depending on the concentration of sample being examined. The data were processed by using NMRPIPE software package (Delaglio et al. 1995).
Acknowledgments
We would like to thank A. Savchencko, A. Yee, J. Northey, A. Kachatryan, A. Dharamsi, J. Gu, and D. Christendat for technical assistance. This work was supported by a grant from the Ontario Research and Development Challenge Fund. A.M.E. and C.H.A. are Canadian Institutes for Health Research (CIHR) Scientists. K.L.M. is supported by a CIHR Fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0393503.
References
- Babbitt, P.C., West, B.L., Buechter, D.D., Kuntz, I.D., and Kenyon, G.L. 1990. Removal of a proteolytic activity associated with aggregates formed from expression of creatine kinase in Escherichia coli leads to improved recovery of active enzyme. Biotechnology 8 945–949. [DOI] [PubMed] [Google Scholar]
- Blattner, F.R., Plunkett III, G., Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The completed genome sequence of Escherichia coli K-12. Science 277 1453–1474. [DOI] [PubMed] [Google Scholar]
- Christendat, D., Yee, A., Dharamsi, A., Kluger, Y., Gerstein, M., Arrowsmith, C.H., and Edwards, A.M. 2000a. Structural proteomics: Prospects for high throughput sample preparation. Prog. Biophys. Mol. Biol. 73 339–345. [DOI] [PubMed] [Google Scholar]
- Christendat, D., Yee, A., Dharamsi, A., Kluger, Y., Savchenko, A., Cort, J.R., Booth, V., Mackereth, C.D., Saridakis, V., Ekiel, I., et al. 2000b. Structural proteomics of an archaeon. Nat. Struct. Biol. 7 903–909. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 4 603–614. [DOI] [PubMed] [Google Scholar]
- Goffeau, A., Barrell, B.G., Bussey, H., Davis, R.W., Dujon, B., Feldmann, H., Galibert, F., Hoheisel, J.D., Jacq, C., Johnston, M., et al. 1996. Life with 6000 genes. Science 274 563–567. [DOI] [PubMed] [Google Scholar]
- Hammarstrom, M., Hellgren, N., van Den Berg, S., Berglund, H., and Hard, T. 2002. Rapid screening for improved solubility of small human proteins produced as fusion proteins in Escherichia coli. Protein Sci. 11 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maachupalli-Reddy, J., Kelley, B.D., and De Bernardez Clark, E. 1997. Effect of inclusion body contaminants on the oxidative renaturation of hen egg white lysozyme. Biotechnol. Prog. 13 144–150. [DOI] [PubMed] [Google Scholar]
- Nelson, K., Clayton, R.A., Gill, S.R., Gwinn, M.L., Dodson, R.J., Haft, D.H., Hickey, E.K., Peterson, J.D., Nelson, W.C., Ketchum, K.A., et al. 1999. Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399 323–329. [DOI] [PubMed] [Google Scholar]
- Savchenko, A., Yee, A., Khachatryan, A., Skarina, T., Evdokimova, E., Pavlova, M., Semesi, A., Northey, J., Beasley, S., Lan, N., et al. 2003. Strategies for structural proteomics of prokaryotes: Quantifying the advantages of studying orthologous proteins and of using both NMR and X-ray crystallography approaches. Proteins 15 392–399. [DOI] [PubMed] [Google Scholar]
- Shih, Y.P., Kung, W.M., Chen, J.C., Yeh, C.H., Wang, A.H., and Wang, T.F. 2002. High-throughput screening of soluble recombinant proteins. Protein Sci. 11 1714–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.R., Doucette-Stamm, L.A., Deloughery, C., Lee, H., Dubois, J., Aldredge, T., Bashirzadeh, R., Blakely, D., Cook, R., Gilbert, K., et al. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum δH: Functional analysis and comparative genomics. J. Bacteriol. 179 7135–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran-Moseman, A., Schauer, N., and De Bernardez Clark, E. 1999. Renaturation of Escherichia coli–derived recombinant human macrophage colony-stimulating factor. Protein Expr. Purif. 16 181–189. [DOI] [PubMed] [Google Scholar]
- Wong, H.H., O’Neill, B.K., and Middelberg, A.P. 1996. Centrifugal recovery and dissolution of recombinant Gly-IGF-II inclusion-bodies: The impact of feedrate and re-centrifugation on protein yield. Bioseparation 6 185–192. [PubMed] [Google Scholar]
- Yee, A., Chang, X., Pineda-Lucena, A., Wu, B., Semesi, A., Le, B., Ramelot, T., Lee, G.M., Bhattacharyya, S., Gutierrez, P., et al. 2002. An NMR approach to structural proteomics. Proc. Natl. Acad. Sci. 99 1825–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]