Abstract
Underacetylation of histone H4 is thought to be involved in the molecular mechanism of mammalian X chromosome inactivation, which is an important model system for large-scale genetic control in eukaryotes. However, it has not been established whether histone underacetylation plays a critical role in the multistep inactivation pathway. Here we demonstrate differential histone H4 acetylation between the X chromosomes of a female marsupial, Macropus eugenii. Histone underacetylation is the only molecular aspect of X inactivation known to be shared by marsupial and eutherian mammals. Its strong evolutionary conservation implies that, unlike DNA methylation, histone underacetylation was a feature of dosage compensation in a common mammalian ancestor, and is therefore likely to play a central role in X chromosome inactivation in all mammals.
Keywords: chromatin, gene silencing, marsupials, evolution, epigenetics
The inactive X hypothesis states that genes on all but a single X chromosome are inactivated in the somatic cells of female mammals (1). Inactivation results in transcriptional repression (2) and serves to equalize X chromosome gene expression between male (XY) and female (XX) mammals. The inactive X chromosome (Xi) is late replicating and forms a heterochromatic body (sex chromatin) on the periphery of the nucleus (3). In eutherian (“placental”) mammals, inactivation occurs at random in early embryogenesis and involves counting of X chromosomes, initiation, spreading, and maintenance of the inactive state (4). X inactivation involves changes to DNA (5), and it is well established that cytosine methylation is involved in the maintenance of X inactivation in humans and mice (6, 7), although it is not the only, or necessarily the primary, change (8, 9). XIST, a gene located in a region of the human and mouse X chromosome shown to be essential for inactivation (the X inactivation center) (10, 11), is expressed only from the inactive X and is an essential component of the initiation of inactivation (12). The non-protein-coding RNA product of XIST is associated with the inactive X, but its mode of action is unclear (13–15).
Acetylation of core histones is a modification of chromatin that has been observed in diverse eukaryotic systems (16–19). Experiments on the silenced mating type genes in the yeast Saccharomyces cerevisiae have shown that hypoacetylation of core histones is directly associated with the silencing mechanism and is not simply a consequence of transcriptional inactivity (16). Reduced histone acetylation may therefore be involved in the transcriptional silencing in regions of the eukaryotic genome that are subject to long-term suppression of transcription. Immunolabeling of human and mouse metaphase chromosomes with antibodies specific for the acetylated isoforms of histone H4 has shown that one X chromosome, as well as the constitutively heterochromatic C banded regions, is depleted in the acetylated forms of histone H4 (20). Experiments with cultured mouse embryonic stem cells have shown that the change in acetylation status of the inactive X in development occurs shortly after XIST expression, the onset of late replication and down-regulation of gene expression, but before the appearance of DNA methylation (21). These results suggest that histone underacetylation is more likely to be involved in the maintenance of the inactive state than in its initiation.
X chromosome inactivation is also observed in marsupial mammals, which diverged from eutherians about 130 million years ago (22). Marsupial X chromosome inactivation differs significantly from that in eutherians, being paternal (23), incomplete, and tissue specific (22) as the result of either piecemeal inactivation (24) or spreading from a putative inactivation center (25). Marsupial X inactivation appears to be less stable, with reactivation of some genes occurring in culture (26). As yet, no marsupial X-linked gene has been identified with significant homology to eutherian XIST (M.J.W. and J.A.M.G., unpublished results). The marsupial inactive X chromosome replicates late in S phase (23, 27), and sex chromatin has been observed, though inconsistently, in some species and tissues (24, 28). Methylation of CpG islands of marsupial X-borne genes has not been detected by either restriction enzyme digestion (26) or bisulfite sequencing (29). Similarities to inactivation in the extraembryonic tissues of eutherian mammals, which is also paternal and incomplete and apparently does not involve stable DNA modification, suggests that marsupial X chromosome inactivation may reflect an ancestral mammalian X inactivation system (22).
We have searched for evidence of differential histone acetylation in the X chromosomes of the Tammar wallaby (Macropus eugenii), a small member of the kangaroo family (Macropodidae) adopted for genetic studies (30). The X chromosome is the only small submetacentric chromosome in the Tammar wallaby complement (2n = 16), and it is distinguished by a prominent interstitial nucleolus organizer region (NOR) on the short arm. Most of the short arm is C-band positive and consists of sequences shared with the Y, which have evidently been added relatively recently to both sex chromosomes (34). This entire constitutively heterochromatic region has been demonstrated to be transcriptionally inert (R. Toder, M.J.W., and J.A.M.G., unpublished results), and replicates synchronously and moderately late in both X chromosomes (27). The long arm (Xq) is differentially replicating (27), and contains the suite of conserved human X-borne genes ascribed to the ancestral mammalian X chromosome (31). Studies of three of these Xq genes in a variety of macropodids have demonstrated that Xq is subject to tissue-specific inactivation, whereas the NOR on the short arm is active on both chromosomes (22, 24, 25). The absence of any inactivation of the NOR despite the absence of corresponding sequences on the Y is consistent with the hypothesis that a recent translocation of the NOR region to the X chromosome has occurred in the macropodid lineage. Although the NOR region has not been recruited into the inactivation pathway, there may be specific regulation of NOR transcript levels that compensates for this difference. The inactive constitutive heterochromatin and the active NOR of the short arm therefore provide internal controls for comparisons of the dosage-compensated long arm for our investigation of the relationship between acetylation level and inactivation.
METHODS
Fibroblast cell cultures were initiated from M. eugenii pouch young (day 14) and adult ear biopsies from captive-bred colonies held by the University of Melbourne, Department of Zoology. Animals were held under permit RP95–015, and samples were received under permit RP90–042 and exported under Australian Nature Conservation Agency permit PWS P952569. Cells were cultured for several weeks in DME media (CSL; Melbourne, Australia) supplemented with 10% fetal calf serum (Cytosystems). Metaphase cells were collected by mitotic shakeoff after a 3-h incubation with Colcemid (0.1 μg/ml; Fluka). Cells were swollen in 0.1 M KCl for 10 min at room temperature. Approximately 5 × 104 unfixed cells were spun at 1800 rpm for 8 min onto washed glass slides using a cytocentrifuge (Shandon, Runcorn, U.K.).
Antiserum R12/8 against histone H4 acetylated at lysine-8 (32) was used in immunolabeling as described previously (33) with minor modifications. Briefly, slides were immersed in KCM buffer (120 mM KCl/20 mM NaCl/10 mM Tris⋅HCl, pH 8/0.5 M EDTA/0.1% Triton X-100) for 10 min at room temperature. Slides were briefly incubated with blocking solution (KCM containing 1% BSA). Areas around the sample were dried and 30 μl of antibody diluted in blocking solution was added. After 1-h incubation at 4°C, slides were washed twice for 5 min in KCM buffer. After incubation with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG second antibody (Sigma) and washing (as above), slides were fixed for 10 min in 4% formaldehyde in KCM. Slides were counter-stained and mounted as described previously, then viewed using a Ziess Axioplan epifluorescence microscope. Black and white photographs were scanned and a colorized composite image was prepared in Adobe Photoshop 3.0. The photographs shown are representative of at least 20 metaphase chromosome spreads. All spreads examined showed similar labeling patterns.
RESULTS AND DISCUSSION
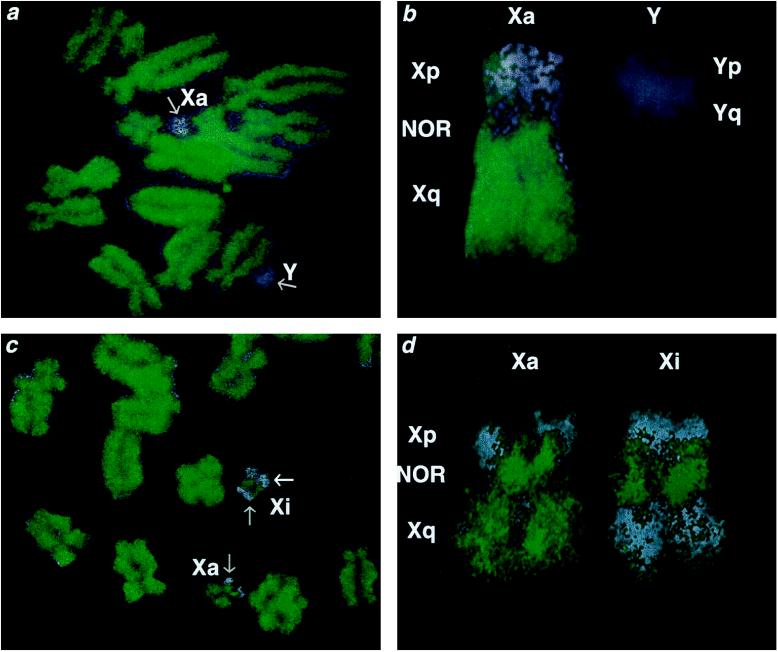
To test the hypothesis that marsupial X inactivation (and therefore ancestral X inactivation) involves histone underacetylation, and to search for molecular correlates of incomplete inactivation, we labeled metaphase chromosomes of Tammar wallaby fibroblasts with an antibody specific to acetylated forms of histone H4 (Fig. 1). Labeling was of approximately equal intensity over all the autosomes, and often showed banding which was less well defined than in human or mouse chromosomes (19, 20). The heterochromatic centromeric regions of some autosomes were consistently unlabeled. Labeling over the X chromosome was different in the late replicating long arm (Xq) and the heterochromatic, NOR-bearing, short arm (Xp). The C-banding distal region of Xp labeled only weakly (i.e., was underacetylated) in both X chromosomes in female cells and on the single X in males. In contrast, the interstitial NOR was strongly labeled on each X chromosome, particularly in the female cell line. NOR labeling has also been described for the mitotic chromosomes of plants (18). The Y chromosome labeled weakly, consistent with its homology to Xp and its heterochromatin content.
Figure 1.
Acetylation status of X chromosomes in M. eugenii fibroblasts. (a and b) A male spread stained with Hoechst 33342 (blue) and labeled with antiserum against acetylated histone H4 (green). An enlargement of the X and Y chromosomes is shown in b. The long arm of the single active X and the NOR label strongly (equivalent to autosomes) and the heterochromatic Xp and Y label weakly (gray arrows in a). (c and d) A female spread stained with Hoechst 33342 (blue) and labeled with antiserum against acetylated histone H4 (green). An enlargement of the two X chromosomes is shown in d. Labeling over Xp is equivalent over both X chromosomes—i.e., weak over the heterochromatin and strong over the NOR (grey arrows in c)—but strikingly different over the long arms. While labeling over one Xq (Xa) is equivalent to that over the autosomes (and the single X in male-derived preparations), labeling over the other Xq (Xi) is very weak (white arrow in c).
In male-derived preparations the long arm of the single active X chromosome was brightly labeled. However, in female-derived preparations strikingly differential labeling was observed between the entire long arms of the two X chromosomes. One Xq was brightly labeled, equivalent to the autosomes, whereas the other showed a distinctly weaker labeling over the whole of its length (Fig. 1), indicating differential underacetylation. The weakly labeled Xq occurred only in females, and must therefore represent the inactive X.
Previous studies indicated that two genes located proximally and distally on Xq of other macropodids escape inactivation in fibroblasts, and it was suggested that only the medial region, around an inactivation center, might be inactive in this tissue (25). However, we observed no evidence for polarity or patterning of underacetylation which might be expected to accompany incomplete inactivation in Tammar wallaby fibroblasts. We conclude that the entire Tammar wallaby Xq region subject to inactivation is underacetylated and that escape from inactivation in fibroblasts must occur either at a level other than acetylation or in small domains undetectable on a chromosomal scale.
Our observation that X chromosome inactivation is accompanied by histone underacetylation in distantly related mammalian infraclasses strongly supports the hypothesis that underacetylation of nucleosome subunits was associated with X chromosome inactivation in an ancestral therian mammal, and it remains a fundamental component of the X inactivation pathway in all mammals. The correlation of histone underacetylation with transcriptional silencing in a wide variety of eukaryotes, including yeast and Drosophila, suggests that it is a ubiquitous control mechanism which was recruited to a function in mammalian dosage compensation. This must have occurred when the X and Y chromosomes began to be differentiated in the common mammalian ancestor more than 130 million years ago, and spread into the recently added region of the eutherian X between 80 and 130 million years ago.
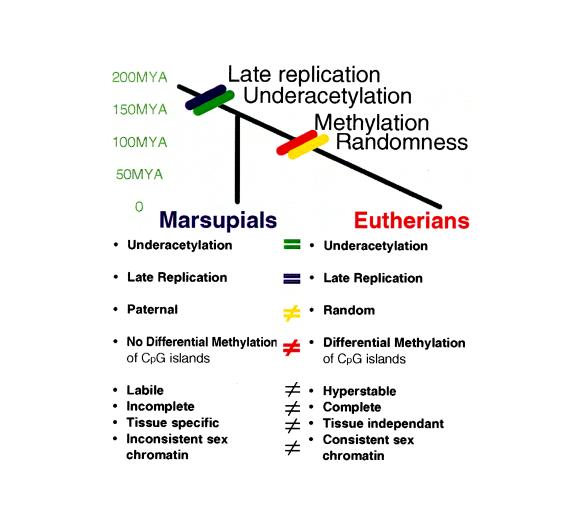
We propose that the more complex, multistep control of X chromosome inactivation in eutherians (8) evolved by the addition of locking mechanisms such as DNA methylation, to produce the complete, hyperstable system in eutherian somatic tissues (Fig. 2). To discover how the histone acetylation differential is established between the active and inactive X chromosomes, and the mechanism by which underacetylation affects expression, marsupials may provide an ideal model system because of the absence of additional levels of regulation.
Figure 2.
Comparison of the features of X chromosome inactivation in marsupial and eutherian mammals indicating that the shared features of late replication and underacetylation must have occurred in a common therian ancestor and the differential features of methylation and random inactivation must have occurred after the marsupial–eutherian split. MYA, million years ago.
Acknowledgments
We thank Jayne Lavender, Rosalia Bruzzese, and Joanne La Rose for their excellent technical assistance, and Prof. Marilyn Renfree, University of Melbourne, Department of Zoology, for the generous provision of material. This work was supported by the Australian Research Council (Grant A09602203) and the Wellcome Trust (Grant 045030/2/95).
ABBREVIATION
- NOR
nucleolus organizer region
References
- 1.Lyon M F. Nature (London) 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 2.Graves J A M, Gartler S M. Somat Cell Mol Genet. 1986;12:275–280. doi: 10.1007/BF01570786. [DOI] [PubMed] [Google Scholar]
- 3.Grumbach M M, Morishima A, Taylor J H. Proc Natl Acad Sci USA. 1963;49:581–589. doi: 10.1073/pnas.49.5.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballabio A, Willard H F. Curr Opin Genet Dev. 1992;2:439–447. doi: 10.1016/s0959-437x(05)80155-8. [DOI] [PubMed] [Google Scholar]
- 5.Liskay R M, Evans R J. Proc Natl Acad Sci USA. 1980;77:4895–4898. doi: 10.1073/pnas.77.8.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohandas T, Sparkes R S, Shapiro L J. Science. 1981;211:393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- 7.Graves J A M. Exp Cell Res. 1982;141:99–105. doi: 10.1016/0014-4827(82)90072-6. [DOI] [PubMed] [Google Scholar]
- 8.Gartler S M, Dyer K A, Graves J A M, Rocchi M. In: Chemistry, Biochemistry and Biology of DNA Methylation. Cantoni G L, Razin A, editors. New York: Liss; 1985. pp. 223–235. [Google Scholar]
- 9.Lock L F, Takagi N, Martin G R. Cell. 1987;48:39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- 10.Brown C J, Ballabio A, Rupert J L, Lafreniere R G, Grompe M, Tonlorenzi R, Willard H F. Nature (London) 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 11.Brockdorff N, Ashworth A, Kay G F, Cooper P, Smith S, McCabe V M, Norris D P, Penny G D, Patel D, Rastan S. Nature (London) 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 12.Penny G D, Kay G F, Sheardown S A, Rastan S, Brockdorff N. Nature (London) 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 13.Brown C J, Hendrich B D, Rupert J L, Lafreniere R G, Xing Y, Lawrence J, Willard H F. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 14.Brockdorff N, Ashworth A, Kay G F, McCabe V M, Norris D P, Cooper P J, Swift S, Rastan S. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 15.Clemson C M, McNeil J A, Willard H F, Lawrence J B. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 17.Turner B M, Birley A J, Lavender J. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 18.Houben A, Belyaev N D, Turner B M, Schubert I. Chromosome Res. 1996;4:191–194. doi: 10.1007/BF02254958. [DOI] [PubMed] [Google Scholar]
- 19.Belyaev N D, Keohane A M, Turner B M. Hum Genet. 1996;97:573–578. doi: 10.1007/BF02281863. [DOI] [PubMed] [Google Scholar]
- 20.Jeppesen P, Turner B M. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 21.Keohane A M, O’Neill L P, Belyaev N D, Lavender J S, Turner B M. Dev Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- 22.Cooper D W, Johnston P G, Watson J M, Graves J A M. Semin Dev Biol. 1993;4:117–128. [Google Scholar]
- 23.Sharman G B. Nature (London) 1971;230:231–232. doi: 10.1038/230231a0. [DOI] [PubMed] [Google Scholar]
- 24.VandeBerg J L, Robinson E S, Samollow P B, Johnston P G. Isozymes Curr Top Biol Med Res. 1987;16:225–253. [PubMed] [Google Scholar]
- 25.Graves J A M, Dawson G W. Genet Res. 1988;51:103–109. doi: 10.1017/s0016672300024113. [DOI] [PubMed] [Google Scholar]
- 26.Kaslow D C, Migeon B R. Proc Natl Acad Sci USA. 1987;84:6210–6214. doi: 10.1073/pnas.84.17.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graves J A M. Exp Cell Res. 1967;46:37–57. doi: 10.1016/0014-4827(67)90407-7. [DOI] [PubMed] [Google Scholar]
- 28.McKay L M, Wrigley J M, Graves J A M. Aust J Biol Sci. 1987;40:397–404. doi: 10.1071/bi9870397. [DOI] [PubMed] [Google Scholar]
- 29.Loebel D A F, Johnston P G. Genome Res. 1996;6:114–123. doi: 10.1101/gr.6.2.114. [DOI] [PubMed] [Google Scholar]
- 30.Hinds L A, Poole W E, Tyndale-Biscoe C H, van Oorschot R A H, Cooper D W. Aust J Zool. 1990;37:223–34. [Google Scholar]
- 31.Graves J A M. BioEssays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- 32.Turner B M, O’Neill L P, Allan I M. FEBS Lett. 1989;253:141–145. doi: 10.1016/0014-5793(89)80947-0. [DOI] [PubMed] [Google Scholar]
- 33.Jeppesen P, Mitchell A, Turner B, Perry P. Chromosoma. 1992;101:322–332. doi: 10.1007/BF00346011. [DOI] [PubMed] [Google Scholar]
- 34.Toder, R., Weinberg, J., O’Brien, P., Maccarone, P. & Graves, J. A. M. (1997) Chromosoma, in press. [DOI] [PubMed]