Abstract
The expression of dipeptidyl peptidase 4 (DP4, CD26) affects T-cell recruitment to lungs in an experimental rat asthma model. Furthermore, the gene of the structural homologous DP10 represents a susceptibility locus for asthma in humans, and the functional homologous DP8/9 are expressed in human leukocytes. Thus, although several mechanisms may account for a role of DP4-like peptidases in asthma, detailed information on their anatomical sites of expression and function in lungs is lacking. Therefore, bronchi and lung parenchyma were evaluated using immunohistochemistry and histochemical/enzymatic activity assays, as well as quantitative real-time PCR for this family of peptidases in naïve and asthmatic rat lungs derived from wild-type F344 and DP4-deficient F344 rat strains. Surprisingly, results show not only that the induction of experimental asthma increases DP4 enzymatic activity in the bronchoalveolar lavage fluid and parenchyma, but also that DP8/9 enzymatic activity is regulated and, as well as the expression of DP10, primarily found in the bronchial epithelium of the airways. This is the first report showing a differential and site-specific DP4-like expression and function in the lungs, suggesting a pathophysiologically significant role in asthma. (J Histochem Cytochem 56:147–155, 2008)
Keywords: dipeptidyl peptidase 4, dipeptidyl peptidases 8/9, dipeptidyl peptidase 10, asthma, lung, bronchi, F344 rat substrains, DP4-like activity
Asthma is a chronic inflammatory disease of the airways, which is characterized by bronchial hyper-responsiveness and airway obstruction and is accompanied by wheezing, coughing, and breathlessness (Busse and Lemanske 2001). Furthermore, an asthmatic response is characterized by an elevated production of IgE, cytokines, and chemokines; mucus hypersecretion; and eosinophilia. Many of these disease parameters are induced or modulated by T-cell recruitment and activation, cell adhesion, and chemokine metabolism. All of these disease-modulating processes are potentially further modulated by dipeptidyl peptidase 4 (DP4) (EC 3.4.14.5, CD26) expression and enzymatic activity (Boonacker and Van Noorden 2003). This might also be the case for the other members of the DP4-like gene family, including the structural homologous DP10 and the functional homologous DP8/9 (Chen et al. 2003). In addition, DP4 has been reported to be expressed in the serosal submucosal glands of the human bronchus (van der Velden et al. 1998) and in human bronchoalveolar lavage (BAL) fluid (van der Velden et al. 1999).
In line with this concept, it was recently demonstrated that lack of DP4/CD26 expression in a deficient rat model remarkably reduces T-cell recruitment to the lungs during experimental asthma (Kruschinski et al. 2005) and that especially DP4-positive, activated T-cells are recruited to the lungs after induction of asthma in F344 rats (Skripuletz et al. 2007). In addition to these preclinical data indicating an important role of DP4 in asthma, large-scale genetic association screenings revealed that the gene of DP10, a structural homolog of DP4 (Qi et al. 2003), is a susceptibility marker of human asthma (Allen et al. 2003). The other functional homologs, DP8 (Abbott et al. 2000) and DP9 (Ajami et al. 2004), show cytoplasmatic expression, but there is no direct evidence yet of a functional involvement during asthma except for their expression in human leukocytes (Maes et al. 2007).
For a better understanding of these important indications of the role of DP4-like peptidases in asthma, the present study investigates in detail their sites of expression in rat lungs with and without an allergic-like inflammation status.
Materials and Methods
Animals
Male wild-type F344/Ztm rats (DP4pos) and male DP4 mutant rats [F344/Crl(Wiga)SvH-Dpp4m] lacking DP4 activity as well as DP4 expression (DP4neg) were used (Karl et al. 2003). All animals were housed at the Central Animal Facility of the Hannover Medical School, maintained in a separate minimal-barrier–sustained facility, and microbiologically monitored according to Federation of European Laboratory Animal Science Associations recommendations (Rehbinder et al. 1996). The temperature was regulated at 21 ± 2C, relative humidity was 60 ± 5% with an air change rate of 15 times per hour, under a 12-hr light 12-hr dark cycle (lights on at 6:00 am). Food and water were available ad libitum. All research and animal care procedures had been approved by the review board of the Landesamt fuer Verbraucherschutz und Lebensmittelsicherheit (Oldenburg, Germany) and were performed according to international guidelines on the use of laboratory animals.
Sensitization and Allergen Challenge
At the age of 12 weeks (260 ± 30 g), five DP4-positive and five DP4-negative rats in the asthma group were sensitized 14 and 7 days before challenge, as previously described (Skripuletz et al. 2007). In brief, sensitization was performed with 1 mg of ovalbumin (OVA; Sigma, Deisenhofen, Germany) and 200 mg of Al(OH)3 (Sigma) in 1 ml 0.9% (sterile, pyrogen-free) NaCl applied subcutaneously in a hind limb. As a second adjuvant, concentrated preparations of 6 × 109 heat-killed Bordetella pertussis bacilli (kindly provided by Chiron Behring, Marburg, Germany) in 0.4 ml 0.9% NaCl were given intraperitoneally at the same time. Animals were challenged with 7.5% of aerosolized OVA using a Pari LC Star nebulizer (Pari; Starnberg, Germany). The control group consisted of five DP4-positive and five DP4-negative naïve rats that were neither sensitized nor challenged.
Dissection of Animals
The animals were dissected under isoflurane anesthesia 22 ± 0.5 hr after challenge, as previously described (Skripuletz et al. 2007). Briefly, the animals were sacrificed by aortic exsanguination, thereby collecting EDTA blood samples. For BAL isolation, a cannula was inserted into the trachea in situ and the lungs were lavaged four times with portions of 5 ml 0.9% NaCl solution. The recovery of fluid was over 90% in all animals. For further analysis, both lungs were excised from the thorax. The trachea, main bronchi, and hilar lymph nodes were removed. For PCR analyses, the inferior lobe of the right lung was frozen in liquid nitrogen. The left lungs were instilled with 3 ml of Tissue-Tek O.C.T. compound (Miles Inc.; Elkhart, IN) mixed 1:4 with PBS and placed on aluminum foil on dry ice.
Synthesis of the Histochemical Substrate H-Gly-l-Pro-1-hydroxy-4-naphthylamide Hydrochloride
On the basis of a method described by Dikov et al. (1999), the synthesis of H-Gly-l-Pro-1-hydroxy-4-naphthylamide hydrochloride was modified according to a two-step procedure. First, the precursor of the substrate Boc-Gly-l-Pro-1-hydroxy-4-naphthylamide was synthesized from Boc-Gly-l-Pro-OH (Bachem; Bubendorf, Switzerland) and 1-hydroxy-4-naphthylamine (Sigma-Aldrich Chemie GmbH; Taufkirchen, Germany) using the mixed-anhydride method composed of isobutyl chloroformate and N-methylmorpholine. After the usual workup, the crude product was purified by flash chromatography (gradient elution, CH3OH/CHCl3: 1/1 → 3/1 → CH3OH) to give a pure compound with a yield of 88%. In a second step, the precursor compound Boc-Gly-l-Pro-1-hydroxy-4-naphthylamide was dissolved in a solution of hydrochloric acid in 1,4-dioxane (8 M) and stirred for 4 hr at room temperature. The deprotection solution was removed under reduced pressure, and the resulting residue was purified by flash chromatography (gradient elution, CH3OH/CHCl3: 1/1 → 3/1 → CH3OH) generating the substrate H-Gly-l-Pro-1-hydroxy-4-naphthylamide hydrochloride, with a yield of 41% and a purity of 100%. In addition to the usual HPLC analysis, the identity and purity of the compound was further characterized by proton nuclear magnetic resonance spectroscopy and mass-spectrometry electrospray ionization.
Histochemical Activity Assay
Lungs were cut on a cryotome, and resulting sections (10 μm) were mounted on poly-l-lysine–coated glass slides and fixed in acetone for 10 min at −20C. Sections were washed in 0.1 M phosphate buffer (pH 7.8) and then incubated for either 25 min or 20 hr at 37C in an incubation solution consisting of 0.25 mM H-Gly-l-Pro-1-hydroxy-4-naphthylamide hydrochloride (synthesized at Probiodrug; Halle/Saale, Germany) and 0.25 mM nitro blue tetrazolium (NBT) (Sigma-Aldrich; Steinheim, Germany) dissolved in a minimal volume (less than 0.6%) of dimethyl sulfoxide (DMSO) (Merck; Darmstadt, Germany) in 0.1 M phosphate buffer (pH 7.8). Control sections were incubated in 0.25 mM NBT dissolved in DMSO in 0.1 M phosphate buffer (pH 7.8). After incubation, the sections were washed in 0.1 M phosphate buffer (pH 7.0), fixed in 4% paraformaldehyde, and stained with methyl green nuclear counterstain (Vector Laboratories; Burlingame, CA). Subsequently, the sections were dehydrated in increasing concentrations of alcohol, cleared in xylene, and covered with Eukitt (O. Kindler GmbH and Co.; Freiburg, Germany). The above histochemical assay is specific for DP4, DP8, DP9, and, to a lesser extent, for DP2, owing to the slightly alkaline pH. However, the activity of DP2 is inhibited by NBT (Dikov et al. 2000). Cleavage of the substrate by these DPs forms a strong reducing agent, which reduces NBT to a diformazan. This diformazan precipitates at the sites of enzymatic activity and is visible as a blue staining. As a further control for specificity and in addition to the use of DP4-deficient animals, this histochemical activity assay was also performed using a DP4-specific inhibitor as previously described (Frerker et al. 2007), which was added to the incubation solution in a final concentration of 2 μM. The sections were incubated for 20 hr in this solution and treated further as described above. As a general rule, during each run of stainings, sections of all groups were incubated in the same cuvette and treated completely identically to ensure the comparability between stainings.
Light microscopy investigations were carried out on a Nikon Eclipse 80i microscope (Nikon GmbH; Duesseldorf, Germany), and representative pictures were taken with a MicroFire digital microscope camera (Optronics; Goleta, CA).
Immunohistochemistry
Two consecutive alkaline phosphatase antialkaline phosphatase (APAAP) stainings (Cordell et al. 1984) were performed on 40 μm acetone-fixed cryostat sections of DP4-positive whole left lungs with Fast Blue (Sigma) as the detection system for labeled T-cells and Fast Red (Sigma) as the detection system for labeled DP4-positive cells. In detail, the sections were incubated with the primary monoclonal antibody (mAb) against the α/β T-cell receptor (mAb R73; Serotec, Duesseldorf, Germany, 1:5000) for 4 hr at room temperature. After washing with TBS-Tween (0.05% Tween 20; Serva, Heidelberg, Germany), the sections were incubated for 30 min with the bridging antibody (Dako; Hamburg, Germany, 1:50 in PBS, with 5% inactivated rat serum), washed again, and incubated with the APAAP complex (Dako; 1:50 in TBS-Tween) for 30 min. The incubations with the bridging antibody and the APAAP complex were repeated once for 15 min. After the addition of the substrate Fast Blue for 30 min, the incubation with the primary antibody against DP4 (mAb 5E8; Cell Sciences, Canton, MA) was performed overnight at 4C, followed by an identical staining procedure, except that Fast Red was the substrate. Finally, the sections were covered with Mowiol (Hoechst AG; Frankfurt/Main, Germany). The DP4-negative lung sections were not stained with an anti-DP4 antibody. As in previous studies, no mAb binding in the knock-out-like model used in this study could be detected (Shingu et al. 2003).
Similarly, rabbit polyclonal antibodies against DP8 (Abcam; Cambridge, United Kingdom, 1:500), DP9 (Abcam; 1:1000), and DP10 (Abcam; 1:250) were incubated on 10-μm lung sections using a mouse anti-rabbit antibody (Dako; 1:50) for 30 min after the 30 min incubation with the primary antibody, and a hemalaun counterstaining (Merck; 1:5 in PBS) for 20 sec after the APAAP staining with Fast Red.
Enzymatic Activity Assay In Vitro
DP4-like enzymatic activity of the different rat strains was determined by incubating EDTA-plasma samples and BAL fluid samples with the substrate H-glycyl-prolyl-4-nitroaniline hydrochloride (H-Gly-Pro-pNA*HCl) (Bachem; Bubendorf, Switzerland) and measuring the release of paranitroaniline (pNA) by an increase in absorbance at 405 nm over time, as described previously (Karl et al. 2003), using the PowerWave XS photometer (Bio-Tek Instruments; Bad Friedrichshall, Germany). The assay was composed of 20 μl of the samples in 40 mM HEPES buffer (pH 7.6) and 0.4 mM H-Gly-Pro-pNA*HCl. Prior to the enzymatic reaction, the samples were incubated with the HEPES buffer for 15 min at 37C. The reaction was initiated by adding the substrate, and the release of pNA was measured up to 10 min. One unit is defined as the amount of enzyme necessary to hydrolyze 1 μM of substrate per minute. The assay is selective for DP4-like activities; however, due to the alkaline pH, the contribution of DP2 is negligible (Frerker et al. 2007).
Quantitative Real-time PCR
RNA was prepared from shock-frozen lobes of the right lungs, trachea, bronchi, parenchyma, and brain by using the RNeasy Mini Kit (Qiagen; Hilden, Germany), and cDNA was synthesized using M–MLV reverse transcriptase (Invitrogen; Karlsruhe, Germany) following the manufacturer's guidelines.
PCR was carried out on a thermal cycler (Eppendorf; Hamburg, Germany). The 50-μl reaction mixture was composed of 75 ng cDNA, 200 μM deoxynucleotide triphosphate, 0.1 μM primer (each), 1× PCR buffer (minus Mg), 1.5 mM MgCl2, and 2.5 U Taq (Invitrogen). The protocol contained a 5-min initial denaturation step at 95C and 35 cycles of the following steps: 30-sec denaturation at 95C, 1-min primer annealing at a primer-specific temperature (Table 1), and 45-sec extension at 72C. A final extension step for 5 min at 72C was performed. To visualize the PCR products, gel electrophoresis (2% agarose in Tris acetate EDTA buffer) was performed, and 15 μl of the final PCR product was applied to the gel. This protocol was used for all primers except DP10. Because of the low amounts of the DP10 PCR product, 90 ng cDNA was used in a 50-μl preparation, 40 cycles were performed, and 30 μl of the final PCR product was applied to the agarose gel.
Table 1.
Primers used for rat RPL13a, DP4, DP8, DP9, and DP10
| Gene | Primer sequence (5′-3′) | Annealing temperature (C) | Size PCR product (bp) |
|---|---|---|---|
| RPL13a | CCTCCACCCTATGACAAGGA (forward) | 58 | 186 |
| TTCCGGTAATGCATCTTTGC (reverse) | |||
| DP4 | TCCCAACTCCAGAGGACAAC (forward) | 57 | 152 |
| CAGGGCTTTGGAGATCTGAG (reverse) | |||
| DP8 | ACAGCAAACCCAAAGGTCAC (forward) | 58 | 152 |
| TCTGGAGTCCATCCAGCTCT (reverse) | |||
| DP9 | AATGACTATGACTGGACGGA (forward) | 60 | 196 |
| CGTAGAGGTGATGTTCCAGG (reverse) | |||
| DP10 | TCATTTCCAGCATTCAGCAG (forward) | 53 | 176 |
| GCAGCACGGATACTTCTTCC (reverse) |
Quantitative real-time PCR was carried out on an iCycler thermal cycler with the iQ5 real-time PCR detection system (Bio-Rad; Munich, Germany) using a SYBR green detection protocol (Qiagen) with 12.5 ng of each cDNA and a final concentration of 0.6 μM of each primer per preparation. A 15-min initial activation step at 95C was performed, followed by 45 cycles of 15-sec denaturation at 95C, annealing for 30 sec at a primer-specific temperature, and 30-sec extension at 72C. Primers for the detection of DP4, DP8, DP9, DP10, and the housekeeping gene ribosomal protein L13a (RPL13a) were designed based on sequences from the National Center for Biotechnology Information database (RPL13a: NM_173340; DP4: NM_012789; DP8: XM_236345; DP9: XM_217309; DP10: NM_001012205) using the Primer3 application (Rozen and Skaletsky 2000). Details are provided in Table 1. Mean normalized expression was calculated using the Q-Gene application (Simon 2003). The sequences of rat DP8 and rat DP9 have not been published until now. Accordingly, primers were designed using predicted sequences derived from genomic sequences by automated computational analyses. DP4-negative rats carry a mutation that causes a rapid intracellular degradation of DP4 without the mutant protein being processed to the mature form (Tsuji et al. 1992). The primers used for DP4 detection are located outside of the mutated region of DP4 in the DP4-negative rats, explaining the positive PCR signals in these rats. A similarity between mutant and wild-type DP4 mRNA levels has been described by Thompson et al. (1991), although the protein of DP4 is functionally inactive and retained inside the cell in DP4-negative F344 rats.
Statistical Analysis
Differences among groups were analyzed using two-way ANOVA. Treatment (control versus asthma) and genetic background (DP4pos versus DP4neg) were the factors, followed by the Fisher protected least-significant difference test for post hoc comparisons, if appropriate. Statistically significant effects between the asthma group and the control group are indicated by asterisks (*p<0.05), and for comparison of DP4neg and DP4pos groups, by rhombs (###p<0.0001). All data are displayed as mean ± SEM.
Results
By means of the histochemical activity assay, the localization of sites exhibiting DP4, DP8, and DP9 enzymatic activity in rat lungs under naïve (control) and asthmatic conditions was documented in DP4-negative and DP4-positive rat lungs. Positive blue precipitates in lungs of DP4-negative rats or of those incubated with a DP4-specific inhibitor were interpreted as DP8/9 specific. Thus, the blue staining of lung sections obtained from wild-type Fischer rats represents the sum of the enzymatic activity of the three dipeptidyl peptidases, DP4, DP8, and DP9, visible even after 25 min of incubation. No staining was detectable on control sections of DP4-positive lungs incubated for 20 hr in NBT without substrate (not shown).
Incubation periods of 25 min and 20 hr appeared well-suited to analyze the enzymatic activity on DP4-positive and DP4-negative lung sections. Although NBT inhibits the activity of DP2, the resulting staining on the lung sections of DP4-positive rats is caused by the cumulative activity of DP4, DP8, and DP9 (Figures 1A–1C and 1G–1I), and the staining on lung sections of DP4-negative rats (Figures 1D–1F and 1J–1L) is based only on DP8 and DP9 enzymatic activity.
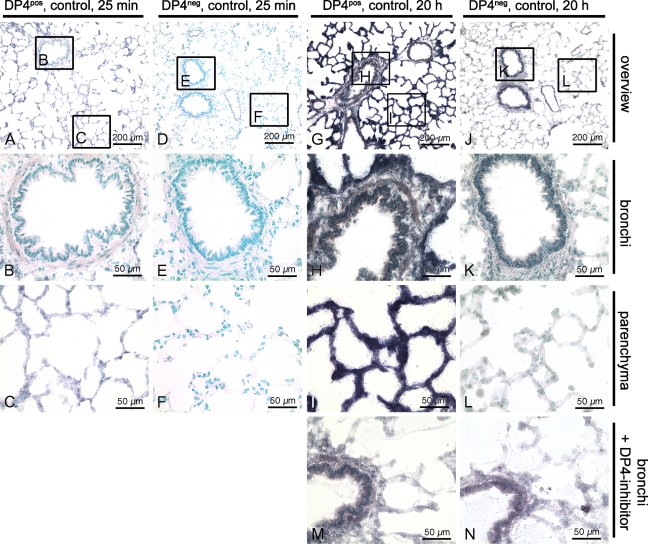
Figure 1.
Detection of DP4-like activities with a histochemical activity assay. (A–C) Lung section of a DP4-positive control rat after an incubation period of 25 min with a slight staining of the lung parenchyma. (D–F) Lung section of a DP4-negative control rat after an incubation period of 25 min with no visible staining. (G–I) Lung section of a DP4-positive control rat after an incubation period of 20 hr with a strong staining of the lung parenchyma and a weaker staining of the bronchi. (J–L) Lung section of a DP4-negative control rat after an incubation period of 20 hr with hardly any staining of the lung parenchyma and a weak staining of the bronchi. (M) Lung section of a DP4-positive control rat after 20 hr incubation in the presence of a DP4-specific inhibitor. (N) Lung section of a DP4-negative control rat after 20 hr incubation in the DP4 inhibitor solution.
The enzymatic reaction product of cumulative DP4-like peptidases in lung sections was much more pronounced compared with the DP8 and DP9 activity. This was clearly demonstrated by comparing DP4-positive and -negative lung sections after an incubation period of 20 hr (Figures 1G and 1J) and was even visible after 25 min in the lung parenchyma (Figures 1A and 1D). Although there was no remaining staining at all after 25 min on DP4-negative sections (Figure 1D), a slight blue staining was visible on DP4-positive sections (Figure 1A). After 20 hr, the DP4-positive sections were intensely blue-colored (Figure 1G), whereas the DP4-negative sections were only faintly blue-stained (Figure 1J).
The comparison of DP4-positive and -negative sections also revealed differential DP4-like activities in different compartments of the lungs. Although the DP4 activity was very pronounced in lung parenchyma (Figure 1G), the activity of DP8 and DP9 was primarily located in the bronchi (Figure 1J) and to a lesser extent in the parenchyma. The bronchi of DP4-positive and -negative sections showed almost no difference after 20 hr (Figures 1H and 1K), which indicated that the activity in the bronchi was mainly derived from DP8 and DP9. This finding was confirmed by the additional use of a DP4-specific inhibitor (Figures 1M and 1N). Incubation of a DP4-positive section in the presence of this inhibitor (Figure 1M) showed the same staining pattern and kinetics as incubation of a DP4-negative section in the solution (Figure 1N), as did DP4-negative sections in the solution without this inhibitor. Thus, the genetic model and pharmacological approach were cross-validated and strongly suggest specificity of findings.
In addition to the different compartmentalization of DP4 and DP8/9 activity, an upregulation of their activities after induction of asthma was observed (Figures 2A–2E). In addition to the enzymatic activity assay on lung sections, the enzymatic activity of both rat substrains was also measured in vitro. Plasma and BAL fluid samples from DP4-negative rats showed only a very low DP4-like enzymatic activity. The DP4-like activity of the plasma samples did not vary between the naïve control group and the asthma group (data not shown), whereas the DP4-like activity of BAL fluid samples showed a significant asthma-specific treatment effect (p=0.01), in addition to a significant effect of the genetic background (p<0.0001) (Figure 2A). In line with the findings in the BAL, the histochemical determination of DP4 enzymatic activity in the lung parenchyma of F344 wild-type rats of the asthma group (Figure 2C) appeared to be upregulated, compared with the control group (Figure 2B) after an incubation period of 25 min. In addition, the activity of DP8/9 appeared to be upregulated in the bronchi after asthma induction (Figure 2E), compared with the control group (Figure 2D) after an incubation period of 20 hr.
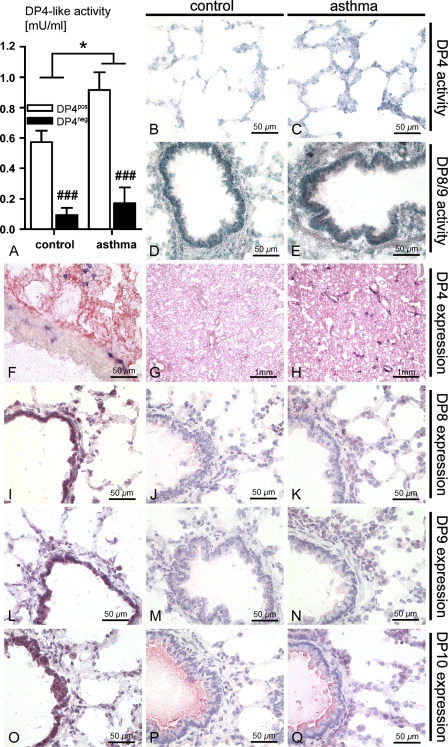
Figure 2.
Comparison of the DP4-like peptidases in the lungs. (A) DP4-like activity of the bronchoalveolar lavage (BAL) fluid measured by the enzymatic activity assay in vitro (*p<0.05; ###p<0.0001). Histochemical activity assay of a DP4-positive control lung (B), of a DP4-positive asthma lung (C), of a DP4-negative control lung (D), and of a DP4-negative asthma lung (E). (F,G) Staining with a DP4 antibody (red) and a T-cell antibody (blue) of a DP4-positive control lung at different magnifications. (H) The same staining of a DP4-positive asthma lung. Staining with a DP8 antibody of a control lung without BAL (I) and after BAL (J), and of an asthma lung after BAL (K). Staining with a DP9 antibody of a control lung without BAL (L) and after BAL (M), and of an asthma lung after BAL (N). Staining with a DP10 antibody of a control lung without BAL (O) and after BAL (P), and of an asthma lung after BAL (Q).
Immunohistochemical staining for DP4-like proteins further confirmed and complemented our findings based on enzymatic assays (Figures 2F–2Q). Staining of DP4-positive lungs from rats of the control group and the asthma group with a monoclonal antibody against DP4 also revealed no antibody binding in the bronchi (Figure 2F) and a more pronounced staining of lungs after asthma induction (Figure 2H) compared with control lungs (Figure 2G), which is indicative of an upregulation of DP4 protein expression and also in accordance with an increase of DP4 enzymatic activity on tissue and in vitro. Immunohistochemical characterization of DP8, DP9, and DP10 protein expression under naïve and asthmatic conditions also provided evidence of an upregulation of these proteins after induction of experimental asthma. In contrast to DP4, these three peptidases appeared to be strongly expressed in the bronchi and in some leukocytes, but were weakly expressed in the parenchyma. This was most noticeable in lungs that were not lavaged (Figures 2I, 2L, and 2O), but was also evident in lungs after the BAL procedure (Figures 2J–K, 2M–2N, and 2P–2Q).
To obtain additional information regarding mRNA expression levels of DP4, DP8, DP9, and DP10, PCR and real-time PCR approaches were performed (Figure 3A). Taken together, the real-time PCR data of DP4, DP8, and DP9 revealed treatment effects between naïve and asthmatic lungs, whereas no differences were observed when comparing DP4-positive and DP4-negative substrains (Figures 3B–3D). Specifically, although the expression of DP4 did not differ after asthma induction (Figure 3B), the expression of DP8 and DP9 (Figures 3C and 3D) was significantly upregulated in the asthma group compared with the control group (p=0.02 for DP8 and p=0.02 for DP9).
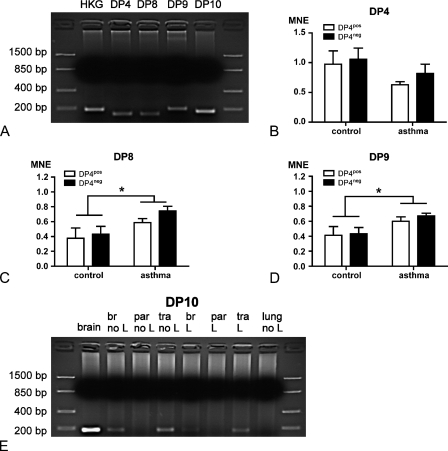
Figure 3.
PCR analyses of DP4, DP8, DP9, and DP10. (A) Overview of the PCR products resulting from different primer pairs. HKG, housekeeping gene. (B) Mean normalized expression (MNE) of DP4 in DP4-positive (DP4pos) and DP4-negative (DP4neg) lungs of control groups and groups after asthma induction. (C) MNE of DP8 in the same groups (*p<0.05). (D) MNE of DP9 in the same groups (*p<0.05). (E) Detection of a DP10 PCR product only in cDNA from brain, trachea, and bronchi of control lungs and lungs after asthma induction. br, bronchi; par, lung parenchyma; tra, trachea; L, sample after BAL; no L, sample without BAL.
Highest mRNA coding for DP10 was found in the brain, followed by trachea and bronchi, but could not be amplified in samples from whole lung and from lung parenchyma (Figure 3E). Apparently, the expression in tissues that were not lavaged (Lanes 3–5) were higher compared with tissues from rats after a BAL (Lanes 6–8), and this was in line with observations on immunohistochemical stainings with a DP10 antibody. In general, because the BAL procedure removes leukocytes and proteins from airways of the lungs, this reduced mRNA expression pattern and faint immunohistochemical-based protein detection suggest significant leukocyte-associated and soluble components of these DP4-like proteins in asthma.
Discussion
In this study, a site-specific and disease-associated expression of the dipeptidyl peptidases DP4, DP8, DP9, and DP10 in lungs was documented in an F344 rat model of bronchial asthma.
Regulation of DP4 expression during experimental asthma provides further evidence for its involvement in the regulation of inflammatory processes. Probably, DP4 is not only an inflammatory marker per se but also exhibits distinct functional properties within this process. For example, it has been shown recently that a genetically induced DP4 deficiency is associated with blunted T-cell recruitment to the lungs in asthma (Kruschinski et al. 2005). Therefore, we were interested in whether this effect was primarily due to a T-cell–specific functional role of DP4 or whether DP4 expression in lungs may also contribute to that observation. The findings presented here suggest not only that T-cell–specific DP4-related functions play a role but also that DP4 expression in the lungs might significantly be involved in this process. One potential mechanism could be that DP4 is also involved in cell adhesion; specifically, that DP4 mediates adhesion to extracellular matrix proteins such as collagen and fibronectin (Hanski et al. 1988; Piazza et al. 1989). In the course of an inflammatory process, such an upregulation of DP4 may be beneficial, because DP4-positive leukocytes are more easily recruited to the sites of inflammation. Because this has to be a rapid response, we found that DP4 mRNA expression at 22 hr after challenge was already on the downslope, whereas the corresponding protein expression and DP4 activity were upregulated in histochemical stainings and in an enzymatic activity assay in vitro. To date, however, such a more general role for DP4 via mediation of leukocyte adhesion has not been documented for leukocyte recruitment during inflammatory processes and has recently been attributed only to tumor cell adhesion (Shingu et al. 2003).
The documented upregulation of DP4-like activity in the BAL fluid of DP4-positive rats after asthma induction is contrary to observations by others (van der Velden et al. 1999). Van der Velden and colleagues did not observe any differences in the peptidase activities between allergic asthmatics and healthy nonsmokers. Because the DP4-like activities in the BAL fluid and the lung might at least be partially released by activated CD4-positive T-cells (Juillerat-Jeanneret et al. 1997), they explained their observations with an unaltered T-cell number in the allergic asthmatics (van der Velden et al. 2000). Corresponding to the above-documented upregulated DP4-like activities in the lung and BAL fluid of rats after asthma induction, our previous studies showed an elevated number of activated T-cells after asthma induction in this rat model (Skripuletz et al. 2007).
The site-specific activity and expression of DP4 and its related peptidases, DP8, DP9, and DP10, indicate their differential functional roles during allergic diseases such as bronchial asthma. The fact that asthma is a disease of the airways, and the observation that DP10 and the activities of DP8 and DP9 are primarily located in the bronchi, strongly suggest an involvement of these peptidases during the clinical course of asthma. In humans, the DP10 gene has already been identified as a locus for asthma susceptibility (Allen et al. 2003), but until now, no information about the functional role of DP10 was available, apart from its association with the Kv4-mediated A-type potassium channels (Zagha et al. 2005). Similarly, until now, it was only known that DP8 and DP9 are intracellularly expressed proteins in leukocytes (Maes et al. 2007) and that they have ubiquitous expression patterns (Abbott et al. 2000; Ajami et al. 2004) with a DP4-like activity. No natural substrates have been identified to date in vivo, although the hydrolysis of glucagon-like peptide 1 (GLP-1), GLP-2, and neuropeptide Y have already been demonstrated in vitro (Bjelke et al. 2006). In this regard, our study should motivate specific research activities to clarify their functional role. A strong DP10 mRNA expression was found in brain samples and a weaker mRNA expression in trachea from F344 rats, which corresponds to observations made by other groups (Allen et al. 2003; Takimoto et al. 2006). Likewise, we could not find any mRNA expression in samples from the whole lung (Chen et al. 2006), whereas in samples from the bronchi, the PCR signal and immunohistochemical protein detection were positive. The strong PCR signal for DP10 in the central nervous system–derived positive control might even suggest that the more precise site of expression of DP10 within the bronchi might be found in the bronchus-associated nervous system, which would then in turn strengthen the concept of an important nervous system–mediated component in the regulation of allergic responsiveness.
The observation that DP8 and DP9 mRNA levels are upregulated during asthma induction and the localization of their activity in the bronchi suggest that these intracellular peptidases specifically respond to the inflammatory stimulus. Elevated DP8 mRNA levels have also been documented in activated lymphocytes (Abbott et al. 2000; Gorrell 2005). Likewise, the contribution of DP8/9 to the overall DP4-like activity and their necessity for T-cell proliferation have been demonstrated using selective inhibitors (Lankas et al. 2005; Maes et al. 2007).
In conclusion, because asthma is a disease of the airways, a site-specific expression and a regulation of DP4 and the DP4-like peptidases DP8, DP9, and DP10 in the whole lung and especially in the bronchi after asthma induction point to their potential role in asthma, which should be further investigated.
Acknowledgments
This study was supported by the German Research Foundation (SFB 587, project B11).
We thank Susanne Kuhlmann, Susanne Fassbender, and Antje Hamann for skillful technical assistance, Tina Beekmann for preparing the cDNA, and Sheila Fryk for the correction of the English.
References
- Abbott CA, Yu DM, Woollatt E, Sutherland GR, McCaughan GW, Gorrell MD (2000) Cloning, expression and chromosomal localization of a novel human dipeptidyl peptidase (DPP) IV homolog, DPP8. Eur J Biochem 267:6140–6150 [DOI] [PubMed] [Google Scholar]
- Ajami K, Abbott CA, McCaughan GW, Gorrell MD (2004) Dipeptidyl peptidase 9 has two forms, a broad tissue distribution, cytoplasmic localization and DPIV-like peptidase activity. Biochim Biophys Acta 1679:18–28 [DOI] [PubMed] [Google Scholar]
- Allen M, Heinzmann A, Noguchi E, Abecasis G, Broxholme J, Ponting CP, Bhattacharyya S, et al. (2003) Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat Genet 35:258–263 [DOI] [PubMed] [Google Scholar]
- Bjelke JR, Christensen J, Nielsen PF, Branner S, Kanstrup AB, Wagtmann N, Rasmussen HB (2006) Dipeptidyl peptidases 8 and 9: specificity and molecular characterization compared with dipeptidyl peptidase IV. Biochem J 396:391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonacker E, Van Noorden CJ (2003) The multifunctional or moonlighting protein CD26/DPPIV. Eur J Cell Biol 82:53–73 [DOI] [PubMed] [Google Scholar]
- Busse WW, Lemanske RF Jr (2001) Asthma. N Engl J Med 344:350–362 [DOI] [PubMed] [Google Scholar]
- Chen T, Ajami K, McCaughan GW, Gai WP, Gorrell MD, Abbott CA (2006) Molecular characterization of a novel dipeptidyl peptidase like 2-short form (DPL2-s) that is highly expressed in the brain and lacks dipeptidyl peptidase activity. Biochim Biophys Acta 1764:33–43 [DOI] [PubMed] [Google Scholar]
- Chen T, Ajami K, McCaughan GW, Gorrell MD, Abbott CA (2003) Dipeptidyl peptidase IV gene family. The DPIV family. Adv Exp Med Biol 524:79–86 [DOI] [PubMed] [Google Scholar]
- Cordell JL, Falini B, Erber WN, Ghosh AK, Abdulaziz Z, MacDonald S, Pulford KA, et al. (1984) Immunoenzymatic labeling of monoclonal antibodies using immune complexes of alkaline phosphatase and monoclonal anti-alkaline phosphatase (APAAP complexes). J Histochem Cytochem 32:219–229 [DOI] [PubMed] [Google Scholar]
- Dikov A, Dimitrova M, Pajpanova T, Krieg R, Halbhuber KJ (2000) Histochemical method for dipeptidyl aminopeptidase II with a new anthraquinonyl hydrazide substrate. Cell Mol Biol (Noisy-le-grand) 46:1213–1218 [PubMed] [Google Scholar]
- Dikov A, Dimitrova M, Stoineva I, Halbhuber KJ (1999) New tetrazolium method for the histochemical localization of dipeptidyl peptidase IV. Cell Mol Biol (Noisy-le-grand) 45:225–231 [PubMed] [Google Scholar]
- Frerker N, Wagner L, Wolf R, Heiser U, Hoffmann T, Rahfeld JU, Schade J, et al. (2007) Neuropeptide Y (NPY) cleaving enzymes: structural and functional homologues of dipeptidyl peptidase 4. Peptides 28:257–268 [DOI] [PubMed] [Google Scholar]
- Gorrell MD (2005) Dipeptidyl peptidase IV and related enzymes in cell biology and liver disorders. Clin Sci (Lond) 108:277–292 [DOI] [PubMed] [Google Scholar]
- Hanski C, Huhle T, Gossrau R, Reutter W (1988) Direct evidence for the binding of rat liver DPP IV to collagen in vitro. Exp Cell Res 178:64–72 [DOI] [PubMed] [Google Scholar]
- Juillerat-Jeanneret L, Aubert JD, Leuenberger P (1997) Peptidases in human bronchoalveolar lining fluid, macrophages, and epithelial cells: dipeptidyl (amino)peptidase IV, aminopeptidase N, and dipeptidyl (carboxy)peptidase (angiotensin-converting enzyme). J Lab Clin Med 130:603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl T, Chwalisz WT, Wedekind D, Hedrich HJ, Hoffmann T, Jacobs R, Pabst R, et al. (2003) Localization, transmission, spontaneous mutations, and variation of function of the Dpp4 (Dipeptidyl-peptidase IV; CD26) gene in rats. Regul Pept 115:81–90 [DOI] [PubMed] [Google Scholar]
- Kruschinski C, Skripuletz T, Bedoui S, Tschernig T, Pabst R, Nassenstein C, Braun A, et al. (2005) CD26 (dipeptidyl-peptidase IV)-dependent recruitment of T cells in a rat asthma model. Clin Exp Immunol 139:17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankas GR, Leiting B, Roy RS, Eiermann GJ, Beconi MG, Biftu T, Chan CC, et al. (2005) Dipeptidyl peptidase IV inhibition for the treatment of type 2 diabetes: potential importance of selectivity over dipeptidyl peptidases 8 and 9. Diabetes 54:2988–2994 [DOI] [PubMed] [Google Scholar]
- Maes MB, Dubois V, Brandt I, Lambeir AM, Van der Veken P, Augustyns K, Cheng JD, et al. (2007) Dipeptidyl peptidase 8/9-like activity in human leukocytes. J Leukoc Biol 81:1252–1257 [DOI] [PubMed] [Google Scholar]
- Piazza GA, Callanan HM, Mowery J, Hixson DC (1989) Evidence for a role of dipeptidyl peptidase IV in fibronectin-mediated interactions of hepatocytes with extracellular matrix. Biochem J 262:327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi SY, Riviere PJ, Trojnar J, Junien JL, Akinsanya KO (2003) Cloning and characterization of dipeptidyl peptidase 10, a new member of an emerging subgroup of serine proteases. Biochem J 373:179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehbinder C, Baneux P, Forbes D, van Herck H, Nicklas W, Rugaya Z, Winkler G (1996) FELASA recommendations for the health monitoring of mouse, rat, hamster, gerbil, guinea pig and rabbit experimental units. Report of the Federation of European Laboratory Animal Science Associations (FELASA) Working Group on Animal Health accepted by the FELASA Board of Management, November 1995. Lab Anim 30:193–208 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386 [DOI] [PubMed] [Google Scholar]
- Shingu K, Helfritz A, Zielinska-Skowronek M, Meyer-Olson D, Jacobs R, Schmidt RE, Mentlein R, et al. (2003) CD26 expression determines lung metastasis in mutant F344 rats: involvement of NK cell function and soluble CD26. Cancer Immunol Immunother 52:546–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon P (2003) Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19:1439–1440 [DOI] [PubMed] [Google Scholar]
- Skripuletz T, Schmiedl A, Schade J, Bedoui S, Glaab T, Pabst R, von Horsten S, et al. (2007) Dose-dependent recruitment of CD25+ and CD26+ T cells in a novel F344 rat model of asthma. Am J Physiol Lung Cell Mol Physiol 292:L1564–L1571 [DOI] [PubMed] [Google Scholar]
- Takimoto K, Hayashi Y, Ren X, Yoshimura N (2006) Species and tissue differences in the expression of DPPY splicing variants. Biochem Biophys Res Commun 348:1094–1100 [DOI] [PubMed] [Google Scholar]
- Thompson NL, Hixson DC, Callanan H, Panzica M, Flanagan D, Faris RA, Hong WJ, et al. (1991) A Fischer rat substrain deficient in dipeptidyl peptidase IV activity makes normal steady-state RNA levels and an altered protein. Use as a liver-cell transplantation model. Biochem J 273(Pt 3):497–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji E, Misumi Y, Fujiwara T, Takami N, Ogata S, Ikehara Y (1992) An active-site mutation (Gly633-->Arg) of dipeptidyl peptidase IV causes its retention and rapid degradation in the endoplasmic reticulum. Biochemistry 31:11921–11927 [DOI] [PubMed] [Google Scholar]
- van der Velden VH, Naber BA, van Hal PT, Overbeek SE, Hoogsteden HC, Versnel MA (1999) Peptidase activities in serum and bronchoalveolar lavage fluid from allergic asthmatics—comparison with healthy non-smokers and smokers and effects of inhaled glucocorticoids. Clin Exp Allergy 29:813–823 [DOI] [PubMed] [Google Scholar]
- van der Velden VH, Naber BA, van Hal PT, Overbeek SE, Hoogsteden HC, Versnel MA (2000) Peptidases in the asthmatic airways. Adv Exp Med Biol 477:413–430 [DOI] [PubMed] [Google Scholar]
- van der Velden VH, Wierenga-Wolf AF, Adriaansen-Soeting PW, Overbeek SE, Moller GM, Hoogsteden HC, Versnel MA (1998) Expression of aminopeptidase N and dipeptidyl peptidase IV in the healthy and asthmatic bronchus. Clin Exp Allergy 28:110–120 [DOI] [PubMed] [Google Scholar]
- Zagha E, Ozaita A, Chang SY, Nadal MS, Lin U, Saganich MJ, McCormack T, et al. (2005) DPP10 modulates Kv4-mediated A-type potassium channels. J Biol Chem 280:18853–18861 [DOI] [PubMed] [Google Scholar]