Abstract
The CLCA family of proteins consists of a growing number of structurally and functionally diverse members with distinct expression patterns in different tissues. Several CLCA homologs have been implicated in diseases with secretory dysfunctions in the respiratory and intestinal tracts. Here we present biochemical protein characterization and details on the cellular and subcellular expression pattern of the murine mCLCA6 using specific antibodies directed against the amino- and carboxy-terminal cleavage products of mCLCA6. Computational and biochemical characterizations revealed protein processing and structural elements shared with hCLCA2 including anchorage in the apical cell membrane by a transmembrane domain in the carboxy-terminal subunit. A systematic light- and electron-microscopic immunolocalization found mCLCA6 to be associated with the microvilli of non-goblet cell enterocytes in the murine small and large intestine but in no other tissues. The expression pattern was confirmed by quantitative RT-PCR following laser-capture microdissection of relevant tissues. Confocal laser scanning microscopy colocalized the mCLCA6 protein with the cystic fibrosis transmembrane conductance regulator CFTR at the apical surface of colonic crypt cells. Together with previously published functional data, the results support a direct or indirect role of mCLCA6 in transepithelial anion conductance in the mouse intestine. (J Histochem Cytochem 56:495–509, 2008)
Keywords: calcium-activated chloride channel, mCLCA6, non-goblet cell enterocytes, mouse, intestine, cystic fibrosis, CFTR, membrane protein
The clca gene family is a heterogeneous multigene family with cell-type-specific expression patterns and diverse protein structures and processing of different family members (Loewen and Forsyth 2005). The term CLCA had initially been coined due to a novel calcium-activated chloride current observed following heterologous expression of several CLCA proteins in various cell lines, implying a function such as calcium-activated chloride channels. However, several other functions have been proposed and include adhesion molecule function (Abdel-Ghany et al. 2003), bicarbonate channel activity (Thevenod et al. 2003), modulation of other channel proteins (Greenwood et al. 2002; Loewen et al. 2002), inhibition of tumor growth by promoting apoptosis (Elble and Pauli 2001), and protease activity (Pawlowski et al. 2006). To date, the CLCA gene family comprises 18 members known in eight mammalian species with six murine and four human homologs reported (Cunningham et al. 1995; Gandhi et al. 1998; Gruber et al. 1998a; Gruber and Pauli 1999; Komiya et al. 1999; Lee et al. 1999; Romio et al. 1999; Elble et al. 2002; Leverkoehne and Gruber 2002; Evans et al. 2004; Anton et al. 2005; Al-Jumaily et al. 2007). The human and murine CLCA proteins are of considerable interest based on their role in diseases with secretory dysfunction including asthma (Nakanishi et al. 2001; Hoshino et al. 2002; Range et al. 2007). Human hCLCA1 and hCLCA4 and murine mCLCA2 and mCLCA3 have also been proposed to modulate the basic chloride secretory defect and the disease phenotype in cystic fibrosis (CF) patients and mouse models of CF (Ritzka et al. 2004; Leverkoehne et al. 2006; Young et al. 2007).
Murine mCLCA6 has recently been discovered as the murine ortholog of human hCLCA4 (Evans et al. 2004). Both proteins are thought to be expressed in the intestine (Agnel et al. 1999; Evans et al. 2004), and the hCLCA1/hCLCA4 gene locus has been shown to play a role as modulator of the basic intestinal chloride secretory defect in CF (Ritzka et al. 2004). However, the cell type expressing these proteins and their subcellular location are as yet unknown. Also, the function and biomedical significance of the mCLCA6 and hCLCA4 proteins have not yet been established in detail. Of particular interest for determining their function would be the establishment of their protein structures and association with the plasma membrane. Specifically, it seems critical to establish whether mCLCA6 is an entirely secreted protein like its homolog mCLCA3 (Mundhenk et al. 2006) or whether it is anchored to the plasma membrane, similar to hCLCA2 (Elble et al. 2006). Moreover, based on the intestinal expression of both mCLCA3 and mCLCA6, it is tempting to speculate that mCLCA6 may be able to compensate for a loss of mCLCA3 function in mCLCA3 knockout mice as these mice do not display any obvious spontaneous pathology (Robichaud et al. 2005).
Here we present detailed studies on the cellular processing, membrane association, and cellular and ultrastructural localization of the mCLCA6 protein. Results suggest that mCLCA6 occupies an epithelial niche distinct from and complementary to that of mCLCA3 in the murine intestine.
Materials and Methods
Bioinformatics and Antibody Generation
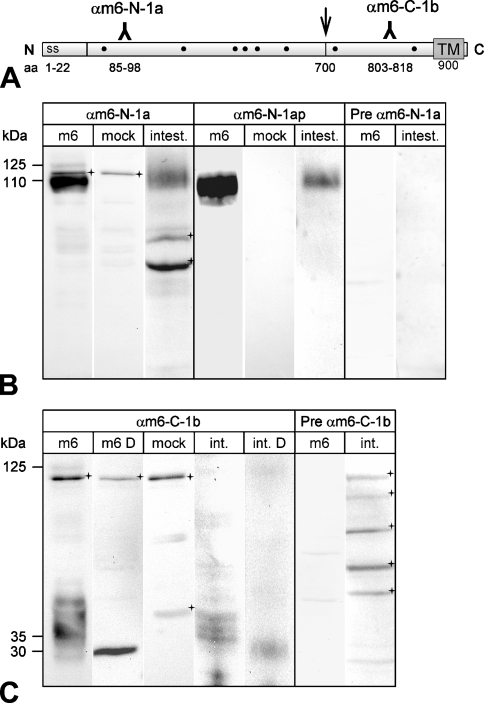
Computational analysis of the mCLCA6 amino acid (aa) sequence (GenBank accession #AAS86332) was performed using the SignalP 3.0 (Nielson et al. 1997), Kyte–Doolittle (Kyte and Doolittle 1982), SOSUI (Hirokawa et al. 1998), HMMtop (Tusnady and Simon 2001), TMPRED (Hofmann 1993), DAS (Cserzo et al. 1997), and PSORT II (Nakai and Horton 1999) algorithms. Peptide sequences of high predicted immunogenicity were selected from the mCLCA6 open reading frame using computer-aided antigenicity analyses. Four oligopeptides were synthesized, two located in the predicted N-terminal cleavage product of mCLCA6 (αm6-N-1 corresponding to aa 85–98, WKDSPQYRRPKQES, and αm6-N-2 corresponding to aa 106–119, VAPPTVEGRDEPYT) and two in the predicted carboxy-terminal cleavage product (αm6-C-1 corresponding to aa 803–818, DNFNNSPRVDTTNLT, and αm6-C-2 corresponding to aa 870–882, QAEPDPDESPSSS). The four oligopeptides were coupled to keyhole limpet hemocyanin (KLH) and used for standard immunization of two rabbits each. Preimmune sera were collected before immunization and used as controls in all immunodetection experiments. The eight antisera were designated αm6-N-1a/αm6-N-1b, αm6-N-2a/αm6-N-2b, αm6-C-1a/αm6-C-1b, and αm6-C-2a/αm6-C-2b. Following initial testings of antiserum reactivities, the anti-N-terminal antibody αm6-N-1a and the anti-carboxy-terminal antibody αm6-C-1b were affinity immunopurified using the corresponding peptides coupled to an EAH–Sepharose column. Immunopurified antisera were designated αm6-N-1ap and αm6-C-1bp (Figure 1A ).
Figure 1.
Generation of antibodies against mCLCA6. (A) Antisera were raised against synthetic peptides corresponding to amino acid (aa) sequences within the amino-(N)-terminal cleavage product (αm6-N-1a) and the carboxy-(C)-terminal cleavage product (αm6-C-1b). Hydrophobicity analyses predicted a signal sequence (ss) at the amino-terminus and a transmembrane domain (TM) at the end of the carboxy-terminal cleavage product. Dots indicate consensus sites for N-linked glycosylation. Arrow shows putative cleavage site. (B) When lysates of heterologously mCLCA6-transfected HEK293 cells were immunoblotted with αm6-N-1a, two specific proteins were detected of 125 and 110 kDa in size, respectively (Lane 1). Lysates of intestinal tissues contained only the 110-kDa protein (Lane 3). Preimmune serum from the same rabbit failed to detect any protein in the same lysates (Lanes 7 and 8). In addition to specific detection of mCLCA6, the unpurified immune serum αm6-N-1a reacted unspecifically with an additional protein of ∼115 kDa in transfected HEK293 cells (Lane 2) and two proteins of ∼70 and 60 kDa (crosses) in the intestinal lysate (Lane 4). However, these proteins were not detected by the immunopurified serum αm6-N-1ap. (C) Anti-carboxy-terminal antibody αm6-C-1b also detected the 125-kDa primary translation product only in overexpressing HEK293 cells (Lane 1), not in intestinal tissue lysates (Lanes 4 and 5). The various protein species of ∼35 kDa were reduced by deglycosylation with PNGase F to a single protein of ∼30 kDa in size (Lane 2, m6 D = cell lysate, deglycosylated; Lane 5, int. D = intestinal lysate, deglycosylated). Vector alone transfected cells served as controls (Lane 3, mock). All antibodies were diluted at 1:500. Crosses = unspecific background staining.
Transient Transfection of HEK293 Cells
HEK293 cells were grown on six-well plates in DMEM containing 10% heat-inactivated fetal calf serum, 2% glutamate, and 1% penicillin/streptomycin (culture medium) at 37C in the presence of 5% CO2. If not mentioned otherwise, cells were washed between medium changes with prewarmed PBS. HEK293 cells were transfected with pcDNA3.1 plasmid containing the mCLCA6 open reading frame (Evans et al. 2004) using Lipofectamine 2000 (Invitrogen; Karlsruhe, Germany) according to the manufacturer's protocol. Cells transfected with pcDNA3.1-vector alone served as negative controls (mock transfected).
Immunoblot Analyses
Forty eight hr after transfection, HEK293 cells were washed three times with PBS followed by incubation in DMEM supplemented with 2% glutamate for 6 hr. Medium was removed, and cells were lysed on ice in 100 μl standard lysis buffer (25 mM Tris–HCl, pH 8.0, 50 mM NaCl, 0.5% DOC, 0.5% Triton X-100) and supplemented with a mixture of protease inhibitors (1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml aprotinin, 5 μg/ml antipain, 5 μg/ml leupeptin, 100 μg/ml trypsin–chymotrypsin inhibitor). To remove any cells, medium samples were spun at 1000 × g for 5 min at 4C, the pellet was discarded, and the remaining supernatant was spun at 10,000 × g for 15 min at 4C. This second supernatant was ethanol precipitated overnight at −20C, and pellets were resuspended in 100 μl of standard lysis buffer. Tissue samples were lysed in standard lysis buffer using a Precellys 24 homogenizer (Peqlab Biotechnologie GmbH; Erlangen, Germany). Medium and lysate samples were boiled in 3-fold SDS-PAGE Laemmli buffer containing 150 mM DTT, each split into two, and separated by 10% SDS-PAGE. Immunoblot analysis was performed as described (Leverkoehne and Gruber 2002) with αm6-N-1a and αm6-C-1b as primary antibodies (1:500).
Acid Release
Forty eight hr after transfection, HEK293 cells transfected with mCLCA6 grown on a 60-mm culture dish were washed three times with prewarmed PBS and split into two equal volumes. Release of non-covalently bound protein by treatment with pH 2.5 was performed as described earlier (Gibson et al. 2005). In brief, cells were pelleted by brief centrifugation and resuspended in either PBS, pH 7.5, or 0.9% NaCl equilibrated to pH 2.5 with acetic acid. Cells were rotated end-over-end for 20 min at 4C and then collected by brief centrifugation. Supernatants were spun at 40,000 × g for 30 min at 4C and then subjected overnight to ethanol precipitation. Precipitates were resuspended in 20 μl standard lysis buffer and boiled in 3-fold SDS-PAGE Laemmli buffer containing 150 mM DTT. Immunoblot analysis was performed as described above.
Endoglycosidase Treatment
Incubation of intestinal lysate, cell lysate, or supernatant with endo H or PNGase F (New England Biolabs; Frankfurt, Germany) prior to SDS-PAGE analysis was performed according to the manufacturer's protocol. Briefly, an aliquot of each sample was incubated with 50 U/μl endo H or 25 U/μl PNGase F, respectively, for 1 hr. One sample was left untreated as a negative control in each experiment.
RT-PCR Assay for Detection of the mCLCA6 Splice Variant
Total RNA was extracted from HEK293 cells transfected with mCLCA6 as described above. Primers used for RT-PCR were designed using the Beacon Designer 3.0 software (Premier Biosoft International; Palo Alto, CA). Primer sequences for mCLCA6 were 5′-AATCAAGGCTGCAATTCAGGTAT-3′ (forward) and 5′-TGTCTTTCACCTCATCAATACAGT-′3 (reverse) with a predicted amplicon size of 122 bp. To discriminate between mCLCA6 and its splice variant mCLCA6sv (Evans et al. 2004), an additional primer pair was designed as follows: 5′-TACAGATGAAGCCCAGAACAAT-3′ (forward) and 5′-TCCAGGTGACAAGAAAGAATGT-3′ (reverse), spanning the portion of exon 10 absent in mCLCA6 with a predicted amplicon size of 191 bp when amplifying mCLCA6 and 126 bp when amplifying mCLCA6sv. The optimized 25-μl reaction mix contained 2.5 μl 10X buffer, 300 nM of each primer, 100 mM probe, 5.5 mM MgCl2, and RNase free water (Roth; Karlsruhe, Germany) to a final volume of 25 μl. Thermal profile used was 2 min at 95C followed by 34 cycles of 50 sec at 95C, 40 sec at 57C, and 20 sec at 72C with a time increment of 2 sec and finally 10 min at 72C.
Animals and Tissue Processing
Five male and five female 12-week-old C57BL/6 mice were sacrificed by cervical dislocation in accordance with national guidelines. The following tissue samples were collected and immersion fixed in 4% neutral-buffered formalin: liver, gallbladder, spleen, kidneys, urinary bladder, pancreas, oral cavity, parotid and sublingual salivary glands, esophagus, stomach, duodenum, jejunum, ileum, cecum, colon, rectum, nasal cavity, trachea, lung, heart, adrenal glands, thyroid glands, ovaries, uterus, mammary glands, testes, epididymides, male accessory sex glands (seminal vesicular glands, coagulating glands, ampullary glands, bulbourethral glands), lymph nodes, brain (cortex, cerebellum, brainstem, medulla), eyes, Harderian gland, skin, adipose tissue, skeletal muscle, and bone.
Immunohistochemistry
Immunohistochemical (IHC) analyses were performed as described previously (Leverkoehne and Gruber 2002). Briefly, fixed tissue samples were embedded in paraffin, cut at 4-μm thickness, and mounted on adhesive glass slides for IHC. Consecutive tissue sections were routinely stained with hematoxylin and eosin (HE) for histological examination. For IHC, the avidin–biotin–peroxidase complex (ABC) method (Vectastain Elite ABC Kit; Vector Laboratories, Burlingame, CA) was applied with antigen retrieval by 15-min microwave heating (750 W) in 10 mM citric acid, pH 6.0, supplemented with 0.05% Triton X-100. Slides were incubated with the purified sera αm6-N-1ap and αm6-C-1bp or the respective preimmune sera (dilutions ranging from 1:100 to 1:40,000) in PBS containing 1% BSA in coverslides at 4C overnight followed by biotinylated secondary goat anti-rabbit antibodies (1:200) and horseradish peroxidase-coupled streptavidin. DAB was used as substrate for color development. Slides were counterstained with hematoxylin, dehydrated through ascending graded ethanol, cleared in xylene, and coverslipped.
Laser-capture Microdissection, RNA Isolation, and Reverse Transcription
For laser-capture microdissection, isolation of total RNA and quantitative real-time polymerase chain reaction (qRT-PCR) tissue samples were shock frozen in liquid nitrogen and kept at −85C until further use. Total RNA was extracted from whole tissue samples using the Trizol method (Invitrogen) and purified using the RNeasy Mini Kit (Qiagen; Hilden, Germany) according to the manufacturer's protocol including a digestion with RNase-free DNase I (Qiagen). Five hundred ng total RNA was reverse transcribed into 20 μl cDNA (Omniscript; Qiagen) using random hexamer primers (Promega; Mannheim, Germany) in the presence of RNase Out (Invitrogen) according to the manufacturer's protocols.
Frozen jejunum samples from three mice were cut into multiple consecutive cross-sections of 6-μm thickness and mounted onto glass slides coated with a polyethylene naphthalate membrane (PALM membrane slides; PALM Microlaser Technologies, Martinsried, Germany). Sections were fixed for 2 min in 95% ethanol at −20C and air dried for 5 min followed by staining with hematoxylin dissolved in diethylpyrocarbonate (DEPC)-treated water for 2 min, rinsing in DEPC-treated water for 2 min, and staining with 1% eosin dissolved in DEPC-treated water for 30 sec. Sections were dehydrated in ascending graded ethanol (30%, 50%, 70%, 100%), air dried at room temperature for 5 min, and stored at −80C. Total RNA was isolated from four different locations: luminal third, medial third, and basal third of the jejunal villi and jejunal smooth muscle. A total area of 1.5 × 106 μm2 cells from each region of interest was separated by laser microdissection and laser-pressure catapulted into the caps of 0.5-ml Eppendorf tubes containing 60 μl RLT buffer (RNeasy Mini Kit; Qiagen). The tubes were closed, inverted, briefly centrifuged, and filled to 350 μl with RLT buffer. Total RNA was extracted and purified according to the manufacturer's protocol including a digestion with RNase free DNase I (Qiagen). Eight μl total RNA was reverse transcribed into 21 μl cDNA (SuperScript III First-Strand Synthesis System; Invitrogen) using random hexamer primers in the presence of RNase Out following the manufacturer's protocol.
Quantitative Real-time Polymerase Chain Reaction
Primers and probes used for quantitative real-time polymerase chain reaction (qPCR) were designed using the Beacon Designer 3.0 software (Premier Biosoft International; Palo Alto, CA). Primer sequences for mCLCA6 were described above. The corresponding Taqman probe sequence was 5′-FAM-TGGCTGTGCTGTCCTCACCATCAC-TAMRA-3′. Primer pairs and probes were designed to specifically discriminate among all known mCLCA homologs and to encompass an intron to exclude amplification of contaminating genomic DNA. No cross-reactions could be detected when linearized cloned cDNA samples of mCLCA1, mCLCA2, mCLCA3, and mCLCA5 or PCR-derived fragments of mCLCA4 were used as templates containing 107 to 109 copies. To correct for variations in RNA amounts and cDNA synthesis efficacy, a segment of the cDNA-encoding murine elongation factor 1α (EF1α) was amplified in parallel in all experiments as internal housekeeping gene standard (Gruber and Levine 1997). Primer sequences for EF1α were 5′-CAAAAACGACCCACCAATGG-3′ (forward) and 5′-GGCCTGGATGGTTCAGGATA-3′ (reverse) with a predicted amplicon size of 68 bp. The corresponding Taqman probe sequence was 5′-FAM-AGCAGCTGGCTTCACTGCTCAGGTG-TAMRA-3′. Gen-Bank accession numbers for mCLCA6 and EF1α were AY560902 and BC003969, respectively. QRT-PCR and data analyses were conducted using the MX 3000P Quantitative PCR System and MX Pro Software (Stratagene; La Jolla, CA). PCR reactions were carried out in 96-well polypropylene plates covered with optical caps (8× strips; Stratagene). The plates contained triplicates of each cDNA sample, duplicates of no-template controls, and duplicates of positive controls. The optimized 25-μl reaction mix contained 12.5 μl qPCR MasterMix Plus Low ROX (Eurogentec; Seraing, Belgium), 300 nM of each primer, 100 mM probe, 1 μl cDNA, 5.5 mM MgCl2 (mCLCA6 only), and RNase free water (Roth) to a final volume of 25 μl. The thermal profile used was 10 min at 95C followed by 40 cycles of 15 sec at 95C and 1 min at 60C. cDNA samples were used with primers and probes for both mCLCA6 and EF1α on the same plate to ensure equal amplification conditions. QRT-PCR protocols were initially established and optimized using 10-fold serial dilutions (ranging from 10−2 to 10−6 ng/μl or 109 to 103 copies of mRNA) of purified mCLCA6-pcDNA3.1 (Evans et al. 2004) or purified (QIAquick PCR Purification Kit; Qiagen) PCR-derived fragments (1114 bp) generated from cDNA templates from mouse colon for EF1α. Positive controls had highly reproducible amplification efficacies of between 95% and 105%.
Quantification of Target Gene Expression
For each sample analyzed, the mean cycle threshold (Ct) value and its corresponding standard deviation (SD) were given by the MX Pro Stratagene analysis software. Ct value was calculated based on the normalized baseline corrected fluorescence (ΔRN). Relative gene expressions were calculated with the ΔCt-method (ratio = 2 Ct EF1α − Ct GOI = 2 ΔCt) (von Smolinski et al. 2005). Ratios represent the copy numbers of the gene of interest (GOI) relative to the internal reference (EF1α) and were thus normalized to the housekeeping gene EF1α as internal standard.
Statistical Analyses
For each cDNA sample analyzed the SD of the triplicates was calculated. Differences of expression levels were considered statistically significant for p values ≤0.05 in a two-group t-test.
Immune Electron Microscopy
Immune electron microscopy was performed as described earlier (Leverkoehne and Gruber 2002). In brief, samples of the small intestine were embedded in gelatin capsules filled with LR White resin containing LR White accelerator (Plano; Wetzlar, Germany), and resin was allowed to polymerize at room temperature overnight. Ultrathin sections cut at 60–90 nm were collected on uncoated 180-mesh nickel grids (Pelco International; Clovis, CA). For immunolabeling, grids were immersed upside down in drops of the respective solution. After blocking of aldehydes with PBS containing 50 mM glycine, sections were blocked in PBS containing 0.5% BSA and 0.1% liquid gelatin (washing buffer) and 5% heat-inactivated normal goat serum. Purified sera αm6-N-1ap and αm6-C-1bp were incubated in washing buffer at 4C overnight in a humid chamber (dilutions ranging from 1:600 to 1:12,000) followed by repeated washes in washing buffer and by incubation for 1 hr with 10-nm gold particle-conjugated goat anti-rabbit immunoglobulins (Sigma–Aldrich; Munich, Germany) diluted at 1:40 in washing buffer. Sections were postfixed, contrasted for 15 min in aqueous uranyl acetate (Merck; Darmstadt, Germany), and washed in distilled water. Grids were air dried and examined with a Zeiss EM 10 CR transmission electron microscope (Zeiss; Oberkochen, Germany). Negative controls were included using immune sera from rabbit against irrelevant antigens as primary antibodies that failed to yield any signals in the relevant cell types at dilutions of 1:2000 or higher.
Co-localization With CFTR
For confocal laser scanning microscopy, 10-μm tissue sections were processed as described above with minor modifications. For double staining, fresh murine intestinal tissue samples were fixed in 3% (w/v) paraformaldehyde for 16 hr, prior to standard paraffin embedding. Paraffin sections were cut at 5 μm and mounted on adhesive glass slides. Deparaffinization and rehydration were followed by antigen retrieval by boiling the sections in 0.01 M sodium citrate buffer in a microwave oven at 750 W for intervals of 7 min, 3 min, and 3 min. Sections were cooled for 45 min and endogenous peroxidase was blocked using 0.6% w/v H2O2 in PBS containing 0.1% w/v sodium azide for 30 min followed by repeated washes in PBS and washes in PBS containing 0.5% BSA. Sections were incubated at room temperature with the primary anti-CFTR-antibody R3195 diluted 1:50 in PBS containing 0.5% BSA for 90 min. Antibody R3195 (Doucet et al. 2003) was a kind gift of Dr. Hugo DeJonge (University of Rotterdam, The Netherlands). After washing in PBS containing 0.5% BSA, sections were incubated with secondary FITC-labeled antibody (Sigma–Aldrich) diluted 1:80 in PBS containing 0.5% BSA for 60 min and washed in PBS containing 0.5% BSA and then in PBS only. Slides were blocked in PBS containing 2% BSA and 20% heat-inactivated normal goat serum followed by incubation with the purified antibody αm6-N-1ap in PBS (1:100) containing 1% BSA in cover slides at 4C overnight. Sections were washed in PBS/Triton X-100 and incubated at room temperature for 1 hr with lissamine rhodamine sulfonyl chloride (LRSC)-labeled fluorescent antibody (Jackson Immunoresearch Europe Ltd; Newmarket, UK) diluted 1:50 in PBS followed by repeated washes in PBS/Triton X-100. Sections were counterstained with To-Pro-3-iodide (Invitrogen; Breda, The Netherlands) and analyzed using a confocal microscope (TCS SP2; Leica, Wetzlar, Germany). FITC was excited at 495 nm, and emission of green fluorescence was detected at 517 nm. LRSC was excited at 570 nm and emission of red fluorescence was detected at 590 nm. Single optical scans were repeated several times and recorded separately to be subsequently combined as an average of three images.
Results
Antibody Generation
A hallmark of virtually all CLCA proteins discovered so far is a posttranslational cleavage of the precursor molecule, resulting in a larger N-terminal and a smaller carboxy-terminal cleavage product of ∼90–110 and 35 kDa, respectively. Previous studies on the biochemistry and expression patterns of other CLCA proteins have either used antibodies directed only against the N-terminal subunit (Leverkoehne and Gruber 2002; Connon et al. 2005; Elble et al. 2006) or epitope-tagged constructs (Gruber et al. 1998b; Gruber and Pauli 1999; Evans et al. 2004; Gibson et al. 2005; Elble et al. 2006; Mundhenk et al. 2006). To study both cleavage products without risk of artifacts associated with epitope tagging, we generated specific antibodies against the amino- and carboxy-terminal cleavage products of the mCLCA6 protein (Figure 1A). Sizes of the proteins detected by these antibodies in lysates of mCLCA6-transfected HEK293 cells and in lysates of mouse intestine are consistent with the general model previously proposed for CLCA protein processing (Gruber et al. 2002). Antibody αm6-N-1a detected a faint 125-kDa and a 110-kDa protein in mCLCA6 overexpressing HEK293 cells, corresponding to the glycosylated precursor molecule and the glycosylated N-terminal cleavage product, respectively (Figure 1B). When applied on intestinal tissue lysates, the same antibody detected only the 110-kDa protein, most likely representing the glycosylated N-terminal cleavage product. Consistently, only the cleavage products but not the precursor protein have previously been detected in tissue lysates of other CLCA proteins (Leverkoehne and Gruber 2002; Mundhenk et al. 2006). Preimmune serum failed to detect any proteins of relevant sizes in the lysates of transfected cells or intestinal tissue lysates, and unspecific reactions with a few other proteins were eliminated when using the immunopurified antiserum αm6-N-1ap (Figure 1B, compare Lanes 2 and 3 with Lanes 5 and 6). Unfortunately, the faint 125-kDa band of the precursor was poorly detected by the immunopurified serum; therefore, the following immunoblot experiments were performed with the non-immunopurified antibody αm6-N-1a. Antibody αm6-C-1b that was raised against the carboxy-terminal cleavage product also faintly detected the 125-kDa precursor molecule in heterologously transfected cells and several poorly defined protein species of ∼35 kDa, most likely representing different glycovariants of the carboxy-terminal cleavage product (Figure 1C). Again, only the cleavage product, but not the full-length polypeptide, was detected in the tissue lysate. The presence of different glycovariants was confirmed by reduction of the various low molecular mass bands to a single band of ∼30 kDa in size (Figure 1C, Lane 2). Immunopurification of the αm6-C-1b antiserum resulted in a complete loss of its reactivity on Western blots, probably because of strong immobilization of high affinity antibodies on the purification column that were insolubilizable by the elution buffer used. Both the additional antibodies αm6-N-2a and αm6-C-2a directed against the amino- and the carboxy-terminal cleavage products, respectively, showed a reaction pattern similar to αm6-N-1a and αm6-C-1b (data not shown). An RT-PCR assay designed to detect the shorter splice variant of mCLCA6 (Evans et al. 2004) detected the full-length open reading frame only but not the splice variant both in transfected HEK293 cells and the intestinal lysates (data not shown).
Cellular Processing and Association With the Plasma Membrane
Distinct cellular processing and associations with the plasma membrane have been reported for different CLCA proteins (Gibson et al. 2005; Elble et al. 2006; Mundhenk et al. 2006). To unravel which of these models may be true for mCLCA6, we performed computer-aided sequence analyses followed by biochemical assays using heterologously transfected HEK293 cells, finally verified on lysates of murine intestinal tissues. The SignalP 3.0 algorithm predicted a signal sequence at the amino-terminus, consistent with the protein entering the secretory pathway. A single potential transmembrane domain was predicted by Kyte–Doolittle, SOSUI, and DAS algorithms at ∼aa 885–911 (Kyte–Doolittle ∼aa 900, SOSUI aa 885–908, and DAS aa 885–908), implying a cytoplasmic tail of no less than 13 hydrophilic aa. In contrast, the TMPred and PSORTII algorithms predicted two potential transmembrane regions, one at aa 436–456 and one close to the carboxy-terminus between aa 889 and 911 (TMPred: aa 889–911 and PSORT/PSORT II: aa 889–905). Thus, computer-aided predictions of potential transmembrane domains for mCLCA6 resulted in a consistently predicted transmembrane region close to the carboxy terminus and a possible additional transmembrane region in the predicted N-terminal cleavage product, predicted by two of the five algorithms used.
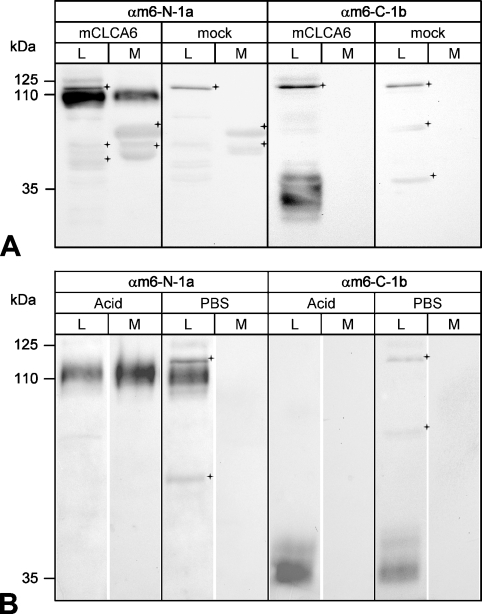
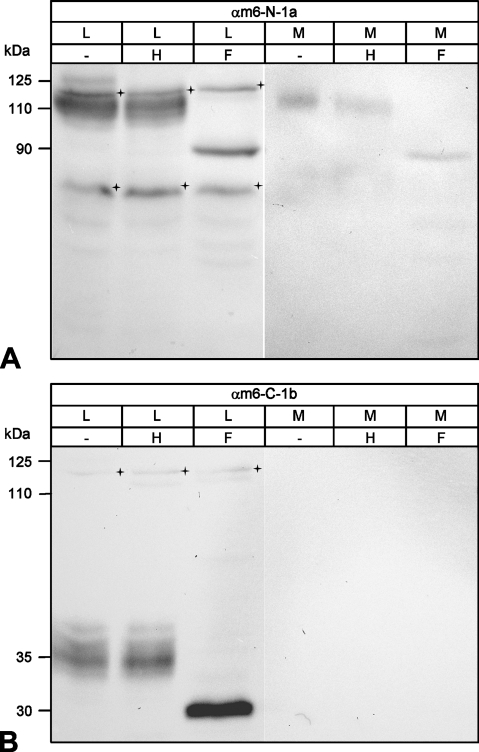
To corroborate these predictions biochemically, the cell lysate and conditioned supernatant of transiently mCLCA6 transfected HEK293 cells were immunoblotted with antibodies αm6-N-1a and αm6-C-1b. A precursor molecule of ∼125 kDa was detected in the cell lysate by antibody αm6-N-1a (Figure 2A ). This precursor molecule was never detected in the supernatant. In contrast, the N-terminal cleavage product of ∼110 kDa was present both in the cell lysate and in the supernatant 6 hr after replacement of the medium (Figure 2A). However, the carboxy-terminal cleavage product of ∼35 kDa in size was detected in the cell lysate only but not in the supernatant (Figure 2A). In the intestinal lysate, both cleavage products were detected when using the respective antibodies (Figures 1B and 1C). An acid-release experiment was performed as described (Gibson et al. 2005) to establish whether mCLCA6, like hCLCA1 and hCLCA2, can be released from the plasma membrane by mild acidic hydrolysis. Similar to hCLCA1 and hCLCA2 (Gibson et al. 2005; Elble et al. 2006), the N-terminal cleavage product of mCLCA6 was released into the medium following treatment with mild acid, whereas the carboxy-terminal cleavage product was not (Figure 2B). We then examined the glycosylation patterns of the mCLCA6 proteins before and after cleavage of the precursor molecule to further characterize the cellular processing and to localize the cellular compartment where protein cleavage occurs. Cell lysates and supernatants were treated with endoglycosidases endo H or PNGase F to identify the extent and kind of glycosylation. The 125-kDa precursor molecule of mCLCA6 was sensitive to both Endo H and PNGase F (Figure 3A ), indicated by its disappearance in Lanes 2 and 3, where the deglycosylated product is likely obscured by lower molecular mass species. Thus, the precursor molecule shows an immature glycoprotein pattern of the high-mannose type. In contrast, the N-terminal subunit found in the cell lysate and in the supernatant and the carboxy-terminal cleavage product found in the cell lysate only were fully resistant to endo H but sensitive to PNGase F (Figure 3). The N-terminal cleavage product was reduced in size to a protein of ∼90 kDa, whereas the carboxy-terminal cleavage product was reduced to an ∼30-kDa product. Glycosylation patterns of both cleavage products detected here are consistent with the six consensus glycosylation sites predicted in the N-terminal cleavage product (Figure 1A) and two glycosylation sites predicted in the carboxy-terminal cleavage product earlier (Evans et al. 2004). In conclusion, results of the deglycosylation experiments indicated that cleavage of the precursor molecule most likely occurs after its exit from the endoplasmic reticulum and that only the two cleavage products have passed the Golgi apparatus.
Figure 2.
Cellular processing of mCLCA6. (A) Immunoblotting analyses identified both the amino- and the carboxy-terminal cleavage products of mCLCA6 in the cell lysate (L, Lanes 1 and 5) but only the N-terminal subunit in the conditioned medium (M, Lane 2). HEK293 cells transiently transfected with mCLCA6 were analyzed by immunoblotting using antibodies αm6-N-1a (1:500) or αm6-C-1b (1:500), respectively. For analysis of the conditioned medium, a medium change was performed 42 hr after transfection followed by harvest and analysis of the medium 6 hr later. Vector alone transfected cells served as controls (mock, Lanes 7 and 8). Crosses = unspecific background staining. (B) When cells were incubated at pH 2.5, only the N-terminal cleavage product of mCLCA6 was partially released into the supernatant. Forty eight hr after transfection, HEK293 cells transiently transfected with mCLCA6 were incubated with NaCl, pH 2.5 (pH adjusted with acetic acid) to release non-covalently associated proteins from the plasma membrane. Treatment with PBS served as control. Antibodies αm6-N-1a and αm6-C-1b were diluted at 1:500. Crosses = unspecific background staining.
Figure 3.
Glycosylation patterns of the amino- and carboxy-terminal cleavage products. (A) Deglycosylation experiments identified the N-terminal cleavage product of mCLCA6 as a complex glycosylated mature protein form that has passed the Golgi apparatus. Aliquots of cell lysate (L) and supernatant (M) of HEK293 cells transiently transfected with mCLCA6 were treated with endo H (H), PNGase F (F), or not treated (−). The N-terminal cleavage product of mCLCA6 was resistant to endo H (Lane 2) but was reduced in size to ∼90 kDa when treated with PNGase F (Lane 3). Crosses = unspecific background staining. (B) The carboxy-terminal cleavage product in the cell lysate (L) was also resistant to endo H (Lane 2) and was reduced in size to appear 30 kDa when treated with PNGase F (Lane 3). Crosses = unspecific background staining.
Tissue Expression Pattern of the mCLCA6 Protein and Quantitation of mRNA Expression Levels
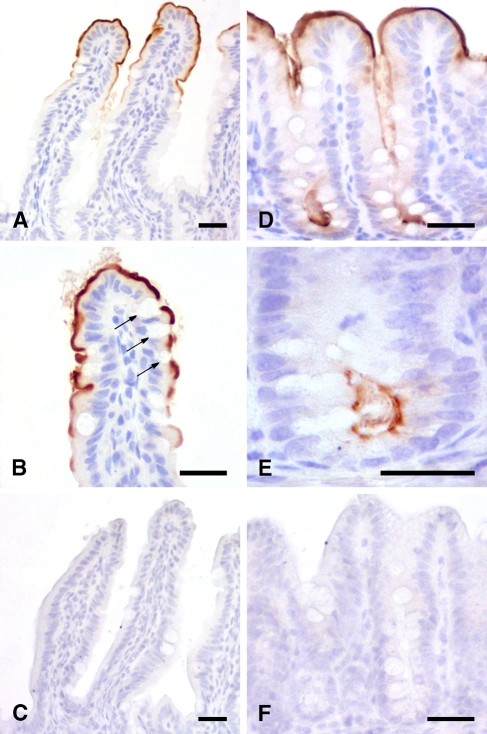
When a systematic IHC analysis was performed including virtually all organs and tissues of a total of five C57BL/6 mice, the mCLCA6 protein was exclusively detected in the intestine (Figure 4 ) but in no other organ or tissue. Only weak staining was observed in the duodenum, with an increasing intensity along the proximal-to-distal axis of the intestinal tract. Similar staining patterns were obtained when the purified antibodies against the amino- or the carboxy-terminal cleavage products of mCLCA6 were used, whereas no staining was observed with the respective preimmune sera. Both the additional antibodies αm6-N-2a and αm6-C-2a directed against the amino- and the carboxy-terminal cleavage product showed a detection pattern similar to αm6-N-1a and αm6-C-1b (data not shown). Interestingly, antisera raised against the amino- and carboxy-terminal cleavage products labeled the same structures in all tissues examined with similar signal strength. Affinity-immunopurified antibodies αm6-N-1ap and αm6-C-1bp were used in all subsequent IHC experiments because of their reduced background staining.
Figure 4.
mCLCA6 is located at the apical surface of non-goblet cell enterocytes. Immunohistochemical analyses localized mCLCA6 at the apical surface of non-goblet cell enterocytes at the villous tips of the jejunum (A,B) and in the crypts of the colon (D,E). Goblet cells stained negative (arrows in B). Tissue sections were incubated with αm6-N-1ap (diluted 1:20,000 in A,B,D,E) or the respective preimmune serum (diluted 1:20,000 in C,F). Color was developed using DAB as substrate (brown) with hematoxylin as counterstain (blue). Bar = 20 μm.
Higher magnifications revealed that the mCLCA6 protein was present only at the apical membrane of enterocytes. In the small intestine, it was expressed only by non-goblet cell enterocytes in the luminal third of the villi, whereas it was present along the entire crypt–lumen axis in the large intestine, with no staining of goblet cells or mucus along the entire intestinal tract. No staining was observed in the stomach.
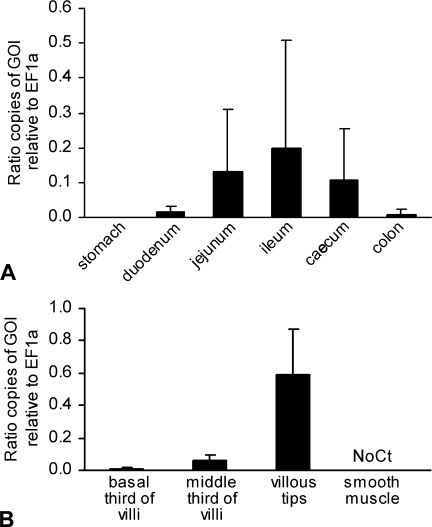
The mCLCA6 expression pattern was verified on the mRNA level. Real-time quantitative reverse transcription–polymerase chain reaction (qRT-PCR) using whole-tissue mRNA extracts confirmed increasing mCLCA6 mRNA expression from the duodenum to the ileum and decreasing levels toward the colon. Maximal average expression levels peaked at 0.2 copies of mCLCA6 mRNA per mRNA copy of the housekeeping gene EF1α (Figure 5A ).
Figure 5.
Quantification of mCLCA6 mRNA expression in the murine intestine using quantitative RT-PCR. (A) Relative copy numbers of mCLCA6 mRNA increased from the duodenum to the ileum and thereafter decreased to the colon, confirming the staining intensities observed using immunohistochemical protein detection. Bars = standard deviations. (B) Laser capture microdissection was used to isolate different segments along the crypt–villus axis of the jejunal villi prior to quantitative RT-PCR. Results confirmed increasing expression levels in the villous tips (compare with Figure 4A). Expression levels are given in ratios of gene of interest (GOI) relative to the copy numbers of the housekeeping gene EF1α (n=5 C57BL/6 mice). No expression was detected in jejunal smooth muscle (No Ct).
Increase of mCLCA6 protein staining toward the luminal third of the crypt–villus axis was verified on the mRNA level using laser-capture microdissection of selected tissue areas followed by qRT-PCR. Results confirmed increasing relative copy numbers of mCLCA6 mRNA from the basal third of the villus to the luminal third (Figure 5B). During initial establishments of the real-time PCR protocols, no cross-reactivity was detected with the murine homologs mCLCA1, mCLCA2, mCLCA3, mCLCA4, and mCLCA5, respectively.
Ultrastructural Localization
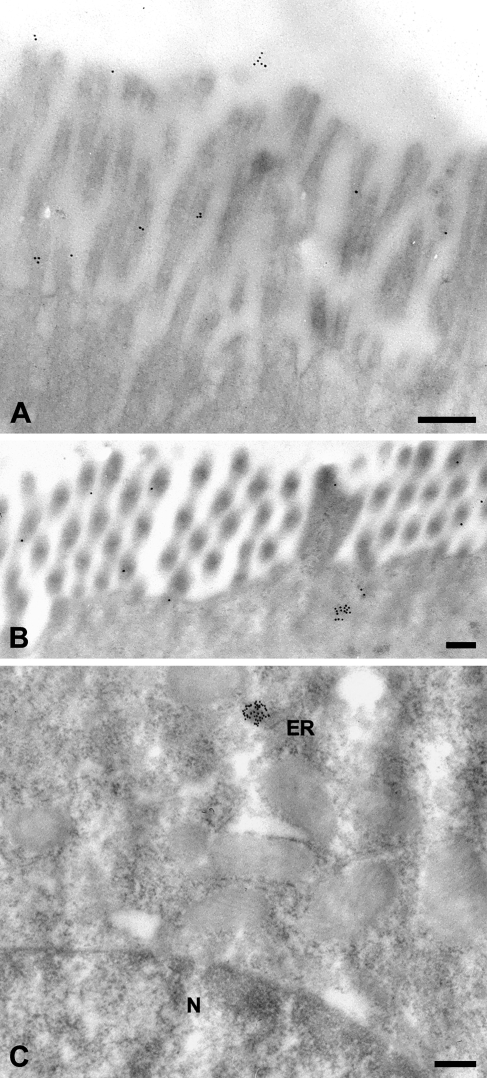
Immune electron microscopic detection of the mCLCA6 protein using 10-nm gold-particle-labeled secondary antibodies identified both the N-terminal and the carboxy-terminal cleavage products of mCLCA6 to be associated with the apical microvilli of enterocytes (Figure 6 ). In addition, small clusters of gold particles were located underneath the terminal actin web consistent with secretory vesicles (Figure 6B) and occasionally associated with the endoplasmic reticulum (Figure 6C). Only for the antibody against the N-terminal cleavage product, considerably fewer signals were observed in the enteric lumen above the microvilli (Figure 6A). No gold particles were detected in the nucleus, mitochondria, or other cellular structures, and no other cell types were labeled.
Figure 6.
Immune electron microscopic localization of the mCLCA6 protein. On the ultrastructural level, the mCLCA6 protein was localized to the brush border of enterocytes (A,B) in small clusters underneath the terminal actin web, in small clusters associated with the endoplasmic reticulum near the nucleus (C), and in small numbers in the lumen of the intestine (A). Tissue sections were incubated with αm6-N-1ap (diluted 1:12,000). Incubation with αm6-C-1bp (1:50) resulted in the same staining pattern except for the lumen, which was negative for the carboxy-terminal cleavage product. Incubation without primary antibody served as negative control. ER, endoplasmic reticulum; N, nucleus. Bar = 200 nm.
Co-localization of mCLCA6 With the CFTR Protein Using Immune Confocal Laser Scanning Microscopy
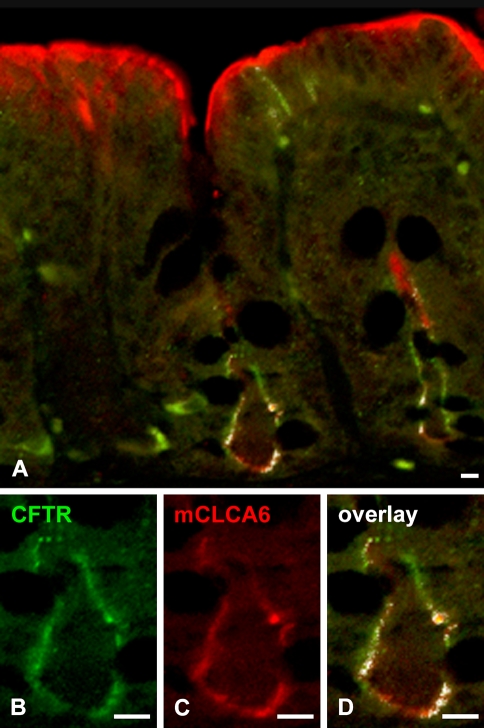
Based on the well-established expression of the CFTR protein in the apical membrane of non-goblet cell enterocytes in the intestinal crypts (Ameen et al. 2000a) and a putative role of CLCA proteins as modulators of the CF phenotype (Ritzka et al. 2004; Leverkoehne et al. 2006; Young et al. 2007), we hypothesized that the mCLCA6 and CFTR proteins may be coexpressed at the apical plasma membrane of enterocytes. IHC detection using antibody R3195 confirmed expression of the CFTR protein in the apical membranes of large intestinal crypt enterocytes with no staining of goblet cells. When these signals were overlaid with the signals identifying the mCLCA6 protein, colocalization was clearly visualized in the apical plasma membranes of crypt epithelial cells in the large intestine (Figure 7 ). However, only mCLCA6, but not CFTR, was strongly expressed at the apical plasma membrane of luminal enterocytes.
Figure 7.
Colocalization of mCLCA6 and CFTR in apical plasma membranes of non-goblet cell enterocytes in crypts of the large intestine. The mCLCA6 protein (red) was visualized along the apical plasma membrane of non-goblet cell enterocytes both at the intestinal lumen and in the crypts (A,C). In contrast, the CFTR protein was only detected at the apical plasma membrane of enterocytes in the deep crypt areas (green in A,B) where it was clearly colocalized with the mCLCA6 protein (A,C). (A,D) Overlay of both signals. (A,B) CFTR staining using antibody R3195 diluted 1:25 with FITC-labeled secondary antibody. (A,C) mCLCA6 staining using antibody αm6-N-1ap diluted 1:100 with LRSC-labeled secondary antibody. Bar = 10 μm.
Discussion
Since their discovery in the mid-1990s, the structure of CLCA proteins has been the subject of considerable debate. The first family member to be characterized biochemically was the bovine endothelial Lu-ECAM-1, alias bCLCA1, which was considered a complex of five proteins of 130, 120, 90, 38, and 32 kDa in size, all being encoded by a single open reading frame (Zhu et al. 1991; Elble et al. 1997). The second family member to be cloned, the bovine Ca–CC, alias bCLCA2, had initially been characterized as a homopolymeric complex of two or four copies of a 38-kDa protein (Ran and Benos 1992; Cunningham et al. 1995). A 2- or 3-fold transmembrane spanning heterodimer of a 90- and 40-kDa protein has been proposed for the human hCLCA1 and hCLCA2 proteins, respectively (Gruber et al. 1998a; Gruber and Pauli 1999). Recently, the latter models have been corrected by further biochemical evidence, and two distinct models now seem to be accepted for different members of the gene family. One class of CLCA proteins consists of a heterodimeric glycoprotein of ∼90–110 and 35-kDa subunits encoded by a single open reading frame. They enter the secretory pathway and are fully secreted by the cell. Prototypes for this model are the human hCLCA1 (Gibson et al. 2005) and the murine gob-5, alias mCLCA3 (Mundhenk et al. 2006). In contrast, a second class with the human hCLCA2 as prototype is clearly anchored to the plasma membrane via a hydrophobic transmembrane domain in the ∼35-kDa carboxy-terminal subunit but otherwise shares many properties of the secreted CLCA proteins (Elble et al. 2006). Computational predictions and biochemical evidence obtained for mCLCA6 in this study clearly show a structural homology with the model proposed for hCLCA2 including plasma membrane association of the 35-kDa carboxy-terminal subunit. Common to both classes of CLCA proteins, a precursor molecule of ∼900 aa is glycosylated and cleaved in the endoplasmic reticulum into amino- and carboxy-terminal cleavage products of ∼110 and 35 kDa in size, respectively, consistent with previous observations (Evans et al. 2004). Most previously proposed models suggested that the two cleavage products remain associated with one another (Gruber et al. 1998a; Mundhenk et al. 2006). Although data obtained here would be consistent with this notion, physical association could not be shown for the two mCLCA6 subunits here because none of the antibodies used was able to immunoprecipitate its target protein (data not shown).
The fate and location of the carboxy-terminal cleavage product of virtually all CLCA proteins have been unclear so far, mostly due to the lack of antibodies specifically directed against this subunit. Several studies have used epitope-tagged versions (Gruber et al. 1998a; Gruber and Pauli 1999; Elble et al. 2006; Mundhenk et al. 2006), but these may yield misleading results by affecting correct protein processing and assembly (Gruber et al. 1998a) and are principally not helpful for in situ detection of natural proteins in their tissues of origin. In this study we were able to localize, for the first time, both the amino- and carboxy-terminal cleavage products of a CLCA family member in their natural microenvironment by IHC and immune electron microscopy using antibodies directed against either subunit. Results clearly indicate that both cleavage products remain colocalized at the apical plasma membrane of non-goblet cell enterocytes at the villous tips and large intestinal crypts. Moreover, IHC staining yielded similar signal strengths in all locations, suggesting equimolarity of the two cleavage products consistent with the notion of a heterodimeric functional protein (Mundhenk et al. 2006).
Similar to hCLCA2 (Elble et al. 2006), only the mCLCA6 carboxy-terminal cleavage product seems to be anchored to the plasma membrane, whereas the N-terminal cleavage product remains associated with the membrane by an as yet unknown mechanism susceptible to mild acid treatment. Although a few signals were observed in the intestinal lumen for the N-terminal cleavage product by immune electron microscopy, the vast majority of this subunit and all of the carboxy-terminal subunit were associated with the membrane. This is at variance with the mCLCA3 protein, which is entirely secreted by intestinal goblet cells (Leverkoehne and Gruber 2002; Gibson et al. 2005; Mundhenk et al. 2006). This difference and the different cellular locations of mCLCA3 and mCLCA6 argue against a compensatory role of mCLCA6 in the mCLCA3 knockout mouse, which has no apparent spontaneous pathology (Robichaud et al. 2005). It remains unknown whether release of the larger N-terminal subunit of mCLCA6 from the membrane as shown here by acid treatment plays any significant role in vivo. Also similar to hCLCA2, computational models predict and biochemical evidence supports the presence of a 13-aa carboxy-terminal cytoplasmic tail, which may be involved in signal transduction pathways or in association with structural cytoplasmic proteins. Clearly, although biochemical data obtained here cast the first light on the structure of the mCLCA6 protein and support the model of Elble et al. (2006), no conclusions can be drawn on the function of the protein in that location. Any role in transepithelial ion conduction would be conceivable for mCLCA6 as a heterodimeric protein at the apical surface of enterocytes, either as part of a more complex channel structure or as a modulator of other channel proteins. Both scenarios would be consistent with the novel calcium-activated chloride current observed in heterologously mCLCA6-transfected cells (Evans et al. 2004). Expression of mCLCA6 at the small intestinal villous tips and along the large intestinal crypts may also suggest a role in absorptive processes that typically take place in these locations. Surprisingly, the small intestinal crypts where secretory processes predominate do not express mCLCA6. Whether this expression pattern may point toward a role in absorption rather than secretion will have to be addressed in future functional studies. Alternatively, although less obvious at this point, a role as adhesion factor or in the control of apoptosis as discussed previously cannot be entirely excluded (Abdel-Ghany et al. 2001; Elble and Pauli 2001).
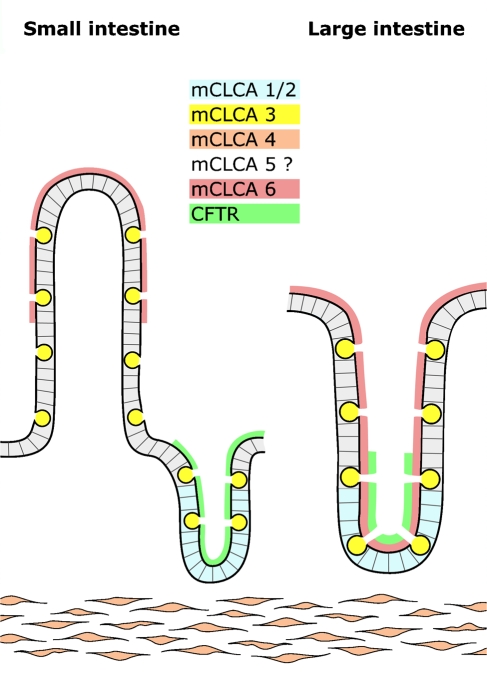
Detection of the mCLCA6 protein on the microvilli of non-goblet cell enterocytes supports the notion of different CLCA proteins filling different functional niches in the intestinal microenvironment (Figure 8 ). Of note, all murine CLCA homologs are expressed in the intestine: mCLCA1/2 has been localized to the crypt epithelial cells (Gruber et al. 1998b), mCLCA3 to the secretory granules of intestinal goblet cells (Leverkoehne and Gruber 2002), and mCLCA4 to smooth muscle cells (Elble et al. 2002). mCLCA5 mRNA has been detected in intestinal RNA with the expressing cell type so far remaining unknown (Evans et al. 2004). Moreover, mCLCA3 is also present in high amounts in the intestinal mucous layer as a secreted glycoprotein (Leverkoehne and Gruber 2002; Mundhenk et al. 2006). Thus, allocation of different murine CLCA family members to different cell types in the intestinal microenvironment supports the model of different evolutionary paths for each homolog and emphasizes their overall relevance. On the other hand, different evolutionary paths seem to exist in different mammalian species for this gene family. For example, murine mCLCA1 and 2 have been found only in the mouse without any orthologs in the human genome (Ritzka et al. 2003). Also, different functional properties have been reported for direct orthologs of different species (Gruber et al. 1998a; Loewen et al. 2004). Therefore, caution is warranted when results on CLCA family members are translated to another species including the model proposed here for the intestine (Figure 8). This seems to be particularly true for the murine mCLCA6 and its human ortholog hCLCA4. The latter, in addition to its intestinal expression, is also strongly expressed in the brain, uterus, and salivary, mammary, and prostate glands as well as several other epithelial tissues (Agnel et al. 1999) where mCLCA6 was not detected in this study, both at the mRNA and protein levels.
Figure 8.
Different structural and functional niches are occupied by distinct CLCA homologs in the murine intestine. The mCLCA1 and mCLCA2 protein (blue) are expressed in basal crypt epithelia of the small and large intestine (Gruber et al. 1998b; Leverkoehne et al. 2006), mCLCA3 (yellow) in secretory vesicles of goblet cells (Romio et al. 1999; Leverkoehne and Gruber 2002), mCLCA4 (orange) in smooth muscle cells (Elble et al. 2002), and mCLCA6 (red) on the apical surface of non-goblet cell enterocytes in the small intestinal villi (left panel) and throughout the large intestinal crypts (right panel). The cell type expressing mCLCA5 in the mouse intestine is so far unknown (Evans et al. 2004). CFTR chloride channel (green) has previously been localized to the apical surface of crypt enterocytes (Ameen et al. 2000a) where it is coexpressed with mCLCA6 in the large intestine.
Relating mRNA expression levels of mCLCA6 to mRNA copy numbers of a standardized housekeeping gene offers the advantage of directly comparing expression levels among different CLCA family members. Because elongation factor 1α has also been used to quantify the expression of other murine CLCA family members in the intestine of the same mouse strain and at the same age (Leverkoehne et al. 2006), the relative mRNA copy numbers of mCLCA6 obtained here can be directly compared with those of other CLCA homologs. This comparison reveals that mCLCA6 is even more strongly expressed than the high-abundant protein mCLCA3 and is only surpassed by mCLCA4 in smooth muscle cells of the tunica muscularis (Leverkoehne et al. 2006). In contrast, mCLCA1/2 and mCLCA4 were found to be expressed ∼100- and 10-fold less, respectively, than mCLCA6. Given the well-established fact that the CFTR protein is a low-abundant protein with only few protein copies on the cell surface in the intestine (Doucet et al. 2003), results of colocalization studies with the CFTR protein in this study should be interpreted with caution. This difference in expression levels may well have obscured coexpression in other cell types due to too low CFTR protein expression to be detected by immunofluorescence microscopy. The low copy numbers of the CFTR protein per cell have already previously yielded confusing results in immunofluorescent assays. Similar to the localization of mCLCA6 in the mouse, Ameen et al. (2000b) also found the CFTR protein to be localized in the microvilli of CFTR high-expressor enterocytes that appear to be well-differentiated enterocytes in the rat intestine. In contrast with our results that confirm previously published data (Doucet et al. 2003; Charizopoulou et al. 2006), Hayden and Carey (1996) found the CFTR protein in intestinal goblet cells but not in enterocytes using a different antibody. In that case, CFTR would also be coexpressed; however, with the mCLCA3 protein. In general, immunolocalization of the CFTR protein both in humans and mice has proven difficult so far due to its low abundance and contradictory signals obtained by different antibodies (Doucet et al. 2003; Mendes et al. 2004).
In any case, colocalization of mCLCA6 with the CFTR protein clearly supports the hypothesis of shared pathways in transepithelial electrolyte secretion and/or absorption (Ritzka et al. 2004; Loewen and Forsyth 2005; Leverkoehne et al. 2006). In particular, the gene locus of the human hCLCA1 and hCLCA4, the ortholog of mCLCA6, has been identified as a modulator of the basic chloride secretory defect in CFTR-deficient CF patients (Ritzka et al. 2004). Questions remain whether CLCA proteins are involved in the alternative calcium-activated chloride conductance that has been proposed to partially compensate for the chloride secretory defect in CF (Rozmahel et al. 1996; Ritzka et al. 2004; Young et al. 2007). Localization of mCLCA6 at the apical surface of non-goblet enterocytes also raises the question of whether the mCLCA6 protein might be colocalized with other ion channels like bestrophins, tweeties, CLC, or the epithelial sodium channel (ENaC). Interestingly, to the best of our knowledge, the distribution pattern observed here for mCLCA6 has not been reported for any other protein so far. Bestrophins are expressed in the apical region of enterocytes in the proximal murine colon (Bakall et al. 2003; Barro Soria et al. in press). Similarly, mRNA of tweety genes has been located to various tissues including large intestine (Suzuki and Mizuno 2004). Within the CLC family of chloride channels, only CLC-2, CLC-6, and CLC-7 are expressed in the intestine (Jentsch et al. 1999). Of note, similar to mCLCA6, the murine CLC-2 protein is expressed by small intestinal epithelial cells only at the villous tips (Gyomorey et al. 2000). However, the protein was localized at the basolateral rather than apical membrane, arguing for a separate function in the same subtype of enterocytes (Catalan et al. 2004; Pena-Munzenmayer et al. 2005). In addition to anion channels, the ENaC sodium channel is also present at the apical membrane of enterocytes (Duc et al. 1994; Renard et al. 1995) where it has recently been shown to be physically associated with the CFTR protein (Berdiev et al. 2007). Thus, localization of mCLCA6 at the apical membrane of enterocytes may not necessarily point toward a functional interaction with only CFTR. Any functional interrelationship with other proteins in this location would be conceivable, including numerous non-channel proteins such as sucrose–isomaltase complex, dipeptidyl peptidase IV, aminopeptidase N, or lactase (Kenny and Maroux 1982). Clearly, colocalization alone of mCLCA6 with CFTR cannot be regarded as proof for shared or mutually compensatory functions. Further work is needed to elucidate any potential functional or spatial interrelationships between CLCA proteins and CFTR or other channel or non-channel proteins at the apical enterocyte membrane.
Acknowledgments
This study was supported by the German Mukoviszidose eV.
Anti-CFTR antibody R3195 was a kind gift from Dr. Hugo DeJonge (University of Rotterdam, The Netherlands). The mCLCA6-pcDNA3.1 clone was a kind gift from Dr. Stella Evans (University of Pennsylvania, Philadelphia, PA). We thank Dr. Marcel Nordhoff and Verena Eckert-Funke for help with confocal laser scanning microscopy and immune electron microscopy, respectively.
References
- Abdel-Ghany M, Cheng HC, Elble RC, Lin H, DiBiasio J, Pauli BU (2003) The interacting binding domains of the β4 integrin and calcium-activated chloride channels (CLCAs) in metastasis. J Biol Chem 278:49406–49416 [DOI] [PubMed] [Google Scholar]
- Abdel-Ghany M, Cheng HC, Elble RC, Pauli BU (2001) The breast cancer β4 integrin and endothelial human CLCA2 mediate lung metastasis. J Biol Chem 276:25438–25446 [DOI] [PubMed] [Google Scholar]
- Agnel M, Vermat T, Culouscou JM (1999) Identification of three novel members of the calcium-dependent chloride channel (CaCC) family predominantly expressed in the digestive tract and trachea. FEBS Lett 455:295–301 [DOI] [PubMed] [Google Scholar]
- Al-Jumaily M, Kozlenkov A, Mechaly I, Fichard A, Matha V, Scamps F, Valmier J, et al. (2007) Expression of three distinct families of calcium-activated chloride channel genes in the mouse dorsal root ganglion. Neurosci Bull 23:293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameen N, Alexis J, Salas P (2000a) Cellular localization of the cystic fibrosis transmembrane conductance regulator in mouse intestinal tract. Histochem Cell Biol 114:69–75 [DOI] [PubMed] [Google Scholar]
- Ameen N, van Donselaar E, Posthuma G, de Jonge H, McLaughlin G, Geuze HJ, Marino C, et al. (2000b) Subcellular distribution of CFTR in rat intestine supports a physiologic role for CFTR regulation by vesicle traffic. Histochem Cell Biol 114:219–228 [DOI] [PubMed] [Google Scholar]
- Anton F, Leverkoehne I, Mundhenk L, Thoreson WB, Gruber AD (2005) Overexpression of eCLCA1 in small airways of horses with recurrent airway obstruction. J Histochem Cytochem 53:1011–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakall B, Marmorstein LY, Hoppe G, Peachey NS, Wadelius C, Marmorstein AD (2003) Expression and localization of bestrophin during normal mouse development. Invest Ophthalmol Vis Sci 44:3622–3628 [DOI] [PubMed] [Google Scholar]
- Barro Soria R, Spitzner M, Schreiber R, Kunzelmann K (In Press) Bestrophin 1 enables Ca2+ activated Cl- conductance in epithelia. J Biol Chem. Published online September 26, 2006 (DOI: 10.1074) [DOI] [PMC free article] [PubMed]
- Berdiev BK, Cormet-Boyaka E, Tousson A, Qadri YJ, Oosterveld-Hut HM, Hong JS, Gonzales PA, et al. (2007) Molecular proximity of cystic fibrosis transmembrane conductance regulator and epithelial sodium channel assessed by fluorescence resonance energy transfer. J Biol Chem 282:36481–36488 [DOI] [PubMed] [Google Scholar]
- Catalan M, Niemeyer MI, Cid LP, Sepulveda FV (2004) Basolateral ClC-2 chloride channels in surface colon epithelium: regulation by a direct effect of intracellular chloride. Gastroenterology 126:1104–1114 [DOI] [PubMed] [Google Scholar]
- Charizopoulou N, Wilke M, Dorsch M, Bot A, Jorna H, Jansen S, Stanke F, et al. (2006) Spontaneous rescue from cystic fibrosis in a mouse model. BMC Genet 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connon CJ, Kawasaki S, Yamasaki K, Quantock AJ, Kinoshita S (2005) The quantification of hCLCA2 and colocalisation with integrin β4 in stratified human epithelia. Acta Histochem 106:421–425 [DOI] [PubMed] [Google Scholar]
- Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A (1997) Prediction of transmembrane α-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng 10:673–676 [DOI] [PubMed] [Google Scholar]
- Cunningham SA, Awayda MS, Bubien JK, Ismailov II, Arrate MP, Berdiev BK, Benos DJ, et al. (1995) Cloning of an epithelial chloride channel from bovine trachea. J Biol Chem 270:31016–31026 [DOI] [PubMed] [Google Scholar]
- Doucet L, Mendes F, Montier T, Delepine P, Penque D, Ferec C, Amaral MD (2003) Applicability of different antibodies for the immunohistochemical localization of CFTR in respiratory and intestinal tissues of human and murine origin. J Histochem Cytochem 51:1191–1199 [DOI] [PubMed] [Google Scholar]
- Duc C, Farman N, Canessa CM, Bonvalet JP, Rossier BC (1994) Cell-specific expression of epithelial sodium channel alpha, beta, and gamma subunits in aldosterone-responsive epithelia from the rat: localization by in situ hybridization and immunocytochemistry. J Cell Biol 127:1907–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble RC, Ji G, Nehrke K, DeBiasio J, Kingsley PD, Kotlikoff MI, Pauli BU (2002) Molecular and functional characterization of a murine calcium-activated chloride channel expressed in smooth muscle. J Biol Chem 277:18586–18591 [DOI] [PubMed] [Google Scholar]
- Elble RC, Pauli BU (2001) Tumor suppression by a proapoptotic calcium-activated chloride channel in mammary epithelium. J Biol Chem 276:40510–40517 [DOI] [PubMed] [Google Scholar]
- Elble RC, Walia V, Cheng HC, Connon CJ, Mundhenk L, Gruber AD, Pauli BU (2006) The putative chloride channel hCLCA2 has a single C-terminal transmembrane segment. J Biol Chem 281:29448–29454 [DOI] [PubMed] [Google Scholar]
- Elble RC, Widom J, Gruber AD, Abdel-Ghany M, Levine R, Goodwin A, Cheng HC, et al. (1997) Cloning and characterization of lung-endothelial cell adhesion molecule-1 suggest it is an endothelial chloride channel. J Biol Chem 272:27853–27861 [DOI] [PubMed] [Google Scholar]
- Evans SR, Thoreson WB, Beck CL (2004) Molecular and functional analyses of two new calcium-activated chloride channel family members from mouse eye and intestine. J Biol Chem 279:41792–41800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Elble RC, Gruber AD, Schreur KD, Ji HL, Fuller CM, Pauli BU (1998) Molecular and functional characterization of a calcium-sensitive chloride channel from mouse lung. J Biol Chem 273:32096–32101 [DOI] [PubMed] [Google Scholar]
- Gibson A, Lewis AP, Affleck K, Aitken AJ, Meldrum E, Thompson N (2005) hCLCA1 and mCLCA3 are secreted non-integral membrane proteins and therefore are not ion channels. J Biol Chem 280:27205–27212 [DOI] [PubMed] [Google Scholar]
- Greenwood IA, Miller LJ, Ohya S, Horowitz B (2002) The large conductance potassium channel β-subunit can interact with and modulate the functional properties of a calcium-activated chloride channel, CLCA1. J Biol Chem 277:22119–22122 [DOI] [PubMed] [Google Scholar]
- Gruber AD, Elble RC, Ji HL, Schreur KD, Fuller CM, Pauli BU (1998a) Genomic cloning, molecular characterization, and functional analysis of human CLCA1, the first human member of the family of Ca2+-activated Cl− channel proteins. Genomics 54:200–214 [DOI] [PubMed] [Google Scholar]
- Gruber AD, Elble RC, Pauli BU (2002) Discovery and cloning of the CLCA gene family. Curr Top Membr 53:367–387 [Google Scholar]
- Gruber AD, Gandhi R, Pauli BU (1998b) The murine calcium-sensitive chloride channel (mCaCC) is widely expressed in secretory epithelia and in other select tissues. Histochem Cell Biol 110:43–49 [DOI] [PubMed] [Google Scholar]
- Gruber AD, Levine RA (1997) In situ assessment of mRNA accessibility in heterogeneous tissue samples using elongation factor-1α (EF-1α). Histochem Cell Biol 107:411–416 [DOI] [PubMed] [Google Scholar]
- Gruber AD, Pauli BU (1999) Molecular cloning and biochemical characterization of a truncated, secreted member of the human family of Ca2+-activated Cl− channels. Biochim Biophys Acta 1444:418–423 [DOI] [PubMed] [Google Scholar]
- Gyomorey K, Yeger H, Ackerley C, Garami E, Bear CE (2000) Expression of the chloride channel ClC-2 in the murine small intestine epithelium. Am J Physiol Cell Physiol 279:C1787–1794 [DOI] [PubMed] [Google Scholar]
- Hayden UL, Carey HV (1996) Cellular localization of cystic fibrosis transmembrane regulator protein in piglet and mouse intestine. Cell Tissue Res 283:209–213 [DOI] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- Hofmann KW (1993) TMbase—A database of membrane spanning protein segments. Biol Chem Hoppe-Seyler 374:A166 [Google Scholar]
- Hoshino M, Morita S, Iwashita H, Sagiya Y, Nagi T, Nakanishi A, Ashida Y, et al. (2002) Increased expression of the human Ca2+-activated Cl− channel 1 (CaCC1) gene in the asthmatic airway. Am J Respir Crit Care Med 165:1132–1136 [DOI] [PubMed] [Google Scholar]
- Jentsch TJ, Friedrich T, Schriever A, Yamada H (1999) The CLC chloride channel family. Pflugers Arch 437:783–795 [DOI] [PubMed] [Google Scholar]
- Kenny AJ, Maroux S (1982) Topology of microvillar membrance hydrolases of kidney and intestine. Physiol Rev 62:91–128 [DOI] [PubMed] [Google Scholar]
- Komiya T, Tanigawa Y, Hirohashi S (1999) Cloning and identification of the gene gob-5, which is expressed in intestinal goblet cells in mice. Biochem Biophys Res Commun 255:347–351 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132 [DOI] [PubMed] [Google Scholar]
- Lee D, Ha S, Kho Y, Kim J, Cho K, Baik M, Choi Y (1999) Induction of mouse Ca2+-sensitive chloride channel 2 gene during involution of mammary gland. Biochem Biophys Res Commun 264:933–937 [DOI] [PubMed] [Google Scholar]
- Leverkoehne I, Gruber AD (2002) The murine mCLCA3 (alias gob-5) protein is located in the mucin granule membranes of intestinal, respiratory, and uterine goblet cells. J Histochem Cytochem 50:829–838 [DOI] [PubMed] [Google Scholar]
- Leverkoehne I, Holle H, Anton F, Gruber AD (2006) Differential expression of calcium-activated chloride channels (CLCA) gene family members in the small intestine of cystic fibrosis mouse models. Histochem Cell Biol 126:239–250 [DOI] [PubMed] [Google Scholar]
- Loewen ME, Bekar LK, Gabriel SE, Walz W, Forsyth GW (2002) pCLCA1 becomes a cAMP-dependent chloride conductance mediator in Caco-2 cells. Biochem Biophys Res Commun 298:531–536 [DOI] [PubMed] [Google Scholar]
- Loewen ME, Bekar LK, Walz W, Forsyth GW, Gabriel SE (2004) pCLCA1 lacks inherent chloride channel activity in an epithelial colon carcinoma cell line. Am J Physiol Gastrointest Liver Physiol 287:G33–41 [DOI] [PubMed] [Google Scholar]
- Loewen ME, Forsyth GW (2005) Structure and function of CLCA proteins. Physiol Rev 85:1061–1092 [DOI] [PubMed] [Google Scholar]
- Mendes F, Doucet L, Hinzpeter A, Ferec C, Lipecka J, Fritsch J, Edelman A, et al. (2004) Immunohistochemistry of CFTR in native tissues and primary epithelial cell cultures. J Cyst Fibros 3(suppl 2):37–41 [DOI] [PubMed] [Google Scholar]
- Mundhenk L, Alfalah M, Elble RC, Pauli BU, Naim HY, Gruber AD (2006) Both cleavage products of the mCLCA3 protein are secreted soluble proteins. J Biol Chem 281:30072–30080 [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24:34–36 [DOI] [PubMed] [Google Scholar]
- Nakanishi A, Morita S, Iwashita H, Sagiya Y, Ashida Y, Shirafuji H, Fujisawa Y, et al. (2001) Role of gob-5 in mucus overproduction and airway hyperresponsiveness in asthma. Proc Natl Acad Sci USA 98:5175–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielson H, Engelbrecht J, Brunack S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10:1–6 [DOI] [PubMed] [Google Scholar]
- Pawlowski K, Lepisto M, Meinander N, Sivars U, Varga M, Wieslander E (2006) Novel conserved hydrolase domain in the CLCA family of alleged calcium-activated chloride channels. Proteins 63:424–439 [DOI] [PubMed] [Google Scholar]
- Pena-Munzenmayer G, Catalan M, Cornejo I, Figueroa CD, Melvin JE, Niemeyer MI, Cid LP, et al. (2005) Basolateral localization of native ClC-2 chloride channels in absorptive intestinal epithelial cells and basolateral sorting encoded by a CBS-2 domain di-leucine motif. J Cell Sci 118:4243–4252 [DOI] [PubMed] [Google Scholar]
- Ran S, Benos DJ (1992) Immunopurification and structural analysis of a putative epithelial Cl− channel protein isolated from bovine trachea. J Biol Chem 267:3618–3625 [PubMed] [Google Scholar]
- Range F, Mundhenk L, Gruber AD (2007) A soluble secreted glycoprotein (eCLCA1) is overexpressed due to goblet cell hyperplasia and metaplasia in horses with recurrent airway obstruction. Vet Pathol 44:901–911 [DOI] [PubMed] [Google Scholar]
- Renard S, Voilley N, Bassilana F, Lazdunski M, Barbry P (1995) Localization and regulation by steroids of the alpha, beta and gamma subunits of the amiloride-sensitive Na+ channel in colon, lung and kidney. Pflugers Arch 430:299–307 [DOI] [PubMed] [Google Scholar]
- Ritzka M, Stanke F, Jansen S, Gruber AD, Pusch L, Woelfl S, Veeze HJ, et al. (2004) The CLCA gene locus as a modulator of the gastrointestinal basic defect in cystic fibrosis. Hum Genet 115:483–491 [DOI] [PubMed] [Google Scholar]
- Ritzka M, Weinel C, Stanke F, Tümmler B (2003) Sequence comparison of the whole murine and human CLCA locus reveals conserved synteny betweenboth species. Genome Lett 2:149–154 [Google Scholar]
- Robichaud A, Tuck SA, Kargman S, Tam J, Wong E, Abramovitz M, Mortimer J, et al. (2005) Gob-5 is not essential for mucus overproduction in preclinical murine models of allergic asthma. Am J Respir Cell Mol Biol 33:303–314 [DOI] [PubMed] [Google Scholar]
- Romio L, Musante L, Cinti R, Seri M, Moran O, Zegarra-Moran O, Galietta LJ (1999) Characterization of a murine gene homologous to the bovine CaCC chloride channel. Gene 228:181–188 [DOI] [PubMed] [Google Scholar]
- Rozmahel R, Wilschanski M, Matin A, Plyte S, Oliver M, Auerbach W, Moore A, et al. (1996) Modulation of disease severity in cystic fibrosis transmembrane conductance regulator deficient mice by a secondary genetic factor. Nat Genet 12:280–287 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Mizuno A (2004) A novel human Cl− channel family related to Drosophila flightless locus. J Biol Chem 279:22461–22468 [DOI] [PubMed] [Google Scholar]
- Thevenod F, Roussa E, Benos DJ, Fuller CM (2003) Relationship between a HCO3−-permeable conductance and a CLCA protein from rat pancreatic zymogen granules. Biochem Biophys Res Commun 300:546–554 [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850 [DOI] [PubMed] [Google Scholar]
- von Smolinski D, Leverkoehne I, von Samson-Himmelstjerna G, Gruber AD (2005) Impact of formalin-fixation and paraffin-embedding on the ratio between mRNA copy numbers of differently expressed genes. Histochem Cell Biol 124:177–188 [DOI] [PubMed] [Google Scholar]
- Young FD, Newbigging S, Choi C, Keet M, Kent G, Rozmahel RF (2007) Amelioration of cystic fibrosis intestinal mucous disease in mice by restoration of mCLCA3. Gastroenterology 133:1928–1937 [DOI] [PubMed] [Google Scholar]
- Zhu DZ, Cheng CF, Pauli BU (1991) Mediation of lung metastasis of murine melanomas by a lung-specific endothelial cell adhesion molecule. Proc Natl Acad Sci USA 88:9568–9572 [DOI] [PMC free article] [PubMed] [Google Scholar]