Summary
The CYP51 family is an intriguing subject for fundamental P450 structure/function studies and is also an important clinical drug target. This review updates information on the variety of the CYP51 family members, including their physiological roles, natural substrates and substrate preferences, and catalytic properties in vitro. We present experimental support for the notion that specific conserved regions in the P450 sequences represent a CYP51 signature. Two possible roles of CYP51 in P450 evolution are discussed and the major approaches for CYP51 inhibition are summarized.
Keywords: sterol 14α-demethylase (CYP51), sterol biosynthesis, substrate preferences, catalytic activity, inhibition
Family overview
Sterol 14α-demethylation as a general part of sterol biosynthetic pathways in eukaryotes [1] has been known and studied for more than 30 years [2-7]. The enzyme catalyzing this reaction was first purified from yeast in 1984 (Sacharomyces cerevisiea [8]), and following determination of its primary structure [9] the cytochrome P450 sterol 14α-demethylases were placed into the CYP51 family, a number reserved for fungal sequences [10]. In 1986 the orthologous mammalian P450 was purified from rat liver microsomes [11], in 1996 the first sterol 14α-demethylase was found in plants (Sorghum bicolor [12]), and in 2000 the orthologous nature of a CYP51-like gene [13] from Mycobacterium tuberculosis to eukaryotic CYP51s was confirmed [14].
Currently the CYP51 family joins proteins found in 82 organisms from all biological kingdoms. In addition, several plants and fungi contain multiple CYP51 genes (e.g. rice (10), black oats (2), tobacco (2), Arabidopsis thaliana (2), Fuzarium graminearum (3) or Aspergillus species: A. fumigatus (2), A.nidulans (2), A. orizae (3)). As a result, the number of known CYP51 sequences exceeds 100. The reasons for the existence of homologous CYP51 genes in the same species or their precise functions remain unknown, though it was reported that only one of the two CYP51 genes from A. thaliana (CYP51A2) is functional, while the other is an expressed pseudogene [15]. Mammalian genomes contain only one CYP51 gene yet sometimes nonfunctional processed CYP51 pseudogenes (e.g. 3 in human and 1 in rat [16, http://drnelson.utmem.edu/biblioC.html]) have been found.
The average amino acid sequence identity in the CYP51 family is about 30%, varying from relatively high between the proteins from evolutionary closely related species (e.g. 95% in mammals) to lower values within the highly diverse kingdoms of lower eukaryote (41%) and decreasing to 23-34% across the biological kingdoms (Figure 1). Interestingly, sequence identity between multiple genes in the same organism can also be rather moderate. Thus, the average identity for the ten putative CYP51s from rice is only 45% ranging form 78 to 38%; the sequences from three Aspergillus species are about 60% identical to each other, and the identity between the two proteins from Arabidipsis thaliana is 72%.
Figure 1.
Amino acid sequence identity in the CYP51 family. Alignment of 71 sequences was performed using ClustalW 1.82, percentage of amino acid identity was calculated in GeneDoc (2.6). *(number of sequences in the alignment/ percentage of identity). Complete alignment of CYP51 family can be found at the web site https://medschool.mc.vanderbilt.edu/watermanlab/index.php
CYP51 function and physiological role in vivo
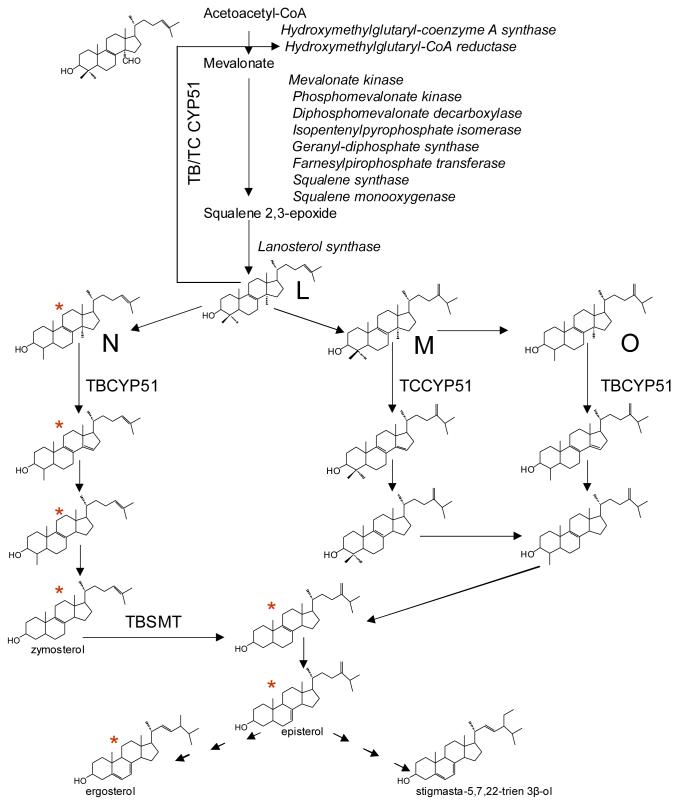
Though the cut off for the level of identity within a P450 family is accepted to be > 40%, sterol 14α-demethylases from all phyla are assigned to the same family because they appear to retain strict catalytic function. All forms studied catalyze one regio- and stereospecific reaction of oxidative removal of the 14α-methyl group from sterol precursors formed in the sterol biosynthetic pathway downstream from cyclization of squalene 2,3-epoxide (Figure 2).
Figure 2.
CYP51 reaction in sterol biosynthesis. D, 24, 25-dihydrolanosterol; L, lanosterol; M, 24-methylenedihydrolanosterol; N, norlanosterol; O, obtusifoliol. *We have found recently that N is likely to be the preferred CYP51 substrate in Trypanosomatidae with the exception of T. cruzi [36]. R1, alkyl or alkylene group; R2, C6 or C7 double bond. The dashed line shows a major site of regulation of sterol biosynthesis.
The CYP51 reaction occurs in three steps, each requiring one molecule of oxygen and two NADPH-derived reducing equivalents [17]. During the first two cycles, which are typical P450-monooxygenations, the 14α-methyl group is converted successively into the 14α-carboxyalcohol and then into the 14α-carboxyaldehyde. In the final step, the 14α-aldehyde group is released as formic acid with concomitant introduction of the Δ14, 15 double bond into the sterol core. The catalysis of the C-C bond cleavage is more complicated and two possible mechanisms (radical reaction or ionic (Beayer-Villiger) process) have been proposed [18, 19].
The 14α-demethylated products are intermediates in the pathways leading to formation of cholesterol in animals, ergosterol in fungi and a variety of 24-alkylated and olefinated sterols in plants, algae and protozoa [1, 20]. De novo sterol biosynthesis takes place in the majority of eukaryotic cells (except for insects and nematodes, which consume sterols from the diet). Generally, the pathway occurs in the endoplasmic reticulum, but there is evidence that Kinetoplastidae might also synthesize sterols in mitochondria [21]. Bulk sterols, such as cholesterol, ergosterol and sitosterol (plants) are ubiquitous components of the plasma membranes, where they play an important structural role to regulate membrane fluidity and permeability and indirectly modulate the activity and distribution of integral membrane proteins, including enzymes, ion channels and components of signal transduction pathways [1, 22]. The sterols serve as precursors for bioactive molecules such as mammalian steroid hormones, plant brassinosteroid hormones and insect ecdysteroid to control developmental processes. Amongst bacteria sterol biosynthesis is known in Methylococcus capsulatus [23], but the function of the CYP51 genes in Mycobacteria and Nocardia remains unclear.
In addition to being an invariant part of an essential biosynthetic pathway, the CYP51 reaction has other role(s) (Figure 2). Thus, in mammals a reaction intermediate (14α-carboxyaldexyde derivative of lanosterol demethylation) is known to downregulate cholesterol production acting as suppressor of HMG-CoA reductase translation. [24-26]. Such a regulatory role of CYP51 might be of special importance in Trypanosomatidae, where the CYP51 reaction aldehyde intermediate of lanosterol can be used by the parasite to switch the pathway from endogenous sterol production to the energetically more favorable consumption of exogenous sterol precursors available from mammalian (host) blood [27].
Finally, in mammals, being expressed ubiquitously in all tissues, CYP51 is overexpressed in germ cells. Interestingly, in sperm in addition to endoplasmic reticulum active sterol 14α-demethylase and its electron donor cytochrome P450 reductase have been shown to be localized in Golgi apparatus and in the outer membrane of Golgi-derived vehicles called acrosomes [28]. In testis and follicular fluid the products of lanosterol 14α-demethylation have been established to act as meiosis inducing substances ((meiosis activating sterols, MAS, see Figure 2) [29, 30]. Detailed analysis of the role of mammalian CYP51 in spermatogenesis can be found in a recent review [31].
CYP51 in vitro
To date (May, 2006), 12 CYP51 have been purified, either from natural sources or by cloning and overexpression in heterologous systems, and characterized. Amongst them 9 are microsomal, eukaryotic enzymes (including 3 mammalian: rat [7], pig [32], and human [33]; 1 plant: S. bicolor [12], 3 fungal: 2 from yeasts (S. cerevisiae [8], Candida albicans [34]), and one from filamentous fungus Ustilago maydis [35] and 2 trypanosomal (Trypanosoma brucei [27] and T. cruzi [36]) while the other 3 are water soluble bacterial orthologs (from M. tuberculosis [14], M. smegmatis [37], and a fusion CYP51-ferredoxin protein from Methylococcus capsulatus [38]).
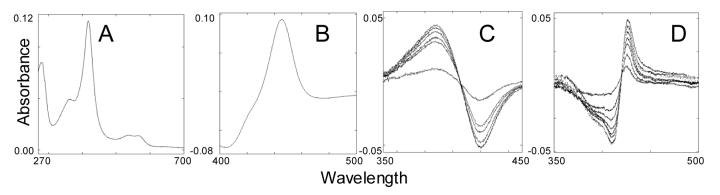
All the P450s are purified in the low-spin form with Soret maximum at 417 nm (Figure 3). In the reduced CO-complexes bacterial enzymes show maximum at 450 nm, while in the spectra of the eukaryotic orthologs the maximum is shifted to 446-447 nm, which might be connected with the formation of aggregates. Similar to other microsomal P450s, the eukaryotic forms of CYP51 require cytochrome P450 reductase as an electron donor partner; bacterial CYP51 from M. tuberculosis, M. smegmatis and M. capsulatus can be enzymatically reduced by flavodoxin/flavodoxin reductase and ferredoxin/ferredoxin reductase systems [14, 37].
Figure 3.
Typical spectra of eukaryotic CYP51 (sterol 14α-demethylase from T. cruzi). A, absolute oxidized; B, reduced CO difference; C, type I spectral response to substrate 24-methylenedihydrolanosterol; D, type II spectral response to azole inhibitor ketoconazole.
Contrary to drug metabolizing P450s, which can accommodate a vast variety of structures, CYP51 has very narrow substrate specificity. The set of substrates is limited to the five naturally occurring 14α-methylsterols (lanosterol, 24, 25-dihydrolanosterol, 24-methylenedihydrolanosterol, obtusifoliol and 4β-desmethyllanosterol (norlanosterol [36]) (see Figure 2)). The reconstituted activity strongly depends on the reaction conditions, the highest reported turnover numbers being summarized in Table 1. Mammalian (human) and yeast (C. albicans) orthologs, which act in vivo directly downstream of formation of the first cyclized compound (lanosterol (Figure 2), can 14α-demethylate all five substrates in vitro with similar rates and under the reconstitution conditions are the fastest sterol 14α-demethylases. The activity of the single characterized CYP51 from filamentous fungi (U. maydis) was reconstituted with its physiological substrate (24, 25-methylenedihydrolanosterol) only [35]. CYP51 from the flowering plant S. bicolor is highly specific toward the C4-monomethylated sterols, the turnover number with the physiological substrate obtusifoliol being the highest (36), which is in good agreement with the results of the earlier work on characterization of this CYP51, yet in this earlier work norlanosterol was not tested [12]. Strict preference toward obtusifoliol was also reported for membrane fractions containing CYP51 orthologs from maize [39] and wheat [40] and thus it was considered a feature general for all plant orthologs until recently, when the ability of CYP51 from S. chacoense (chaco potato) to metabolize lanosterol (though at a rate considerably lower than that with obtusifoliol) was reported [41]. Strong specificity toward the C4-monomethylated substrates of CYP51 from the protozoan parasite T. brucei is similar to the CYP51 from S. bicolor while the ortholog from T. cruzi clearly prefers C4-dimethylated 24, 25-methylenedihydrolanosterol [36]. It is not excluded that there is a correlation between the strict substrate specificity of a CYP51 and the number of steps separating the first stylized sterol of the pathway from the CYP51 site in the biosynthetic scheme (Figure 2), the possible reason being to prevent the pathway from multiple bifurcations. M. tuberculosis CYP51 is able to demethylate all five sterols (turnover of obtusifoliol being the highest); the substrate preferences of CYP51 from M. smegmatis and M. capsulatus have not been reported. Overall, the bacterial orthologs (at least with heterologous electron donor partners) operate one to two orders of magnitude slower than the characterized eukaryotic CYP51s but a larger number of CYP51 orthologs needs to be studied at comparable experimental conditions and preferably with their endogenous reductase systems to evaluate their relative turnover numbers more precisely.
Table 1.
Reconstituted CYP51 activity
| Species/ phylum | Turnover* (nmol substrate/nmol P450/min) |
Reference | ||||
|---|---|---|---|---|---|---|
| 24,25-Dihydrolanosterol | Lanosterol | 24-Methylenedihydrolanosterol (eburecol) | Norlanosterol | Obtusifoliol | ||
| Eukaryote | ||||||
| Human/Mammal | 30 | 29 | 27 | 23 | 29 | [36] |
| Rat/ Mammal | 20 | 14 | ns | ns | ns | [46] |
| C. albicans/Yeast | 25 | 24 | 29 | 17 | 27 | [36] |
| S. cerevisiae/ Yeast | 3.9 | 6.2 | 6.8 | ns | ns | [80] |
| U. maydis/ Filamentous Fungi | ns | ns | 7.7 | ns | ns | [35] |
| T. cruzi/ Kinetoplastida | 1.6 | 0.6 | 2.4 | 0.03 | 0.06 | [36] |
| T. brucei/ Kinetoplastida | <0.01 | <0.01 | <0.01 | 3.0 | 6.2 | [36] |
| S. bicolor/ Flowering plant | <0.01 | <0.01 | <0.01 | 0.9 | 5.1 | [36] |
| Prokaryote | ||||||
| M. tuberculosis/ Bacteria | 0.18 | 0.15 | 0.12 | 0.18 | 0.45 | [36] |
| M. smegmetis/ Bacteria | ns | 0.7 | ns | ns | ns | [37] |
| M. capsulatus/ Bacteria | ns | 0.26 | ns | ns | ns | [38] |
The highest published values are presented. The reasons for the differences in the published calculated rates of the CYP51 turnovers are likely in part connected with differences in the final P450 concentrations in the reconstituted reaction mixture. ns-not studied.
CYP51 signature, structural basis of conservation in the CYP51 family
The complex mechanism of the CYP51 reaction suggests that critical geometry of the substrate is necessary to ensure its proper orientation during the three steps of catalysis in the enzyme active center. Therefore, CYP51s should have a common configuration of their substrate binding pockets, which in turn, regardless of low sequence identity in the family, requires certain amino acid conservation.
Yet, with the increasing number of known CYP51 sequences, the number of the invariant amino acid residues in the family within the last 3 years has dropped from 40 [42] to 29. Seven of them are glycines aligned with those in the M. tuberculosis CYP51 structure, which separate secondary structural elements. Based on the results of site-directed mutagenesis of these glycines in M. tuberculosis CYP51, we have hypothesized that they must be essential in the CYP51 structure to ensure functionally important conformational flexibility of the enzyme [43].
Most of the other conserved CYP51 residues are clustered into six regions, representing substrate recognition sites (SRS) 1-5 and that surrounding the heme-coordinating Cys. While conservation in the heme binding region is general amongst P450 families, high conservation within the SRSs (the sequences of highest variability in the drug metabolizing CYP2 family [44]) is a specific feature of CYP51 [45]. We believe that two of these regions, the most conserved and therefore the most thoroughly studied (SRS1 and SRS4) can be considered as a CYP51 signature (Figures 4, 5).
Figure 4.

Conservation in the CYP51 SRS1 and 4. A CYP51 signature is written below the alignment. Phyla-specific residues are shadowed in gray.
Figure 5.
Location of the CYP51 signature in the P450 structure. A, upper view, B, distal view. The conserved regions within the B'helix/BC loop and in the I-helix are colored in red. Heme is shown in green.
SRS1 (B' helix/B'/C loop) forms the upper surface of a P450 substrate binding cavity (Figure 5). Substitution of the conserved Y, F and G in the B' helix results in complete loss of sterol 14α-demethylase activity of M. tuberculosis and human CYP51 [42] and was shown to strongly affect activity of the rat ortholog (the corresponding Y131 and F139 were studied [46]). Another example provides mutation of conserved P81 in M. tuberculosis CYP51 (there is always P in this position with the exception of one sequence from filamentous fungi Cunninghamella elegans (Y)). In this case, the effect strongly depends on the substitution (Figure 6): thus alanine in this position decreases P450 expression but hardly influences the activity, but short polar G which does not affect P450 expression yet causes a 10-fold decrease in the activity and 5-fold decrease in the amplitude of type I spectral response. On the contrary, the long hydrophobic side chain of L increases two-times the activity and substrate binding but lowers P450 expression 5-fold, most likely as a result of strong destabilization. An example of the important role of phyla specific residues in the CYP51 B' helix is seen from our studies of CYP51 from T. cruzi. Expressing substrate preferences toward the C4-dimethylated 24, 25-methylenedihydrolanosterol, the enzyme has animal/fungi like I in the position where like all plant orthologs, CYP51 from T. brucei and CYP51s found in five other Trypanosomatidae contain F. Substitution of this I to F (I105F) in T. cruzi alters substrate preferences of the mutant more than two orders of magnitude increasing its ability to bind and metabolize C4-monomethyl sterols [36].
Figure 6.

Mutagenesis of conserved P81 in the B' helix of M. tuberculosis CYP51.
In the B'C loop, mutation of V (87/143 (M. tuberculosis/human)) to A prevents expression of M. tuberculosis CYP51 in the P450 form and strongly (more than 10 fold) reduces P450 expression of the human enzyme [42]. The effect of the substitution of two other residues F89 [47] and D90 [42] in this loop is probably phylum-specific. F corresponding to F89 in M. tuberculosis CYP51 is present in this position in all bacterial and fungal CYP51 but in the other species this position is occupied by Y. Mutation of F89 inactivates M. tuberculosis CYP51 but substitution of the corresponding Y in human or C. albicans orthologs has no effect [47] on their activities and interaction with the substrate. Similarly, mutation D90A inactivates M. tuberculosis CYP51 but the D132A mutant of the human ortholog partially retains its sterol 14α-demethylase activity. Later it was found that this D, which is conserved across the rest of the CYP51 family, is replaced with A in all trypanosomal sequences, suggesting that the mutation studied in M. tuberculosis and human CYP51 has occurred in nature. It is possible that in eukaryotic and prokaryotic CYP51, involvement of the BC loop in catalysis is different, especially taking into account that mutagenesis of a number of other, nonconserved residues in the B'C loop of M. tuberculosis CYP51 also strongly affect its catalytic activity (Lepesheva, G.I., Seliskar, M., Waterman, M.R., unpublished data). M. tuberculosis CYP51 is the only known P450 structure where the B'C loop is in an open conformation. Probably, this open conformation represents the channel [48] through which the sterol enter the active site. Thus in the traditional view of P450 structure, M. tuberculosis CYP51 is the only example to date where the substrate enters from the top rather than from the front. It is possible that in the eukaryotic CYP51 the B'C loop is positioned differently so that the role of the specific set of amino acid residues discussed here becomes functionally less crucial because the substrate enters the active site from the front rather than the top. This is only speculation until the structure of a eukaryotic form of CYP51 is determined.
SRS4 is located in the C-terminal part of the P450 I-helix (Figure 5) forming the right wall of the distal surface of the substrate binding cavity. This region contains the unique triplet of the CYP51 family –HT/sS-. The role of each of these three residues has been thoroughly studied on rat CYP51 [46, 49]. The first and the last residues in the triplet (Figure 4) are conserved throughout the CYP51 family, while the middle T, corresponding to the “conserved threonine” [50] in most P450s, is replaced with S in several sequences from filamentous fungi. Mutagenesis in rat CYP51 shows that substitution of T315 to S has little effect on the enzyme, but the other substitutions (V, K or N) cause loss of detectable activity. On the contrary, S316 which is conserved throughout the family can be easily substituted with nonpolar A in rat CYP51 but longer, especially hydrophobic side chains placed here have a strongly negative influence on the activity. The position H in the triplet, invariant amongst the CYP51 family, in most other P450s is occupied by an acidic amino acid. In CYP101 (P450 cam) the corresponding D (D251) has been found to play a key role in proton transfer [51]. Mutation H314D in rat CYP51, however, causes more than a 6-fold decrease in the activity. In our studies of mutagenesis of M. tuberculosis CYP51 the corresponding H259D mutant retained only 5% of the wild type protein activity and the H259N mutant was inactive. Mutation of the conserved G257 (preceding the triplet) which disrupts the I-helix into two helical segments also inactivated M. tuberculosis CYP51 as a sterol 14α-demethylase (Lepesheva, G.I., Waterman, M.R, unpublished data).
Thus, the two motifs, -YxxF/L(i)xxPxFGxxVxF/YD/a- in SRS1 and –GQ/hHT/sS- in SRS4, can be used to identify a P450 sequence as a CYP51 family member (CYP51 signature) and the phyla-specific residue F/L(i), in the B' helix (F in plant and L in animal/fungal CYP51) allows one to predict substrate preferences (mono or dimethyl at C4) of newly identified sterol 14α-demethylases.
CYP51 and P450 evolution
Sterol biosynthesis is known to be a very ancient metabolic pathway, arising during the latest stages of microbial evolution after the introduction of molecular oxygen into the atmosphere [1]. Once aerobic conditions developed, formation of squalene 2, 3-epoxide became possible. Contrary to the anaerobic cyclization of squalene in the bacterial hopanoid pathway, squalene 2, 3-epoxide is cyclized into the steroid precursors (lanosterol or cycloartenol (Figure 2)). Further conversions of the precursors, including oxidative removal of three methyl groups from the core, produce sterols known today. Insertion of sterols into cellular membranes may have paved the way toward the development of eukaryotic organisms [52]. The prokaryotic origin of the sterol biosynthetic pathway implies a common prokaryotic ancestor for all currently existing sterol 14α-demethylases, the only invariant P450 in every known sterol biosynthetic pathway, and in turn provides a very logical explanation for the presence of CYP51 orthologs in all evolutionarily younger biological kingdoms.
Supported by multiple sequence alignment analysis, the idea of the prokaryotic origin of sterol 14α-demethylase might indicate that it is evolutionarily the oldest P450 recognizable today [45, 53-55]. Even though an option of horizontal CYP51 gene transfer from plants to currently existing bacteria [31, 56] (convolutionary process with loss of function) is not excluded, the CYP51 family remains an intriguing subject to study from two key evolutionary aspects: 1) evolution into different phyla with preserved metabolic role yet sometimes slightly different substrate preferences and 2) possible diversification into other P450 species with altered catalytic function.
Upon evolution within the family, alterations in the CYP51 sequences had to occur generally by mutagenesis in the regions which do not play crucial functional roles. As a result of such processes amino acid sequences of CYP51 from phylogenetically distant species became very diverse (e.g. CYP51 orthologs from B. fuckeliana (filamentous fungi) and M. truncatula (plant) or T. cruzi (protozoa) and onion (plant) have only 21 % identity and 42% similarity), yet the set of functionally essential amino acids (part of which are described above) must remain conserved.
Interesting from the evolutionarily viewpoint is identification of phyla-specific residues, switching CYP51 preferences from C4-double to C4-monomethylated substrates [36. Thus, in ancient Euglena CYP51 preferences toward C4-monomethylated obtusifoliol (F as the phyla specific residue in the B' helix (Figure 4)) could have developed with cycloartenol formation. However, the origin for the same preferences in Trypanosomatidae is likely different. Trypanosomatidae evolved independently from Euglena [57. Besides, the first cyclized product of their sterol biosynthesis is lanosterol and not cycloartenol. It suggests that either CYP51 from early photosynthetic organisms and Kinetoplastida share a common, more ancient (most likely prokaryotic), ancestor or that mutation in this key position (L to F) occurred repetitively. Of course the latter option is possible (e.g. in CYP51 from T. cruzi F in this position is substituted to I, as in CYP51 from M. capsulatus, both enzymes favoring C4-dimethylated sterols as substrates). On the other hand, it is not excluded that at first sterol 14α-demethylase could have contained F in this position but (similar to M. tuberculosis CYP51 or I105F mutant of CYP51 from T. cruzi or CYP51 from S. chacoense) was able to metabolize both C4 mono- and C4 dimethylated substrates. If so, then F in this position might be evolutionarily older than L (or I).
Diversification of CYP51 into other P450 species could result from gene duplications followed by mutations in the substrate binding cavity and other areas which influence substrate binding. Potentially, three possibilities exist: if a mutation did not prevent binding of the sterol core, the mutant P450s could have developed the ability to metabolize other sterol molecules and to be included into evolutionarily newer metabolic pathways downstream of the CYP51 reaction Such divergence could have occurred after the separation of the ancestors of animals from the fungal/plant branches [58] (sterol 22 desaturase (CYP61 in fungi [58] and CYP710 in plants [59]. Mammalian steroidogenic CYPs (e.g. CYPs 7, 8, 11, 17, 19, 21, 27, 39, 46) might be another example of this type of diversification. Sterol metabolizing P450s often preserve narrow substrate specificity and among them there are other examples which catalyze multiple step reactions, e.g. CYP11A1, CYP17, CYP19. The last two were found to have essentially the same reaction mechanism as CYP51 [19], sequence identity/similarity to human CYP51 being 12/29, 16/37 and 14/33 %, respectively. If mutations caused loss of ability to bind sterols, the resultant protein either was not required, which could lead to formation of pseudogenes with future elimination or it could have acquired the ability to metabolize completely different structures [60] e.g. exogenous) and thus remain important for the organism
In general, detailed investigation of CYP51 evolution could be of great help for better understanding evolutionary processes in the P450 superfamily, which would possibly provide insight into creation of artificial P450 enzymes with desired properties and to uncover general directions of physiology accommodating the changing environment.
Inhibition of CYP51
Being an interesting subject for fundamental P450 studies, CYP51 is also of great practical importance as a drug target. Inhibition of sterol 14α-demethylase activity blocks sterol biosynthesis, which is lethal in unicellular organisms, influences growth and developmental processes in plants, and lowers endogenous cholesterol production in animals. Though CYP51 inhibitors are being thoroughly studied as potential herbicides and cholesterol lowering drugs, they are most widely used as fungicides to treat human mycoses (clinical antifungals) and prevent food infections (agricultural antifungals). The demands for CYP51 inhibitors are increasing continuously because of drug resistance, worldwide increase in the incidence of opportunistic fungal infections as a consequence of the rising number of immunocompromised hosts (HIV-infections, cancer chemotherapy, organ and bone marrow transplantation) and patients with primary infections such as tuberculosis, etc. Quite a new aspect for CYP51 inhibitors is opened by the findings that they might be helpful in treatment of deadly diseases caused by trypanosomal parasites.
Azoles (imidazoles and triazoles) are the most broadly known CYP51 inhibitors. They coordinate to the heme iron through a basic nitrogen and inhibit activity preventing substrate binding and metabolism. Azoles play a pivotal role in the treatment of systemic and dermal mycoses [61] and have been found effective to cure murine models of leishmaniases and Chagas disease [62-64]. Azoles are less toxic than other antifungal and anti-trypanosomal drugs, inexpensive and broadly available, yet have several disadvantages. Their long term usage can inhibit other P450 enzymes [65] and leads to resistance allowing the drugs tolerance in the pathogen. Amongst possible reasons for resistance are 1) mutations in fungal CYP51 (dozens of mutations which might be connected with azole resistance have been reported [e.g. 65-69]) 2) increase in the CYP51 gene expression and 3) faster azole efflux or decreased permeability [70-72]. To overcame this problem (or at least to make it less severe), in vitro testing of each particular inhibitor with the target fungal enzyme might be useful, because it is well known that affinities of different CYP51 orthologs to the same azole drug may vary significantly (up to 3-4 orders of magnitude [61, 73]) . Also, it is well known that the lower drug susceptibility a fungal strain has, the more frequently and rapidly resistance develops [70, 74]. Besides, selecting the most powerful CYP51 inhibitors would help to shorten treatment time and lower the necessary doses.
An alternative approach to avoid azole resistance might be inhibition of CYP51 with substrate analogs. Contrary to azole derivatives sterols would have longer life-time in water solution and better membrane permeability. Attempts to use CYP51 substrate analogs to block cholesterol biosynthesis in humans have described that 7-oxo, 15-keto, 15-oxime, 15-hydroxy, 26-oxo derivatives of lanosterol are effective [24, 75-77]; derivatives of the 14α-carboaldehyde intermediate of lanosterol are known as hypocholesterolemic agents having dual effect in vivo (competitive CYP51 inhibitors and suppressors of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase) [76-78]. Finally, 14α-aminoderivatives of lanosterol were found effective to inhibit C. albicans and T. cruzi CYP51 orthologs [36, 79]. Another option might be random search for other types of highly selective CYP51 inhibitors through high throughput screening of chemical libraries and selecting the compounds of interest based on their binding to the P450 and subsequently inhibition of its reaction.
CONCLUSION
Sterol 14α-demethylase is a very important enzyme throughout biology because of its essential role in sterol biosynthesis. It is a primary drug target for a number of microbial infections in both animals and plants. CYP51 targeted drugs may also prove useful in treatment of other human conditions such as elevated cholesterol levels and as a herbicide in raising crops around the world. Since CYP51 is the most widely distributed member of the cytochrome P450 superfamily it also plays significant roles in our understanding of these monooxygenases. We have presented here a summary of the biochemical features of CYP51. We expect that this information will continue to expand in the coming years, particularly more detailed analysis of CYP51 structure/function relationships, a better understanding of why some plants and microbes contain what appear to be multiple copies of functional CYP51 genes and development of species specific inhibitors. We encourage readers to keep their eyes on developments in understanding CYP51 because they will continue to provide important insights into the P450 superfamily.
Acknowledgement
The authors acknowledge financial support from the National Institutes of Health (GM067871 and ES00267-32 to M.R.W) and from the American Heart Association (0535121N to G.I.L). They also appreciate the collaboration of many colleagues over the past 13 years of study of CYP51.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Nes WR, McKean MR. Biochemistry of Steroids and Other Iisopentenoids. University Park Press; Baltimore: 1977. [Google Scholar]
- 2.Mitropoulos KA, Gibbons GF, Reeves BE. Lanosterol 14alpha-demethylase. Similarity of the enzyme system from yeast and rat liver. Steroids. 1976;6:821–829. doi: 10.1016/0039-128x(76)90141-0. [DOI] [PubMed] [Google Scholar]
- 3.Akhtar M, Alexander K, Boar RB, McGhie JF, Barton DH DH. Chemical and enzymic studies on the characterization of intermediates during the removal of the 14alpha-methyl group in cholesterol biosynthesis. The use of 32-functionalized lanostane derivatives. Biochem. J. 1978;169:449–463. doi: 10.1042/bj1690449b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiecchi A, Galli-Kienle M, Scala A, Galli G, Grossi Paoletti E, Cattabeni F, Paoletti R. Hydrogen exchange and double bond formation in cholesterol biosynthesis. Proc. R. Soc. Lond. B. Biol. Sci. 1972;180:147–165. doi: 10.1098/rspb.1972.0011. [DOI] [PubMed] [Google Scholar]
- 5.Galli-Kienle M, Anastasia M, Cighetti G, Galli G, Fiecchi A. Studies on the 14 alpha-demethylation mechanism in cholesterol biosynthesis. Eur. J. Biochem. 1980;110:93–105. doi: 10.1111/j.1432-1033.1980.tb04844.x. [DOI] [PubMed] [Google Scholar]
- 6.Trzaskos JM, Bowen WD, Shafiee A, Fischer RT, Gaylor JL. Cytochrome P-450-dependent oxidation of lanosterol in cholesterol biosynthesis. Microsomal electron transport and C-32 demethylation. J. Biol. Chem. 1984;259:13402–13412. [PubMed] [Google Scholar]
- 7.Trzaskos JM, Fischer RT, Favata m.F. Mechanistic studies of lanosterol C-32 demethylation. Conditions which promote oxysterol intermediate accumulation during the demethylation process. J. Biol. Chem. 1986;261:16937–16942. [PubMed] [Google Scholar]
- 8.Yoshida Y, Aoyama Y. Yeast cytochrome P-450 catalyzing lanosterol 14 alpha-demethylation. I. Purification and spectral properties. J. Biol. Chem. 1984;259:1655–1660. [PubMed] [Google Scholar]
- 9.Kalb VF, Loper JC, Dey CR. Isolation of a cytochrome P-450 structural gene from Saccharomyces cerevisiae. Gene. 1986;45:237–245. doi: 10.1016/0378-1119(86)90021-1. [DOI] [PubMed] [Google Scholar]
- 10.Nebert DW, Nelson DR, Coon MJ, Estabrook RW, Feyereisen R, Fujii-Kuriyama Y, Gonzalez FJ, Guengerich FP, Gunsalus IC, Johnson EF, et al. The P450 superfamily: update on new sequences, gene mapping, and recommended nomenclature. DNA Cell. Biol. 1991;10:1–14. doi: 10.1089/dna.1991.10.1. [DOI] [PubMed] [Google Scholar]
- 11.Trzaskos J, Kawata S, Gaylor JL. Microsomal enzymes of cholesterol biosynthesis. Purification of lanosterol 14 alpha-methyl demethylase cytochrome P-450 from hepatic microsomes. J. Biol. Chem. 1986;261:14651–14657. [PubMed] [Google Scholar]
- 12.Kahn RA, Bak S, Olsen CE, Svendsen I, Moller BL. Isolation and reconstitution of the heme-thiolate protein obtusifoliol 14alpha-demethylase from Sorghum bicolor (L.) Moench. J. Biol. Chem. 1996;271:32944–32950. doi: 10.1074/jbc.271.51.32944. [DOI] [PubMed] [Google Scholar]
- 13.Aoyama Y, Horiuchi T, Gotoh O, Noshiro M, Yoshida Y. CYP51-like gene of Mycobacterium tuberculosis actually encodes a P450 similar to eukaryotic CYP51. J. Biochem. (Tokyo) 1998;124:694–696. doi: 10.1093/oxfordjournals.jbchem.a022167. [DOI] [PubMed] [Google Scholar]
- 14.Bellamine A, Mangla AT, Nes WD, Waterman MR. Characterization and catalytic properties of the sterol 14alpha-demethylase from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8937–8942. doi: 10.1073/pnas.96.16.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HB, Schaller H, Goh CH, Kwon M, Choe S, An CS, Durst F, Feldmann KA, Feyereisen R. Arabidopsis cyp51 mutant shows postembryonic seedling lethality associated with lack of membrane integrity. Plant Physiol. 2005;138:2033–2047. doi: 10.1104/pp.105.061598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rozman D, Strömstedt M, Waterman MR. The three human cytochrome P450 lanosterol 14 alpha-demethylase (CYP51) genes reside on chromosomes 3, 7, and 13: structure of the two retrotransposed pseudogenes, association with a line-1 element, and evolution of the human CYP51 family. Arch. Biochem. Biophys. 1996;333:466–474. doi: 10.1006/abbi.1996.0416. [DOI] [PubMed] [Google Scholar]
- 17.Waterman MR, Lepesheva GI. Sterol 14 alpha-demethylase, an abundant and essential mixed-function oxidase. Biochem. Biophys. Res. Commun. 2005;338:418–422. doi: 10.1016/j.bbrc.2005.08.118. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida Y. Sterol biosynthesis. In: Omura T, Oshimura Y, Fujii-Kuriyama Y, editors. Cytochrome P450. 2nd Ed. 1992. pp. 93–101. [Google Scholar]
- 19.Shyadehi AZ, Lamb DC, Kelly SL, Kelly DE, Schunck WH, Wright JN, Corina D, Akhtar M. The mechanism of the acyl-carbon bond cleavage reaction catalyzed by recombinant sterol 14 alpha-demethylase of Candida albicans (other names are: lanosterol 14 alpha-demethylase, P-45014DM, and CYP51. J. Biol. Chem. 1996;271:12445–12450. doi: 10.1074/jbc.271.21.12445. [DOI] [PubMed] [Google Scholar]
- 20.Berriman M, Ghedin E, Hertz-Fowler C, et al. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 21.Pena-Diaz J, Montalvetti A, Flores CL, Constan A, Hurtado-Guerrero R, De Souza W, Gancedo C, Ruiz-Perez LM, Gonzalez-Pacanowska D. Mitochondrial localization of the mevalonate pathway enzyme 3-Hydroxy-3-methyl-glutaryl-CoA reductase in the Trypanosomatidae. Mol. Biol. Cell. 2004;15:1356–1363. doi: 10.1091/mbc.E03-10-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaller H. The role of sterols in plant growth and development. Prog. Lipid Res. 2003;42:163–175. doi: 10.1016/s0163-7827(02)00047-4. [DOI] [PubMed] [Google Scholar]
- 23.Jackson CJ, Lamb DC, Marczylo TH, Warrilow AG, Manning NJ, Lowe DJ, Kelly DE, Kelly SL. A novel sterol 14alpha-demethylase/ferredoxin fusion protein (MCCYP51FX) from Methylococcus capsulatus represents a new class of the cytochrome P450 superfamily. J. Biol. Chem. 2002;277:46959–46965. doi: 10.1074/jbc.M203523200. [DOI] [PubMed] [Google Scholar]
- 24.Aoyama Y, Yoshida Y, Sonoda Y, Sato Y. 7-Oxo-24,25-dihydrolanosterol: a novel lanosterol 14 alpha-demethylase (P-45014DM) inhibitor which blocks electron transfer to the oxyferro intermediate. Biochim. Biophys. Acta. 1987;922:270–277. [PubMed] [Google Scholar]
- 25.Burton PM, Swinney DC, Heller R, Dunlap B, Chiou M, Malonzo E, Haller J, Walker KM, Salari A, Murakami S, Mendizabal G, Azalanstat LT. (RS-21607), a lanosterol 14 alpha-demethylase inhibitor with cholesterol-lowering activityAzalanstat. Biochem. Pharmacol. 1995;50:529–544. doi: 10.1016/0006-2952(95)00152-p. [DOI] [PubMed] [Google Scholar]
- 26.Trzaskos JM, Favata MF, Fischer RT, Stam SH. In situ accumulation of 3 beta-hydroxylanost-8-en-32-aldehyde in hepatocyte cultures. A putative regulator of 3-hydroxy-3-methylglutaryl-coenzyme A reductase activity. J. Biol. Chem. 1987;262:12261–12268. [PubMed] [Google Scholar]
- 27.Lepesheva GI, Nes WD, Zhou W, Hill GC, Waterman MR. CYP51 from Trypanosoma brucei is obtusifoliol-specific. Biochemistry. 2004;43:10789–10799. doi: 10.1021/bi048967t. [DOI] [PubMed] [Google Scholar]
- 28.Cotman M, Jezek D, Fon Tacer K, Frangez R, Rozman D. A Functional Cytochrome P450 Lanosterol 14α-Demethylase CYP51 Enzyme in the Acrosome: Transport through the Golgi and Synthesis of Meiosis-Activating Sterols. Endocrinology. 2004;145:1419–1426. doi: 10.1210/en.2003-1332. [DOI] [PubMed] [Google Scholar]
- 29.Byskov AG, Andersen CY, Nordholm L, Thogersen H, Xia G, Wassmann O, Andersen JV, Guddal E, Roed T. Chemical structure of sterols that activate oocyte meiosis. Nature. 1995;6522:559–562. doi: 10.1038/374559a0. [DOI] [PubMed] [Google Scholar]
- 30.Stromstedt M, Waterman MR, Haugen TB, Tasken K, Parvinen M, Rozman D. Elevated expression of lanosterol 14alpha-demethylase (CYP51) and the synthesis of oocyte meiosis-activating sterols in postmeiotic germ cells of male rats. Endocrinology. 1998;139:2314–2121. doi: 10.1210/endo.139.5.5984. [DOI] [PubMed] [Google Scholar]
- 31.Debeljak N, Fink M, Rozman D. Many facets of mammalian lanosterol 14alpha-demethylase from the evolutionarily conserved cytochrome P450 family CYP51. Arch. Biochem. Biophys. 2003;409:159–171. doi: 10.1016/s0003-9861(02)00418-6. [DOI] [PubMed] [Google Scholar]
- 32.Sono H, Sonoda Y, Sato Y. Purification and characterization of cytochrome P-45014DM (lanosterol 14 alpha-demethylase) from pig liver microsomes. Biochim. Biophys. Acta. 1991;1078:388–394. doi: 10.1016/0167-4838(91)90161-r. [DOI] [PubMed] [Google Scholar]
- 33.Stromstedt M, Rozman D, Waterman MR. The ubiquitously expressed human CYP51 encodes lanosterol 14 alpha-demethylase, a cytochrome P450 whose expression is regulated by oxysterols. Arch. Biochem. Biophys. 1996;329:73–81. doi: 10.1006/abbi.1996.0193. [DOI] [PubMed] [Google Scholar]
- 34.Hitchcock CA, Dickinson K, Brown SB, Evans EG, Adams DJ. Purification and properties of cytochrome P-450-dependent 14 alpha-sterol demethylase from Candida albicans. Biochem. J. 1989;263:573–579. doi: 10.1042/bj2630573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamb DC, Kelly DE, Manning NJ, Hollomon DW, Kelly SL. Expression, purification, reconstitution and inhibition of Ustilago maydis sterol 14 alpha-demethylase (CYP51; P450(14DM)) FEMS Microbiol. Lett. 1998;169:369–373. doi: 10.1111/j.1574-6968.1998.tb13342.x. [DOI] [PubMed] [Google Scholar]
- 36.Lepesheva GI, Zaitseva NG, Nes WD, Zhou W, Arase M, Liu J, Hill GC, Waterman MR. CYP51 from Trypanosoma cruzi: a phyla-specific residue in the B' helix defines substrate preferences of sterol 14alpha-demethylase. J. Biol. Chem. 2006;281:3577–3585. doi: 10.1074/jbc.M510317200. [DOI] [PubMed] [Google Scholar]
- 37.Jackson JC, Lamb DC, Marczylo TH, Parker JE, Manning NL, Kelly DE, Kelly SL. Conservation and cloning of CYP51: a sterol 14 alpha-demethylase from Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 2003;301:558–563. doi: 10.1016/s0006-291x(02)03078-4. [DOI] [PubMed] [Google Scholar]
- 38.Jackson CJ, Lamb DC, Marczylo TH, Warrilow AG, Manning NJ, Lowe DJ, Kelly DE, Kelly SL. A novel sterol 14alpha-demethylase/ferredoxin fusion protein (MCCYP51FX) from Methylococcus capsulatus represents a new class of the cytochrome P450 superfamily. J. Biol. Chem. 2002;277:46959–46965. doi: 10.1074/jbc.M203523200. [DOI] [PubMed] [Google Scholar]
- 39.Taton M, Rahier A. Properties and structural requirements for substrate specificity of cytochrome P-450-dependent obtusifoliol 14 alpha-demethylase from maize (Zea mays) seedlings. Biochem. J. 1991;277:483–492. doi: 10.1042/bj2770483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cabello-Hurtado F, Taton M, Forthoffer N, Kahn R, Bak S, Rahier A, Werck-Reichhart D. Optimized expression and catalytic properties of a wheat obtusifoliol 14alpha-demethylase (CYP51) expressed in yeast. Complementation of erg11Delta yeast mutants by plant CYP51. Eur. J. Biochem. 1999;262:435–446. doi: 10.1046/j.1432-1327.1999.00376.x. [DOI] [PubMed] [Google Scholar]
- 41.O'Brien M, Chantha SC, Rahier A, Matton DP. Lipid signaling in plants. Cloning and expression analysis of the obtusifoliol 14alpha-demethylase from Solanum chacoense Bitt., a pollination- and fertilization-induced gene with both obtusifoliol and lanosterol demethylase activity. Plant Physiol. 2005;139:734–749. doi: 10.1104/pp.105.066639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lepesheva GI, Virus C, Waterman MR. Conservation in the CYP51 family. Role of the B' helix/BC loop and helices F and G in enzymatic function. Biochemistry. 2003;42:9091–9101. doi: 10.1021/bi034663f. [DOI] [PubMed] [Google Scholar]
- 43.Lepesheva GI, Waterman MR. CYP51--the omnipotent P450. Mol. Cell. Endocrinol. 2004;215:165–170. doi: 10.1016/j.mce.2003.11.016. M. R. [DOI] [PubMed] [Google Scholar]
- 44.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 45.Yoshida Y, Aoyama Y, Noshiro M, Gotoh O. Sterol 14-demethylase P450 (CYP51) provides a breakthrough for the discussion on the evolution of cytochrome P450 gene superfamily. Biochem. Biophys. Res. Commun. 2000;273:799–804. doi: 10.1006/bbrc.2000.3030. [DOI] [PubMed] [Google Scholar]
- 46.Nitahara Y, Kishimoto K, Yabusaki Y, Gotoh O, Yoshida Y, Horiuchi T, Aoyama Y. The amino acid residues affecting the activity and azole susceptibility of rat CYP51 (sterol 14-demethylase P450) J. Biochem. (Tokyo) 2001;129:761–768. doi: 10.1093/oxfordjournals.jbchem.a002917. [DOI] [PubMed] [Google Scholar]
- 47.Bellamine A, Lepesheva GI, Waterman MR. Fluconazole binding and sterol demethylation in three CYP51 isoforms indicate differences in active site topology. J. Lipid Res. 2004;45:2000–2007. doi: 10.1194/jlr.M400239-JLR200. [DOI] [PubMed] [Google Scholar]
- 48.Podust LM, Poulos TL, Waterman MR. Crystal structure of cytochrome P450 14alpha -sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc. Natl. Acad. Sci. U S A. 2001;98:3068–3073. doi: 10.1073/pnas.061562898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aoyama Y. Recent progress in the CYP51 research focusing on its unique evolutionary and functional characteristics as a diversozyme P450. Front. Biosci. 2005;10:1546–1557. doi: 10.2741/1639. [DOI] [PubMed] [Google Scholar]
- 50.Raag R, Martinis SA, Sligar SG, Poulos TL. Crystal structure of the cytochrome P-450CAM active site mutant Thr252Ala. Biochemistry. 1991;48:11420–11429. doi: 10.1021/bi00112a008. [DOI] [PubMed] [Google Scholar]
- 51.Vidakovic M, Sligar SG, Li H, Poulos TL. Understanding the role of the essential Asp251 in cytochrome p450cam using site-directed mutagenesis, crystallography, and kinetic solvent isotope effect. Biochemistry. 1998;26:9211–9219. doi: 10.1021/bi980189f. [DOI] [PubMed] [Google Scholar]
- 52.Rohmer M, Bouvier P, Ourisson G. Molecular evolution of biomembranes: structural equivalents and phylogenetic precursors of sterols. Proc. Natl. Acad. Sci. U S A. 1979;76:847–851. doi: 10.1073/pnas.76.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida Y, Noshiro M, Aoyama Y, Kawamoto T, Horiuchi T, Gotoh O. Structural and evolutionary studies on sterol 14-demethylase P450 (CYP51), the most conserved P450 monooxygenase: II. Evolutionary analysis of protein and gene structures. J. Biochem. (Tokyo) 1997;122:1122–1128. doi: 10.1093/oxfordjournals.jbchem.a021870. [DOI] [PubMed] [Google Scholar]
- 54.Nelson DR. Cytochrome P450 and the individuality of species. Arch. Biochem. Biophys. 1999;369:1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- 55.Lamb DC, Fowler K, Kieser T, Manning N, Podust LM, Waterman MR, Kelly DE, Kelly SL. Sterol 14alpha-demethylase activity in Streptomyces coelicolor A3(2) is associated with an unusual member of the CYP51 gene family. Biochem. J. 2002;364:555–562. doi: 10.1042/BJ20011380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rezen T, Debeljak N, Kordis D, Rozman D. New aspects on lanosterol 14alpha-demethylase and cytochrome P450 evolution: lanosterol/cycloartenol diversification and lateral transfer. J. Mol. Evol. 2004;59:51–58. doi: 10.1007/s00239-004-2603-1. [DOI] [PubMed] [Google Scholar]
- 57.El-Sayed NM, Myler PJ, Blandin G, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 58.Kelly SL, Lamb DC, Baldwin BC, Corran AJ, Kelly DE. Characterization of Saccharomyces cerevisiae CYP61, sterol delta22-desaturase, and inhibition by azole antifungal agents. J. Biol. Chem. 1997;272:9986–9988. doi: 10.1074/jbc.272.15.9986. [DOI] [PubMed] [Google Scholar]
- 59.Morikawa T, Mizutani M, Aoki N, Watanabe B, Saga H, Saito S, Oikawa A, Suzuki H, Sakurai N, Shibata D, Wadano A, Sakata K, Ohta D. Cytochrome P450 CYP710A encodes the sterol C-22 desaturase in Arabidopsis and tomato. Plant Cell. 2006;4:1008–1022. doi: 10.1105/tpc.105.036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Omura T. Forty years of cytochrome P450. Biochem. Biophys. Res. Commun. 1999;266:690–698. doi: 10.1006/bbrc.1999.1887. [DOI] [PubMed] [Google Scholar]
- 61.Zarn JA, Bruschweiler BJ, Schlatter JR. Azole fungicides affect mammalian steroidogenesis by inhibiting sterol 14 alpha-demethylase and aromatase. Environ. Health Perspect. 2003;111:255–261. doi: 10.1289/ehp.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Urbina JA, Payares G, Molina J, Sanoja C, Liendo A, Lazardi K, Piras M, Piras R, Perez N, Wincker P, Ryley JF. Cure of short- and long-term experimental Chagas' disease using D0870. Science. 1996;273:969–971. doi: 10.1126/science.273.5277.969. [DOI] [PubMed] [Google Scholar]
- 63.Araujo MS, Martins-Filho OA, Pereira ME, Brener Z. A combination of benznidazole and ketoconazole enhances efficacy of chemotherapy of experimental Chagas' disease. J. Antimicrob. Chemother. 2000;45:819–824. doi: 10.1093/jac/45.6.819. [DOI] [PubMed] [Google Scholar]
- 64.Buckner F, Yokoyama K, Lockman J, Aikenhead K, Ohkanda J, Sadilek M, Sebti S, Van Voorhis W, Hamilton A, Gelb MH. A class of sterol 14-demethylase inhibitors as anti-Trypanosoma cruzi agents. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15149–15153. doi: 10.1073/pnas.2535442100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang W, Ramamoorthy Y, Kilicarslan T, Nolte H, Tyndale RF, Sellers EM. Inhibition of cytochromes P450 by antifungal imidazole derivatives. Drug Metab. Dispos. 2002;30:314–318. doi: 10.1124/dmd.30.3.314. [DOI] [PubMed] [Google Scholar]
- 66.Vanden Bossche HV, Marichal P, Gorrens J, Bellens D, Moereels H, Janssen PAJ. Mutation in cytochrome P-450-dependent 14α-demethylase results in decreased affinity for azole antifungals. Biochem. Soc. Trans. 1990;18:56–59. doi: 10.1042/bst0180056. [DOI] [PubMed] [Google Scholar]
- 67.Marichal P, Koymans L, Willemsens S, Bellens D, Verhasselt P, Luyten W, Borgers M, Ramaekers FC, Odds FC, Bossche HV. Contribution of mutations in the cytochrome P450 14alpha-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology. 1999;145:2701–2713. doi: 10.1099/00221287-145-10-2701. [DOI] [PubMed] [Google Scholar]
- 68.Asai K, Tsuchimori N, Okonogi K, Perfect JR, Gotoh O, Yoshida Y. Formation of azole-resistant Candida albicans by mutation of sterol 14-demethylase P450. Antimicrob. Agents. Chemother. 1999;43:1163–1169. doi: 10.1128/aac.43.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wyand RA RA, Brown JK. Sequence variation in the CYP51 gene of Blumeria graminis associated with resistance to sterol demethylase inhibiting fungicides. Fungal, Genet. Biol. 2005;42:726–735. doi: 10.1016/j.fgb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 70.Lupetti A, Danesi R, Campa M, Del Tacca M, Kelly S. Molecular basis of resistance to azole antifungals. Trends Mol. Med. 2002;2:76–81. doi: 10.1016/s1471-4914(02)02280-3. [DOI] [PubMed] [Google Scholar]
- 71.Brun S, Berges T. Mechanisms of azole resistance in petite mutants of Candida glabrata. Antimicrobial Agents and Chemotherapy. 2004;48:1788–1796. doi: 10.1128/AAC.48.5.1788-1796.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hankins EG, Gillespie JR, Aikenhead K, Buckner FS. Upregulation of sterol C14-demethylase expression in Trypanosoma cruzi treated with sterol biosynthesis inhibitors. Mol. Biochem. Parasitol. 2005;144:68–75. doi: 10.1016/j.molbiopara.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 73.Ortiz de Montellano PR, Correia MA. Inhibition of cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. 2nd ed. Plenum Publishing Corp.; New York: 1995. pp. 305–364. [Google Scholar]
- 74.Matsumoto M, Ishida K. Strong antifungal activity of SS750, a new triazole derivative, is based on its selective binding affinity to cytochrome P450 of fungi. Antimicrob. Agents Chemother. 2002;46:308–314. doi: 10.1128/AAC.46.2.308-314.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morisaki M, Sonoda Y, Makino T, Ogihara N, Ikekawa N, Sato Y. Inhibitory effect of 15-oxygenated sterols on cholesterol synthesis from 24,25-dihydrolanosterol. J. Biochem. (Tokyo) 1986;99:597–600. doi: 10.1093/oxfordjournals.jbchem.a135516. [DOI] [PubMed] [Google Scholar]
- 76.Frye LL, Leonard DA. Lanosterol analogs: dual-action inhibitors of cholesterol biosynthesis. Crit. Rev. Biochem. Mol. Biol. 1999;34:123–140. doi: 10.1080/10409239991209246. [DOI] [PubMed] [Google Scholar]
- 77.Trzaskos JM, Fischer RT, Ko SS, Magolda RL, Stam S, Johnson P, Gaylor JL. Substrate-based inhibitors of lanosterol 14 alpha-methyl demethylase: II. Time-dependent enzyme inactivation by selected oxylanosterol analogs. Biochemistry. 1995;30:9677–9681. doi: 10.1021/bi00030a004. [DOI] [PubMed] [Google Scholar]
- 78.Sonoda Y, Obi N, Onoda M, Sakakibara Y, Sato Y. Effects of 32-oxygenated lanosterol derivatives on 3-hydroxy-3-methylglutaryl coenzyme A reductase activity and cholesterol biosynthesis from 24,25-dihydrolanosterol. Chem. Pharm. Bull. (Tokyo) 1992;40:2796–2799. doi: 10.1248/cpb.40.2796. [DOI] [PubMed] [Google Scholar]
- 79.Cooper AB, Wright IJ, Ganguly AK, Desai J, Loebenberg D, Parmegiani R, Feingold DS, Sud IJ. Synthesis and antifungal properties of 14-aminomethyl-substituted lanosterol derivatives. J. Chem. Soc. Chem. Commun. 1989;12:898–900. doi: 10.1111/j.1749-6632.1988.tb40394.x. [DOI] [PubMed] [Google Scholar]
- 80.Aoyama Y, Yoshida Y. Different substrate specificities of lanosterol 14a-demethylase (P-45014DM) of Saccharomyces cerevisiae and rat liver for 24-methylene-24,25-dihydrolanosterol and 24,25-dihydrolanosterol. Biochem. Biophys. Res. Commun. 1991;178:1064–1071. doi: 10.1016/0006-291x(91)91000-3. [DOI] [PubMed] [Google Scholar]