Abstract
The Xlim-1 gene is activated in the late blastula stage of Xenopus embryogenesis in the mesoderm, and its RNA product becomes concentrated in the Spemann organizer at early gastrula stage. A major regulator of early expression of Xlim-1 is activin or an activin-like signal. We report experiments aiming to identify the activin response element in the Xlim-1 gene. The 5′ flanking region of the gene contains a constitutive promoter that is not activin responsive, whereas sequences in the first intron mediate repression of basal promoter activity and stimulation by activin. An intron-derived fragment of 212 nt is the smallest element that could mediate activin responsiveness. Nodal and act-Vg1, factors with signaling properties similar to activin, also stimulated Xlim-1 reporter constructs, whereas BMP-4 did not stimulate or repress the constructs. The mechanism of activin regulation of Xlim-1 and the sequence of the response element are distinct from activin response elements of other genes studied so far.
Dorsoventral polarity in the embryo of Xenopus laevis is preset by cytoplasmic rotation before first cleavage (1), leading to the formation of a dorsalizing center named the Nieuwkoop center (1–4). Signaling through the Wnt pathway, at least its downstream component β-catenin, is critical in Nieuwkoop center function (5–7). In addition to the dorsalizing role of Wnt/β-catenin, certain factors in the transforming growth factor β (TGF-β) superfamily, including Vg1, activin, and nodal, mediate dorsal mesoderm-inducing signals (reviewed in refs. 3, 4, 8, and 9). The molecular consequences of these signals include activation of multiple genes in temporally and spatially regulated fashion, generating complex relationships between domains of expression of different regulatory genes by the gastrula stage. The embryonic body plan emerges as a result of interactions among the gastrula dorsalizing center, the Spemann organizer, and opposing influences present in ventrolateral regions of the embryo (10–17).
A hierarchy of gene activation appears to control gene expression and patterning of the Xenopus embryo. The early dorsally expressed homeobox gene siamois (18) is induced by the Wnt signaling pathway, but not by dorsal mesoderm-inducing factors of the TGF-β family (19–21). The slightly later organizer-specific gene goosecoid is activated both by Wnt-like and activin-like signals (22, 23), whereas the more broadly expressed genes Mix.1 or Mix.2 (24) are controlled directly by an activin-like signal mediated by XMAD2/Smad2 (25). The Xlim-1 gene belongs to a group of dorsal genes that are activated slightly later by an activin-like signal (26). Xlim-1 is expressed in chordal and prechordal mesoderm (27), has a role in dorsalization of mesoderm and the induction and patterning of the neural plate (28, 29), and its mouse ortholog, Lim-1 or Lhx-1, is essential for the formation of the head (30). Part of the role of Xlim-1 during gastrulation could be mediated by activation of the signaling molecule chordin whose expression is not directly stimulated by activin (29–32). In addition to homeobox genes, the activin-like signal stimulates the expression of the forkhead gene XFD-1/XFKH-1/pintallavis during gastrulation (33–35).
The mechanism of transcriptional activation of regionally restricted genes in early development is of interest because it will help explain how pattern emerges during embryogenesis. The distinct nature of the Wnt and activin-like signals is seen in the gsc promoter where separate sequence elements in the 5′ upstream region mediate Wnt and activin responses (23). A distinct activin response element (ARE) has been identified in the in the 5′ upstream region of the Mix.2 promoter (36) whose activation is mediated by XMAD2 in combination with the forkhead class factor FAST-1 (25). The AREs of the XFD-1′ (37) and HNF1α genes (38) also reside in the respective 5′ upstream regions. A remarkable feature of these AREs is the fact that they share very little sequence similarity (see ref. 37).
In this paper we describe the sequences required for activin responsiveness of the Xlim-1 gene. Not only did we fail to detect substantial similarity between the previously reported AREs and the relevant element in Xlim-1, but in addition the control element is located in the first intron rather than in the 5′ upstream region.
MATERIALS AND METHODS
Genomic clones for Xlim-1 were isolated by screening a genomic library (39) with a cDNA probe (26). Reporter constructs were generated by inserting genomic fragments into the luciferase vector pGL2-basic (Promega). The exonuclease III method (40) was used to generate deletions. To modify intron sequences, the intron was cut at the unique BamHI site and resected with ExoIII to different extent. A construct from which much of intron I was removed (nt 878–2,284) was modified by insertion of the following polylinker (intron in lower case, added nucleotides are in uppercase letters; see Figs. 1 and 3): aggaccATGTCCATGGGACGTCAGTTGGATCCAGGCCTAAGCTTAGTCATGCATGCGGCCGCAGATCCCGCCTTAACTGgctaga. The luciferase construct containing the resected intron I and polylinker is named Ex-2Δ3, or Δ3 (see Fig. 1). Portions of the intron generated by PCR were reintroduced into Δ3 at the StuI site in the polylinker.
Figure 1.
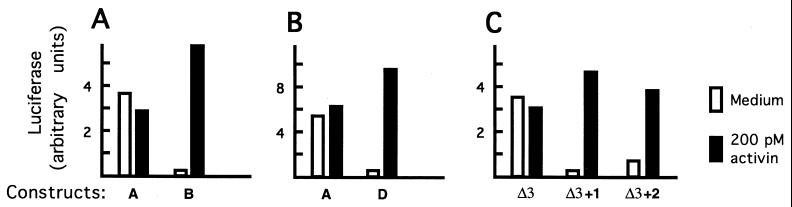
Activin responsiveness of Xlim-1-luciferase fusion constructs. The top line depicts the organization of the Xlim-1 gene. Lines represent noncoding regions, boxes are protein coding (filled) or untranslated (open) regions; introns III and IV are not to scale, their approximate size in kb is indicated. Constructs are symbolized below and identified both by a descriptive term (e.g., Ex-1 means fusion in exon 1) and a letter identifier. The Ex-2Δ3 (Δ3 for short) construct was used as a vector in which to test different regions of intron I that is shown expanded in the lower part of the figure. The numbers correspond to nucleotide positions in the Xlim-1 sequence (Fig. 3). The constructs labeled Δ3+1, etc., were derived from Δ3 by the addition of the intron sequence indicated; thus, Δ3+1 contains 300 nt, from 1,837 to 2,137, inserted into the polylinker between positions 887 and 2,285; see also Materials and Methods and text. The activin response of the various constructs is summarized as follows: negative constructs had responses very close to unity; construct Δ3+3 gave a low response (1.3- to 4-fold) and is therefore listed as +/−; the response in constructs labeled + varied with embryo batch, but was never lower than 4 and usually much higher (see Fig. 2). N is the number of experiments; in the cases identified as 1a, additional deletions with close-by endpoints were tested, confirming the results.
Figure 3.
Sequence of the Xlim-1 gene including a portion of the 5′ flanking region through exon 1, intron I, and part of exon 2. Exon sequences are in uppercase letters, flanking and intron sequences are in lowercase letters. The start site of transcription was determined by primer extension (M. Rebagliati, M.L.R., and I.B.D., unpublished work) and is consistent with the longest cDNA clones that were obtained; the start of transcription is termed position +1. The initiation codon in underlined (599–601). These sequences have been deposited with GenBank under accession number AF013242. The previously published Xlim-1 cDNA sequence (26) has accession number X63889.
A luciferase construct containing the basal thymidine kinase (TK) promoter (41) was modified by insertion of Xlim-1 intron sequences upstream of the promoter.
Transcriptional activity was assayed after injection of 50 pg plasmid DNA into the animal region of two-cell embryos. Animal explants were dissected at stage 8.5–9.5 (42) and cultured to equivalent stage 11 (about 4 hr) with or without 200 pM activin A, and luciferase activity was assayed in triplicate.
RESULTS
Activin Response in Xlim-1 Is Controlled by a Site in the First Intron.
As a start in unraveling the mechanism of regulation of Xlim-1 expression we focused on its response to activin in early embryogenesis. In animal explants, the Xlim-1 gene is not expressed but can be turned on by activin independent of protein synthesis (26). To analyze this activation, we injected Xlim-1-luciferase fusions into two-cell embryos, cultured animal explants with and without activin, and assayed for luciferase activity. Reporter constructs with upstream regions fused to luciferase in frame in the first exon were active but not stimulated by activin (Figs. 1 and 2 A and B), irrespective of the length of the flanking region up to 5 kb. We have not analyzed the upstream region in detail, but a construct starting at position −109 still supported reporter activity, whereas deletion of all flanking sequences and fusion at position +477 resulted in an inactive construct. The sequence of the 5′ flanking region (Fig. 3) does not contain a TATA box.
Figure 2.
Activin response of Xlim-1–luciferase fusion constructs. Constructs identified in Fig. 1 were injected into the embryo, and animal explants were cultured in control medium or in the presence of 200 pM activin and assayed for luciferase.
To test for a possible ARE in an intron we generated a construct of Xlim-1 containing all four introns, fused in-frame to luciferase just upstream of the stop codon; construct B was weakly active in the absence of activin and strongly stimulated by it (Figs. 1 and 2A). Experiments with different batches of embryos are not directly comparable in quantitative terms (see also ref. 23); therefore we summarize the behavior of constructs in Fig. 1 and illustrate individual experiments in Fig. 2. Resection of the construct from the 3′ end showed that the relevant sequences were located in intron I; construct C, fused in exon 2, was at least as responsive to activin as construct B (Fig. 1). Activin responsiveness did not depend on a long 5′ flanking region, as seen by gradual resection (data not shown); construct D, containing 350 nt of 5′ flanking region, was fully responsive to activin (Figs. 1 and 2B). From these experiments we conclude that the Xlim-1 gene carries a constitutive promoter in its proximal 5′ flanking region and an activin-sensitive silencer in its first intron.
Deletion Analysis of the ARE in Intron I.
Fig. 3 presents the sequence of a portion of the Xlim-1 gene, including intron I, which is just under 2 kb in length. To localize the ARE we generated deletions with exonuclease starting from the BamHI site at position 1,598. The most informative of several deletions tested are shown in Fig. 1. Construct Ex-2Δ1 (nt 1,373–1,932 deleted) was unresponsive, whereas Ex-2Δ2 (1,412–1,777) did respond to activin; this and additional deletions indicated that sequences downstream of about position 1,800 are required for the activin response. To test for ARE function, a vector was generated from construct D by replacing the central 1.4 kb of intron I by a polylinker, preserving the ends of the intron to allow splicing (construct Ex-2Δ3; Fig. 1; see Materials and Methods). The Δ3 construct is activin unresponsive, like construct A (Figs. 1 and 2C). Different regions of the intron were then generated by PCR and inserted into the polylinker within intron I (Fig. 1). Insertion of a region of 300 nt, positions 1,837–2,137, into the Δ3 construct (Δ3+1) resulted in strong activin responsiveness (Fig. 2C). A fragment of 212 nt internal to the 300-nt fragment (construct Δ3+2) also supported an activin response, but generally with a lower stimulation ratio due to higher unstimulated activity (Fig. 2C). Both the 300- and 212-nt fragments were inserted into Δ3 in reverse orientation; these constructs were as responsive to activin as those with the original orientation (data not shown). Further deletion beyond the boundaries of the 212-nt fragment greatly reduced or abolished the response (Fig. 1). The Xlim-1 intron ARE as defined by these experiments is rather long compared with transcriptional control elements reported in other systems. We attempted to check for possible substructure in the element by deleting sequences in the middle of the 212-nt fragment; such deletions abolished the ARE function. Thus, the 212-nt fragment is the shortest we could identify as a functional ARE.
Transfer of the ARE to a Heterologous Promoter.
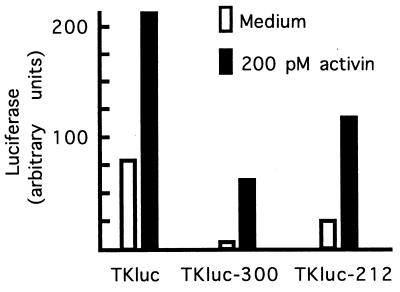
A luciferase gene driven by a basic TK promoter was highly active in the embryo, and its expression was stimulated about 2-fold by activin (Fig. 4); the basis of this modest stimulation is not understood. Insertion of the 300- or 212-nt fragment from Xlim-1 intron I on the 5′ side of the TK promoter yielded constructs that were less active than the basic TK reporter but responded more strongly to activin (Fig. 4). Thus, the ARE in intron I behaved similarly in this heterologous context as in the Xlim-1 gene itself: it led to an inhibition of basal activity which was largely relieved by activin. Whereas control by the ARE appeared less tight in the TK promoter context, these results support the view that the activin-mediated effects are transcriptional.
Figure 4.
The activin response can be transferred to the TK promoter. A luciferase reporter containing the basic TK promoter (41) was modified by insertion of the 300 nt (1,837–2,137) or 212 nt (1,862–2,974) fragment from intron I, and assayed as described in Materials and Methods and Fig. 2.
The sequence in the 300- and 212-nt intron I fragments was compared with that of AREs previously reported in other genes. No meaningful regions of similarity were found in the AREs of Mix.2 (36), goosecoid (23), HNF1α (38), or XFD-1′ (37), all of which are located in the 5′ flanking regions of their respective genes.
Responsiveness to Other Inducing Factors.
While activin is found in the embryo (43), its role in development is not fully understood, even though recent work strongly supports such a role (44). Other TGF-β superfamily members such as Vg1 and nodal have similar signaling properties as activin and are likely involved in dorsal mesoderm induction (45–48). The TGF-β superfamily member BMP-4 acts as a ventralizing factor which can antagonize dorsal influences in mesodermal patterning and neuralization (10, 11, 16, 17). We tested the effect of these factors on the Xlim-1 luciferase reporter D by RNA injection.
Injection of BMP-4 RNA did not stimulate reporter activity and did not inhibit the activin response in animal explants (Fig. 5A). This is different from the results obtained with constructs from the XFD-1′ gene, where BMP-4 abolished the activin response (37). The truncated form of the BMP receptor (ΔBR) acts as a dominant-negative form in the embryo, leading to duplication of dorsal structures by inhibiting BMP-mediated ventralization (49, 50). Whereas ΔBR might thus be expected to activate a dorsal marker gene like Xlim-1, it did not stimulate reporter expression (Fig. 5A); ΔBR also failed to inhibit activin stimulation of the reporter. In contrast, a truncated type II activin receptor, ΔAR (or ΔXAR; refs. 51 and 52) completely blocked stimulation of the Xlim-1 reporter by activin (Fig. 5A).
Figure 5.
Response of construct D, containing the entire intron I, to different factors. The DNA construct was injected together with RNA encoding different proteins, as indicated. In A, animal explants were cultured in control medium or with 200 pM activin; in B all explants were cultured in control medium. The following RNAs were injected at the levels per embryo indicated: globin (control), 400 pg (64); BMP-4, 400 pg (65); ΔBR, truncated BMP receptor type I, 400 pg (49); ΔAR (or ΔXAR), truncated AR type II, 2 ng (51, 52); activin βB, 40 pg (66); nodal, 400 pg (53, 55); Act-Vg1, a chimeric construct between the pro region of activin and the mature region of Vg1, 400 pg (46).
Nodal, known to be required for mesoderm formation in the mouse (53, 54) and able to function in a heterologous system (55), stimulated the activity of the Xlim-1 reporter; this stimulation was blocked by ΔAR (Fig. 5B). Likewise, Act-Vg1, a processing enhanced form of Vg1 (46), stimulated reporter activity and was sensitive to inhibition by ΔAR (Fig. 5B).
The data in Fig. 5 indicate that the Xlim-1 reporter D is stimulated by dorsal mesoderm inducers of the TGF-β superfamily and does not respond to the ventralizing factor BMP-4. We have also tested Xwnt-8 and the dominant-negative form of GSK-3β (3β-KM), both known to be strong dorsalizing factors (7, 56), and basic fibroblast growth factor, a general mesoderm inducing factor (9, 57), for stimulation of the Xlim-1 reporter; none of these factors activated the reporter (data not shown), consistent with the fact that these agents fail to induce the resident Xlim-1 gene in animal explants. The situation is different with retinoic acid, which does induce Xlim-1 in explants and in the whole embryo (26, 27), but failed to induce the reporters tested including construct B. It is possible that the retinoic acid response element for the Xlim-1 gene is located in its 3′ flanking region, as is the case for HoxA1 (58). Thus, the sequences located in intron I of the Xlim-1 gene respond to activin and factors with activin-like inducing potential.
DISCUSSION
Establishment of Differential Expression Domains in the Xenopus Embryo.
The establishment of the body plan in the Xenopus embryo involves multiple inductive events that determine dorsal-ventral polarity, mesoderm induction and patterning, and specification of the neural plate (1, 3, 4, 7–9, 16, 17). These interactions lead to transcriptional readouts beginning at the midblastula transition, which rapidly divide the embryo into regions of distinct gene expression. Dorsalization through a Wnt-like pathway that involves accumulation and nuclear translocation of β-catenin (7) leads in an apparently direct way to the expression of the siamois gene in the dorsovegetal region of the embryo (18). The early dorsalizing or Nieuwkoop center, characterized by β-catenin action and siamois expression, gives rise to the Spemann organizer at the gastrula stage. Organizer-specific genes encoding transcription factors or signaling molecules have been described, whose action is believed to endow the organizer with its ability to establish the embryonic axis (3, 4, 16, 17). Formation of the organizer, as distinct from the Nieuwkoop center, may require an activin-like mesoderm-inducing signal in addition to a β-catenin-derived dorsalizing signal. One reason for this view is the fact that the early organizer-specific gene goosecoid that apparently is expressed slightly later than siamois, has both Wnt and activin responsive elements in its promoter (23).
Xlim-1 may represent the next wave of organizer-specific genes that do not respond directly to dorsalizing cues of the β-catenin type. There are at least two reasons for such a view. First, the initial activation of Xlim-1 is not tightly restricted to the dorsal mesoderm but includes lateral expression (26), as is particularly apparent in the zebrafish (55). Both in frog and fish embryos, the expression pattern rapidly sharpens to become organizer-specific by early gastrula. This pattern suggests that the Xlim-1 gene is activated in response to broader mesodermal cues rather than a strictly dorsal signal. The second point is the presence of an ARE but not of a Wnt response element in the Xlim-1 gene. The fact that our reporter constructs do not respond to Wnt signals could be dismissed by assuming that the relevant sequences were excluded from the constructs, were it not for the excellent correspondence with the results of Carnac et al. (19), who showed that expression of siamois, a major mediator of the Wnt-like signal, activates goosecoid and chordin but not Xlim-1 in animal explants.
If Xlim-1 is activated in dorsal and lateral regions by an activin-like signal, what restricts its expression to the organizer during subsequent development? Our results provide no support for a direct inhibitory role of BMP-4, as was found in the case of the XFD-1′ gene (37). It is possible that BMP-4 affects Xlim-1 expression through other molecules that are present in the marginal zone, where regulation normally takes place, but not in the animal explants which we used as test system.
Heterogeneity of AREs.
Activin or an activin-like signal has a major influence on axis formation and differentiation of the mesoderm in Xenopus. Thus it is not surprising that activin responsiveness has been studied in several genes that are expressed in a temporally and spatially regulated way in Xenopus embryogenesis. The remarkable fact that has emerged from studies so far is the lack of a consensus sequence for the ARE (see ref. 37). The Xlim-1 ARE continues this pattern, having no discernable sequence similarity to previously reported AREs. In fact, Xlim-1 goes even further in establishing a distinct manner of regulation. In the previously reported cases, Mix.2 (36), goosecoid (23), HNF1α (38) and XFD-1′ (37), the AREs are located in the 5′ flanking regions of the respective genes and appear to function as classical enhancers: without the ARE the promoters are quite weak, with the ARE they show a similar low basal level that can be stimulated by activin. Xlim-1 behaves differently in that the 5′ flanking region contains a constitutive promoter that functions well in animal explants but is entirely unresponsive to activin. The ARE, located in intron I, appears to act as an activin-responsive silencer. Constructs that contain the flanking region promoter and a minimum of 212 nt from intron I are transcriptionally inhibited compared with constructs containing only flanking region, but can be activated by the addition of activin. Thus, the basic mechanism of activin regulation of Xlim-1 is quite different from the mechanism of the other genes that have been studied in this context.
How does activin or a similar factor regulate gene expression through AREs of such divergent sequence and, in the case of Xlim-1, divergent mechanism of regulation? The signal of TGF-β factors is transmitted to the cell through a class of heterodimeric serine/threonine kinase receptors that regulate the function of downstream effectors, the Smad proteins (25, 59–63). Smad proteins are translocated to the nucleus where they participate in DNA binding and the regulation of gene activity. The elegant work of Chen et al. (25) has shown that Smad2 (or XMAD2) forms a complex with the newly discovered forkhead-class protein FAST-1 to bind to and activate the promoter of the Mix.2 gene in Xenopus embryos. This work may provide a paradigm for activation of AREs of divergent sequence: while the participation of Smad2 or a homolog is likely in each case, the active complex probably includes different partner proteins in different cases, accounting for the multiple sequences that have been found to act as AREs. In the case of the Xlim-1 gene the situation may be somewhat different. Our results suggest that the intron I ARE is bound to a protein or protein complex in the absence of an activin-like signal, leading to the suppression of the constitutive activity of the upstream promoter. The activin signal, likely mediated by Smad2, is able to relieve this inhibition. Whether this simply means that the inhibitory protein or protein complex is released, or whether it is transformed into an activating complex, is not known. In many of our experiments the activin-stimulated activity of the intron-containing constructs C or D was about equal to the activity of the intron-less construct A (see Fig. 1). However, our experiments showed some quantitative variations, and in several cases the activin-stimulated activity was considerably higher than the constitutive activity of the intron-less construct; this was especially true when activin RNA was injected, a particularly effective way to generate a high local concentration of the factor. These latter experiments suggest that the intron I ARE can mediate net stimulation of transcription beyond the relief of the inhibition that it exerts in the absence of an activin signal.
In conclusion, an activin-like signal has a major role in controlling the expression of the Xlim-1 gene in the organizer region of the Xenopus gastrula embryo. This regulation is mediated through a sequence in the first intron which acts as a silencer in the absence of the signal. The highly variable sequences and manner of regulation of different activin-responsive genes in the early embryo imply that combinatorial signals achieve the complex and highly ordered patterns of gene regulation that characterize the gastrula embryo.
Acknowledgments
We thank D. Melton, D. Kessler, and S. Minucci for plasmids, Genentech for activin, and L. Kodjabachian and M. Rebagliati for critical reading of the manuscript.
ABBREVIATIONS
- AR
activin receptor
- ARE
activin response element
- BR
BMP receptor
- TGF-β
transforming growth factor β
- TK
thymidine kinase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF013242).
References
- 1.Gerhart J, Danilchik M, Doniach T, Roberts S, Rowning B, Stewart R. Development (Cambridge, UK) . 1989;107 Suppl.: 37–51. doi: 10.1242/dev.107.Supplement.37. [DOI] [PubMed] [Google Scholar]
- 2.Gimlich R L, Gerhart J C. Dev Biol. 1984;104:117–130. doi: 10.1016/0012-1606(84)90042-3. [DOI] [PubMed] [Google Scholar]
- 3.Dawid I B. J Biol Chem. 1994;269:6259–6262. [PubMed] [Google Scholar]
- 4.Lemaire P, Kodjabachian L. Trends Genet. 1996;12:525–531. doi: 10.1016/s0168-9525(97)81401-1. [DOI] [PubMed] [Google Scholar]
- 5.Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, Kintner C, Noro C Y, Wylie C. Cell. 1994;79:791–803. doi: 10.1016/0092-8674(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 6.Wylie C, Kofron M, Payne C, Anderson R, Hosobuchi M, Joseph E, Heasman J. Development (Cambridge, UK) 1996;122:2987–2996. doi: 10.1242/dev.122.10.2987. [DOI] [PubMed] [Google Scholar]
- 7.Miller J R, Moon R T. Genes Dev. 1996;10:2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- 8.Kimelman D, Christian J L, Moon R T. Development (Cambridge, UK) 1992;116:1–9. doi: 10.1242/dev.116.Supplement.1. [DOI] [PubMed] [Google Scholar]
- 9.Slack J M. Curr Biol. 1994;4:116–126. doi: 10.1016/s0960-9822(94)00027-8. [DOI] [PubMed] [Google Scholar]
- 10.Jones C M, Lyons K M, Lapan P M, Wright C V, Hogan B L. Development (Cambridge, UK) 1992;115:639–647. doi: 10.1242/dev.115.2.639. [DOI] [PubMed] [Google Scholar]
- 11.Dale L, Howes G, Price B M, Smith J C. Development (Cambridge, UK) 1992;115:573–585. doi: 10.1242/dev.115.2.573. [DOI] [PubMed] [Google Scholar]
- 12.Zimmerman L B, De Jesus-Escobar J M, Harland R M. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 13.Piccolo S, Sasai Y, Lu B, De Robertis E M. Cell. 1996;86:589–598. doi: 10.1016/s0092-8674(00)80132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holley S A, Neul J L, Attisano L, Wrana J L, Sasai Y, O’Connor M B, De Robertis E M, Ferguson E L. Cell. 1996;86:607–617. doi: 10.1016/s0092-8674(00)80134-8. [DOI] [PubMed] [Google Scholar]
- 15.Wilson P A, Hemmati-Brivanlou A. Nature (London) 1995;376:331–333. doi: 10.1038/376331a0. [DOI] [PubMed] [Google Scholar]
- 16.Hemmati-Brivanlou A, Melton D. Cell. 1997;88:13–17. doi: 10.1016/s0092-8674(00)81853-x. [DOI] [PubMed] [Google Scholar]
- 17.Sasai Y, De Robertis E M. Dev Biol. 1997;182:5–20. doi: 10.1006/dbio.1996.8445. [DOI] [PubMed] [Google Scholar]
- 18.Lemaire P, Garrett N, Gurdon J B. Cell. 1995;81:85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- 19.Carnac G, Kodjabachian L, Gurdon J B, Lemaire P. Development (Cambridge, UK) 1996;122:3055–3065. doi: 10.1242/dev.122.10.3055. [DOI] [PubMed] [Google Scholar]
- 20.Brannon M, Kimelman D. Dev Biol. 1996;180:344–347. doi: 10.1006/dbio.1996.0306. [DOI] [PubMed] [Google Scholar]
- 21.Fagotto F, Guger K, Gumbiner B M. Development (Cambridge, UK) 1997;124:453–460. doi: 10.1242/dev.124.2.453. [DOI] [PubMed] [Google Scholar]
- 22.Cho K W, Morita E A, Wright C V, De Robertis E M. Cell. 1991;65:55–64. doi: 10.1016/0092-8674(91)90407-p. [DOI] [PubMed] [Google Scholar]
- 23.Watabe T, Kim S, Candia A, Rothbacher U, Hashimoto C, Inoue K, Cho K W. Genes Dev. 1995;9:3038–3050. doi: 10.1101/gad.9.24.3038. [DOI] [PubMed] [Google Scholar]
- 24.Rosa F M. Cell. 1989;57:965–974. doi: 10.1016/0092-8674(89)90335-8. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Rubock M J, Whitman M. Nature (London) 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 26.Taira M, Jamrich M, Good P J, Dawid I B. Genes Dev. 1992;6:356–366. doi: 10.1101/gad.6.3.356. [DOI] [PubMed] [Google Scholar]
- 27.Taira M, Otani H, Jamrich M, Dawid I B. Development (Cambridge, UK) 1994;120:1525–1536. doi: 10.1242/dev.120.6.1525. [DOI] [PubMed] [Google Scholar]
- 28.Taira M, Otani H, Saint-Jeannet J P, Dawid I B. Nature (London) 1994;372:677–679. doi: 10.1038/372677a0. [DOI] [PubMed] [Google Scholar]
- 29.Taira M, Saint-Jeannet J P, Dawid I B. Proc Natl Acad Sci USA. 1997;94:895–900. doi: 10.1073/pnas.94.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shawlot W, Behringer R R. Nature (London) 1995;374:425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- 31.Sasai Y, Lu B, Steinbeisser H, De Robertis E M. Nature (London) 1995;376:333–336. doi: 10.1038/376333a0. [DOI] [PubMed] [Google Scholar]
- 32.Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont L K, De Robertis E M. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dirksen M L, Jamrich M. Genes Dev. 1992;6:599–608. doi: 10.1101/gad.6.4.599. [DOI] [PubMed] [Google Scholar]
- 34.Knöchel S, Lef J, Clement J, Klocke B, Hille S, Koster M, Knöchel W. Mech Dev. 1992;38:157–165. doi: 10.1016/0925-4773(92)90007-7. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz i Altaba A, Jessell T M. Development (Cambridge, UK) 1992;116:81–93. doi: 10.1242/dev.116.Supplement.81. [DOI] [PubMed] [Google Scholar]
- 36.Huang H C, Murtaugh L C, Vize P D, Whitman M. EMBO J. 1995;14:5965–5973. doi: 10.1002/j.1460-2075.1995.tb00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufmann E, Paul H, Friedle H, Metz A, Scheucher M, Clement J H, Knöchel W. EMBO J. 1996;15:6739–6749. [PMC free article] [PubMed] [Google Scholar]
- 38.Weber H, Holewa B, Jones E A, Ryffel G U. Development (Cambridge, UK) 1996;122:1975–1984. doi: 10.1242/dev.122.6.1975. [DOI] [PubMed] [Google Scholar]
- 39.Rosa F, Sargent T D, Rebbert M L, Michaels G S, Jamrich M, Grunz H, Jonas E, Winkles J A, Dawid I B. Dev Biol. 1988;129:114–123. doi: 10.1016/0012-1606(88)90166-2. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 41.Minucci S, Zand D J, Dey A, Marks M S, Nagata T, Grippo J F, Ozato K. Mol Cell Biol. 1994;14:360–372. doi: 10.1128/mcb.14.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nieuwkoop P D, Faber J. Normal Table of Xenopus laevis (Daudin) Amsterdam: North Holland; 1967. [Google Scholar]
- 43.Asashima M, Nakano H, Uchiyama H, Sugino H, Nakamura T, Eto Y, Ejima D, Nishimatsu S, Ueno N, Kinoshita K. Proc Natl Acad Sci USA. 1991;88:6511–6514. doi: 10.1073/pnas.88.15.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dyson S, Gurdon J B. Curr Biol. 1997;7:81–84. doi: 10.1016/s0960-9822(06)00030-3. [DOI] [PubMed] [Google Scholar]
- 45.Thomsen G H, Melton D A. Cell. 1993;74:433–441. doi: 10.1016/0092-8674(93)80045-g. [DOI] [PubMed] [Google Scholar]
- 46.Kessler D S, Melton D A. Development (Cambridge, UK) 1995;121:2155–2164. doi: 10.1242/dev.121.7.2155. [DOI] [PubMed] [Google Scholar]
- 47.Smith W C, McKendry R, Ribisi S J, Harland R M. Cell. 1995;82:37–46. doi: 10.1016/0092-8674(95)90050-0. [DOI] [PubMed] [Google Scholar]
- 48.Jones C M, Kuehn M R, Hogan B L, Smith J C, Wright C V. Development (Cambridge, UK) 1995;121:3651–3662. doi: 10.1242/dev.121.11.3651. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki A, Thies R S, Yamaji N, Song J J, Wozney J M, Murakami K, Ueno N. Proc Natl Acad Sci USA. 1994;91:10255–10259. doi: 10.1073/pnas.91.22.10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graff J M, Thies R S, Song J J, Celeste A J, Melton D A. Cell. 1994;79:169–179. doi: 10.1016/0092-8674(94)90409-x. [DOI] [PubMed] [Google Scholar]
- 51.Hemmati-Brivanlou A, Melton D A. Cell. 1994;77:273–281. doi: 10.1016/0092-8674(94)90319-0. [DOI] [PubMed] [Google Scholar]
- 52.Hemmati-Brivanlou A, Melton D A. Nature (London) 1992;359:609–614. doi: 10.1038/359609a0. [DOI] [PubMed] [Google Scholar]
- 53.Zhou X, Sasaki H, Lowe L, Hogan B L, Kuehn M R. Nature (London) 1993;361:543–547. doi: 10.1038/361543a0. [DOI] [PubMed] [Google Scholar]
- 54.Conlon F L, Lyons K M, Takaesu N, Barth K S, Kispert A, Herrmann B, Robertson E J. Development (Cambridge, UK) 1994;120:1919–1928. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- 55.Toyama R, O’Connell M L, Wright C V, Kuehn M R, Dawid I B. Development (Cambridge, UK) 1995;121:383–391. doi: 10.1242/dev.121.2.383. [DOI] [PubMed] [Google Scholar]
- 56.He X, Saint-Jeannet J P, Woodgett J R, Varmus H E, Dawid I B. Nature (London) 1995;374:617–622. doi: 10.1038/374617a0. [DOI] [PubMed] [Google Scholar]
- 57.Slack J M, Isaacs H V. Biochem Soc Trans. 1994;22:585–589. doi: 10.1042/bst0220585. [DOI] [PubMed] [Google Scholar]
- 58.Langston A W, Gudas L J. Mech Dev. 1992;38:217–227. doi: 10.1016/0925-4773(92)90055-o. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Musci T, Derynck R. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]
- 60.Lagna G, Hata A, Hemmati-Brivanlou A, Massagué J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 61.Nakao A, Roijer E, Imamura T, Souchelnytskyi S, Stenman G, Heldin C H, ten Dijke P. J Biol Chem. 1997;272:2896–2900. doi: 10.1074/jbc.272.5.2896. [DOI] [PubMed] [Google Scholar]
- 62.Derynck R, Zhang Y. Curr Biol. 1996;6:1226–1229. doi: 10.1016/s0960-9822(96)00702-6. [DOI] [PubMed] [Google Scholar]
- 63.Massagué J. Cell. 1996;85:947–950. doi: 10.1016/s0092-8674(00)81296-9. [DOI] [PubMed] [Google Scholar]
- 64.Krieg P A, Melton D A. Methods Enzymol. 1987;155:397–415. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- 65.Nishimatsu S, Suzuki A, Shoda A, Murakami K, Ueno N. Biochem Biophys Res Commun. 1992;186:1487–1495. doi: 10.1016/s0006-291x(05)81574-8. [DOI] [PubMed] [Google Scholar]
- 66.Thomsen G, Woolf T, Whitman M, Sokol S, Vaughan J, Vale W, Melton D A. Cell. 1990;63:485–493. doi: 10.1016/0092-8674(90)90445-k. [DOI] [PubMed] [Google Scholar]