Abstract
Fluorescence in situ hybridization (FISH) banding approaches are standard for the exact characterization of simple, complex, and even cryptic chromosomal aberrations within the human genome. The most frequently applied FISH banding technique is the multicolor banding approach, also abbreviated as m-band, MCB, or in its whole genomic variant multitude MCB (mMCB). MCB allows the differentiation of chromosome region–specific areas at the GTG band and sub-band level and is based on region-specific microdissection libraries, producing changing fluorescence intensity ratios along the chromosomes. The latter are used to assign different pseudocolors to specific chromosomal regions. Here we present the first bacterial artificial chromosome (BAC) array comparative genomic hybridization (aCGH) mapped, comprehensive, genome-wide human MCB probe set. All 169 region-specific microdissection libraries were characterized in detail for their size and the regions of overlap. In summary, the unique possibilities of the MCB technique to characterize chromosomal breakpoints in one FISH experiment are now complemented by the feature of being anchored within the human DNA sequence at the BAC level. (J Histochem Cytochem 56:487–493, 2008)
Keywords: multicolor banding approach, array comparative genomic hybridization, microdissection, fluorescence in situ hybridization banding
The characterization of chromosomal rearrangements and the corresponding breakpoint regions by simple, rapid, and reliable approaches is one of the main interests in human cytogenetics. The GTG banding (G-bands by Trypsin using Giemsa; Seabright 1971) technique is a well-established approach that is easy to perform and relatively inexpensive. Although it is still used as the cytogenetic gold standard, it has some profound technical limitations (Liehr et al. 2006b). The fluorescence in situ hybridization (FISH) technique, first introduced in the 1980s, was a great step forward in overcoming the restrictions of banding cytogenetics (Pinkel et al. 1986). The new field of “molecular cytogenetics” was born (Chang and Mark 1997) and further advanced during the last decades with the development of approaches (Liehr et al. 2002a) such as multicolor FISH using all human whole chromosome painting probes with 24 colors and/or multicolor spectral karyotyping (Schröck et al. 1996; Speicher et al. 1996). FISH-based banding methods were recently defined as any kind of molecular cytogenetic technique that is used to simultaneously characterize several chromosomal subregions smaller than a chromosome arm (Liehr et al. 2006b). Although FISH banding methods may have quite different characteristics and approaches, they share the ability to produce DNA-specific chromosomal banding (Liehr et al. 2006a). In contrast to standard chromosome banding techniques, which are based on an enzymatic reaction (i.e., trypsin), FISH banding methods are DNA specific. FISH banding methods recently developed include interspersed PCR multiplex-FISH, cross-species color banding (Rx-FISH), the somatic cell hybrid-based chromosome bar code (CBC), and the microdissection-based multicolor banding (MCB or m-band) and multitude MCB (mMCB) (Liehr et al. 2006b). FISH banding methods that are still under development for the entire human genome include the yeast artificial chromosome (YAC)/bacterial artificial chromosome (BAC)-based CBC, the YAC/BAC-based multicolor banding (Y/B-MCB), and the spectral color banding (SCAN) (Liehr et al. 2006b).
FISH banding methods were applied successfully not only in evolution and radiation biology but also in studies of the nuclear architecture and diagnostic purposes in prenatal, postnatal, and tumor cytogenetics (Liehr et al. 2006a). As summarized on the m-FISH homepage (http://www.med.uni-jena.de/fish/mFISH/mFISHlit.htm), these FISH banding studies led to contributions in almost 200 international publications. Thus, FISH banding methods based on chromosome region–specific probes are already an integral part of cytogenetic research. Among them, the mMCB technique has shown its capability to detect cryptic rearrangements in otherwise “normal” GTG banding metaphase spreads (Weise et al. 2003; Karst et al. 2006).
However, until now, all FISH banding approaches, except for the YAC/BAC-based ones (Liehr et al. 2002b), have had no connection to the relative map position in the human genome sequence. This problem was overcome by hybridizing DNA from region-specific libraries, on which the MCB approach is based, to BAC-based array comparative genomic hybridization (aCGH). Interestingly, the previously published MCB probe set with 138 region-specific libraries (Liehr et al. 2002a) was found to have 47 gaps not covered by this set of probes. Thus, 44 new region-specific probes were generated by microdissection, aCGH mapped, and integrated in the existing probe set. Here we present the first human genome-wide FISH banding approach aligned with the human DNA sequence referred to as probe set array-proven MCB (aMCB).
Materials and Methods
BAC-based aCGH
For BAC-based aCGH, cyanine 5 (Cy5)-labeled MCB libraries were hybridized to a genome-wide array together with cyanine 3 (Cy3)-labeled normal human DNA. This array consisted of 21,658 RP11 BAC clones (average BAC clone size, 142.8 kb) with unique sequences at both ends and tightly distributed insert size that completely covers the entire human genome, thereby minimizing the repeat sequence content (Li et al. 2003). A modified computer program (Biodiscovery; El Segundo, CA) was adapted for the analysis of the library borders. In contrast to the standard aCGH, which uses dye reverse experiments as controls, there is no need for this when using microdissected DNA of interest because of the clear profiles at the expected regions. The rational of this approach was previously described for mapping of marker chromosome–derived DNA (Pietrzak et al. 2007).
Microdissection
Genomic regions that were, according to aCGH, not covered by the established MCB probe set (Liehr et al. 2002a) were subjected to microdissection. Region-specific partial chromosome paints were generated by glass-needle–based microdissection as described previously (Liehr et al. 2002a). Each of the microdissection-derived probes is based on 15–20 chromosomal fragments, which were excised according to the scheme depicted in Figures 1 and 2.
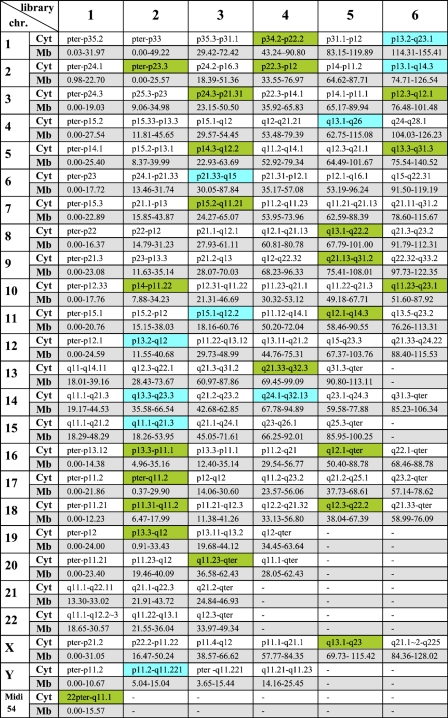
Figure 1.
Overview of all 169 microdissection-derived multicolor banding approach (MCB) libraries with cytogenetic (cyto) and array comparative genomic hybridization (aCGH)-defined borders in megabase pairs (Mb, in respect to UCSC version March 2006) for all human chromosomes. New libraries covering gaps of the previous MCB set (Liehr et al. 2002a) are highlighted in green. New libraries that replaced previous shorter libraries are highlighted in blue. Midi 54 covers the short arms of all acrocentric chromosomes (Mrasek et al. 2001) and is counted as one library.
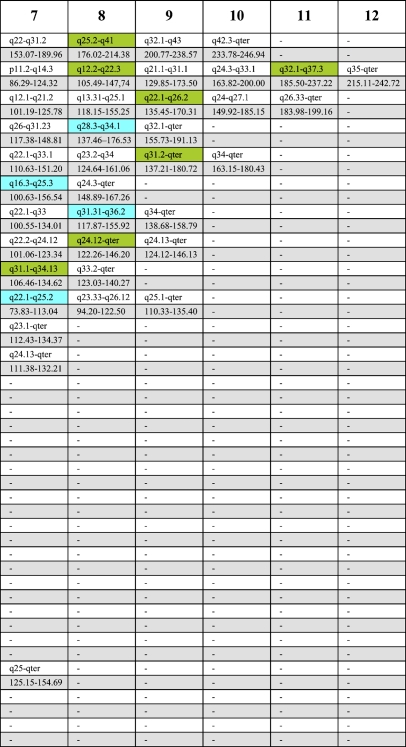
Figure 2.
Graphical overview and labeling scheme for all 169 array-mapped MCB libraries. Double labeling of a single MCB library is indicated with two vertical color lines connected by a horizontal black line. The fluorochrome color key is at the bottom of the legend.
Multicolor Banding Method
Between 3 and 12 microdissection libraries were combined per chromosome. Three to five different fluorochromes were used to label the partial chromosome painting (pcp) probes: SpectrumOrange, SpectrumGreen, TexasRed, Cy5 (Cy5 coupled to avidin; detection of biotinylated probes), and diethylaminocoumarin (DEAC). The probe Midi 54 for the acrocentric chromosomes was added specifically to cover the corresponding short arms of these acrocentric chromosomes (Mrasek et al. 2001). Probe labeling, hybridization, postwashing, signal detection, and image acquisition were performed as previously reported (Chudoba et al. 1999; Liehr et al. 2002a).
Database
All human genome mapping calculations and assignment to cytogenetic bands were determined according to the UCSC database version March 2006 (http://genome.ucsc.edu/cgi-bin/hgGateway).
Results
The first-generation MCB probe set was based on 138 microdissection-derived region-specific pcp probes (Liehr et al. 2002a). No gaps were visible after FISH on normal chromosomes. After aCGH mapping, 47 gaps within this probe set with gaps varying in size between 0.03 and 15.12 Mb were identified. Respectively, a total of 240.86 Mb was not covered by the original MCB probe set. The average size of the gaps was 6.5 Mb. Forty-four new pcps were created by glass-needle–based microdissection to close the gaps. The size of the new pcps was confirmed by FISH and aCGH. Fourteen of those pcps were created to replace others that were too small (Figure 1, labeled in blue). Thirty were newly added to the MCB probe set (Figure 1, labeled in green). Through the principle of overlapping microdissection libraries this new aMCB (array-anchored MCB) set covers 1857.29 Mb of the genome twice, resulting in a 1.63 times coverage of the whole genome. Therefore, the mean overlap of the 169 single libraries was 11 Mb.
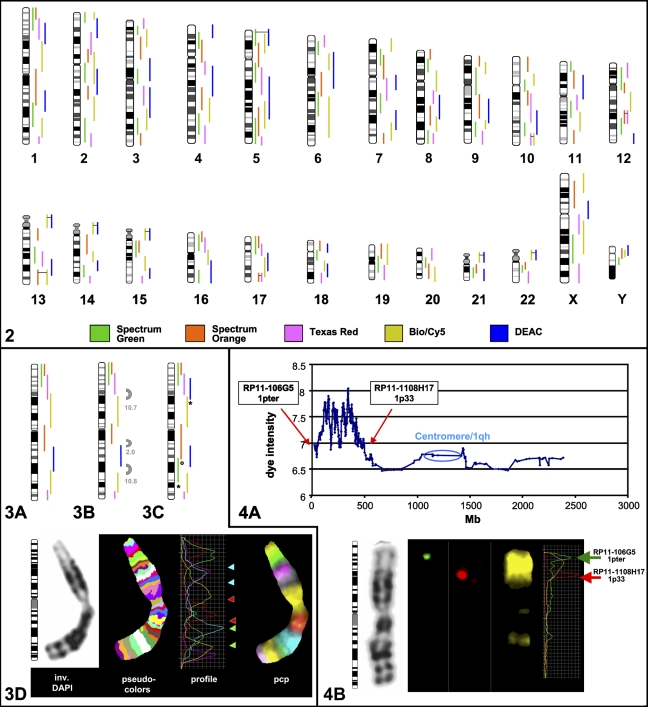
As an example, chromosome 1 is depicted in Figure 3 . The size of the eight overlapping original pcps mapped according to FISH is shown in Figure 3A. aCGH showed three gaps with sizes of 2, 10.7, and 10.8 Mb (Figure 3B). To overcome this limitation, three new pcps were created by microdissection (Figure 3C). aCGH mapping showed that one gap was filled by replacing the old pcp library marked with a circle. The other two gaps were filled by adding new pcps to the old probe set (marked with asterisk).
Figure 3.
Label scheme for chromosome 1: initial set from 2002 (A); three gaps were identified after aCGH ranging in size from 2 to 10.8 Mb (B); asterisk indicates newly microdissected libraries and the circle marks a replacement with a longer library for chromosome 1 (C). Hybridization example for the new MCB set for chromosome 1 (D). From left to right: ideogram, inverted DAPI, pseudocolors, fluorochrome profiles where arrows indicate boundaries of the three new libraries (according to the scheme in Figure 3C), and partial chromosome paints (pcps) of MCB 1 in hybridization colors.
An example of the aCGH is given for the first library of MCB1 (labeled in green in Figures 3A and 3B) in Figure 2A. As previously described (Pietrzak et al. 2007), microdissected and degenerated oligonucleotide primer-PCR amplified DNA was labeled in Cy5 and hybridized to the array together with normal human DNA labeled in Cy3, resulting in a clear plot that indicates where the MCB library was located and also where homologous sequences could be found. For the depicted library, sequence homology around the centromere (small peaks) was also visible by FISH (Figure 4B ) and as previously described (Weise et al. 2005). Expecting only genomic gains, no dye reverse experiments were needed. Therefore the y-axis only indicates the dye intensity.
Figure 4.
aCGH mapping for MCB library 1-2 (A) shows a clear peak where the library is located on chromosome 1 starting from BAC clone RP11-106G5 to RP11-1108H17. Cross-hybridization caused by sequence homology around the centromere and 1q12 heterochromatin can also be detected at the aCGH and fluorescence in situ hybridization (FISH) level (B). Hybridization of MCB 1-2 with the array-mapped BAC clones that mark the borders of this library clearly shows the flaring effect of the FISH technique that covers a broader area than mapped by aCGH (B).
FISH using the most proximal and terminal BAC clones of the region confirmed the results of aCGH and also showed the effect of flaring for the pcp (Figure 4B), especially as shown by the fluorochrome intensity profiles on the right.
Discussion
The initial MCB probe set published in 2002 consisted of 138 microdissection-derived region-specific pcp probes (Liehr et al. 2002a). According to the available FISH-based mapping in 2002, this dissected probe set covered the entire human genome in an overlapping fashion. The reliability of the probe set for characterization of chromosomal breakpoints was proven in >1000 cases (Weise et al. 2002; Manvelyan et al. 2007; unpublished data). Thus, it was surprising to find that the aCGH mapping showed the presence of 47 gaps. The inability to previously detect these gaps is likely because of the flaring effect of the DNA probes used in FISH. This flaring may cause the false-positive formation of pseudocolors when two previously not colocalized regions come together (e.g., because of an inversion) (Schröck et al. 1996). Additionally, flaring leads to apparently larger signals on the chromosomes than the target sequence really is as shown in Figures 3A, 3B, and 4B.
Previously, there was only one case with a complex translocation involving chromosomes 1, 4, and 14 where MCB failed to recognize the inserted region because of either a gap or under coverage in the region. An insertion of 4q27-4q34.1 material in a derivative chromosome 14 was not detectable by MCB but was detectable by SKY (Grasshoff et al. 2003). aCGH confirmed the presence of a 2-Mb gap in the region of 4q34.1 (data not shown).
Forty-four new pcps were created to overcome these limitation and to establish a genome-wide array-based molecularly defined MCB probe set (aMCB) without any apparent sequence gaps (Figure 2) compared with the former one (Liehr et al. 2002a).
In conclusion, the new aMCB offers the same unique possibilities of the MCB technique to characterize chromosomal breakpoints in one FISH experiment but is now complemented by the molecular assignment to the human reference DNA sequence at the BAC level.
Moreover, the suitability of MCB to study the biological origin of chromosomal sub-bands (Lehrer et al. 2004) is now more convenient and straight forward.
Acknowledgments
This work was supported in part by Deutsche Forschungsgemeinschaft Grants WE3617/2-1, LI820/11-1, 436 RUS 17/135/03, 436 RUS 17/109/04, and 436 RUS 17/22/06, Boehringer Ingelheim Fonds, Interdisziplinäres Zentrum für Klinische Forschung Jena (Start-up S16), Stiftung Leukämie, and the Evangelische Studienwerk e.V. Villigst.
We thank Amber Pursley, MS, for comments on the manuscript.
References
- Chang SS, Mark HFL (1997) Emerging molecular cytogenetic techniques. Cytobios 90:7–22 [PubMed] [Google Scholar]
- Chudoba I, Plesch A, Lörch T, Lemke J, Claussen U, Senger G (1999) High resolution multicolor-banding: a new technique for refined FISH analysis of human chromosomes. Cytogenet Cell Genet 84:156–160 [DOI] [PubMed] [Google Scholar]
- Grasshoff U, Singer S, Liehr T, Starke H, Fode B, Schöning M, Dufke A (2003) A complex chromosomal rearrangement with a translocation 4;10;14 in a fertile male carrier: ascertainment through an offspring with partial trisomy 14q13→q24.1 and partial monosomy 4q27→q28. Cytogenet Genome Res 103:17–23 [DOI] [PubMed] [Google Scholar]
- Karst C, Gross M, Haase D, Wedding U, Hoffken K, Liehr T, Mkrtchyan H (2006) Novel cryptic chromosomal rearrangements detected in acute lymphoblastic leukemia detected by application of new multicolor fluorescent in situ hybridization approaches. Int J Oncol 28:891–897 [PubMed] [Google Scholar]
- Lehrer H, Weise A, Michel S, Starke H, Mrasek K, Heller A, Kuechler A, et al. (2004) The hierarchically organized splitting of chromosome bands into sub-bands analyzed by multicolor banding (MCB). Cytogenet Genome Res 105:25–28 [DOI] [PubMed] [Google Scholar]
- Li J, Jiang T, Bejjani B, Rajcan-Separovic E, Cai WW (2003) High-resolution human genome scanning using whole-genome BAC arrays. Cold Spring Harb Symp Quant Biol 68:323–329 [DOI] [PubMed] [Google Scholar]
- Liehr T, Gross M, Karst C, Glaser M, Mrasek K, Starke H, Weise A, et al. (2006a) FISH banding in tumor cytogenetics. Cancer Genet Cytogenet 164:88–89 [DOI] [PubMed] [Google Scholar]
- Liehr T, Heller A, Starke H, Rubtsov N, Trifonov V, Mrasek K, Weise A, et al. (2002a) Microdissection based high resolution multicolor banding for all 24 human chromosomes. Int J Mol Med 9:335–339 [PubMed] [Google Scholar]
- Liehr T, Starke H, Heller A, Kosyakova N, Mrasek K, Gross M, Karst C, et al. (2006b) Multicolor fluorescence in situ hybridization (FISH) applied to FISH-banding. Cytogenet Genome Res 114:240–244 [DOI] [PubMed] [Google Scholar]
- Liehr T, Weise A, Heller A, Starke H, Mrasek K, Kuechler A, Weier HU, et al. (2002b) Multicolor chromosome banding (MCB) with YAC/BAC-based probes and region-specific microdissection DNA libraries. Cytogenet Genome Res 97:43–50 [DOI] [PubMed] [Google Scholar]
- Manvelyan M, Schreyer I, Hols-Herpertz I, Kohler S, Niemann R, Hehr U, Belitz B, et al. (2007) Forty-eight new cases with infertility due to balanced chromosomal rearrangements: detailed molecular cytogenetic analysis of the 90 involved breakpoints. Int J Mol Med 19:855–864 [DOI] [PubMed] [Google Scholar]
- Mrasek K, Heller A, Rubtsov N, Trifonov V, Starke H, Rocchi M, Claussen U, et al. (2001) Reconstruction of the female Gorilla gorilla karyotype using 25-color FISH and multicolor banding (MCB). Cytogenet Cell Genet 93:242–248 [DOI] [PubMed] [Google Scholar]
- Pietrzak J, Mrasek K, Obersztyn E, Stankiewicz P, Kosyakova N, Weise A, Cheung SW, et al. (2007) Molecular cytogenetic characterization of eight small supernumerary marker chromosomes originating from chromosomes 2, 4, 8, 18, and 21 in three patients. J Appl Genet 48:167–175 [DOI] [PubMed] [Google Scholar]
- Pinkel D, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83:2934–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröck E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, et al. (1996) Multicolor spectral karyotyping of human chromosomes. Science 273:494–497 [DOI] [PubMed] [Google Scholar]
- Seabright M (1971) A rapid banding technique for human chromosomes. Lancet 30:971–972 [DOI] [PubMed] [Google Scholar]
- Speicher MR, Gwyn Ballard S, Ward DC (1996) Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nat Genet 12:368–375 [DOI] [PubMed] [Google Scholar]
- Weise A, Heller A, Starke H, Mrasek K, Kuechler A, Pool-Zobel BL, Claussen U, et al. (2003) Multitude multicolor chromosome banding (mMCB): a comprehensive one-step multicolor FISH banding method. Cytogenet Genome Res 103:34–39 [DOI] [PubMed] [Google Scholar]
- Weise A, Starke H, Heller A, Tonnies H, Volleth M, Stumm M, Gabriele S, et al. (2002) Chromosome 2 aberrations in clinical cases characterised by high resolution multicolour banding and region specific FISH probes. J Med Genet 39:434–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise A, Starke H, Mrasek K, Claussen U, Liehr T (2005) New insights into the evolution of chromosome 1. Cytogenet Genome Res 108:217–222 [DOI] [PubMed] [Google Scholar]