Abstract
Aims
To investigate the role of DNA repair proteins and their prognostic significance in non-small-cell lung cancer (NSCLC).
Methods and results
A retrospective analysis of 108 cases of stage I–II NSCLC was undertaken. Immunohistochemical expression of DNA repair proteins MLH1, MSH2 and MGMT was assessed using tissue microarrays of paraffin-embedded samples of invasive carcinoma and precursor lesions. Results were analysed in relation to clinicopathological parameters and patient survival. Reduced expression of MLH1 was found in 58.5% of tumours and occurred less frequently in poorly differentiated tumours (P = 0.044) and large cell carcinomas (P = 0.004). MSH2 and MGMT expression was reduced in 18.1% and 77.8% of cases, respectively. There was an inverse relationship between MLH1 and MSH2 expression (P = 0.012). Normal expression of MLH1, MSH2 and MGMT was found in all cases of squamous metaplasia and squamous dysplasia. Only a single case of carcinoma in situ (12.5%) showed reduced MLH1, none showed reduced MSH2 and 25% showed reduced MGMT. Survival analyses showed no prognostic significance based on expression of MLH1 (P = 0.92), MSH2 (P = 0.78) or MGMT (P = 0.57).
Conclusions
Reduction in expression of DNA repair proteins MLH1, MSH2 and MGMT is relatively common in NSCLC, appears to be a late event in the development of invasive malignancy and does not influence survival in this patient cohort.
Cooper W A, Kohonen-Corish M R J, Chan C, Kwun S Y, McCaughan B, Kennedy C, Sutherland R L & Lee C-S (2008) Histopathology52, 613–622
Prognostic significance of DNA repair proteins MLH1, MSH2 and MGMT expression in non-small-cell lung cancer and precursor lesions
Keywords: DNA mismatch repair proteins, immunohistochemistry, MGMT, MLH1, MSH2, non-small-cell lung cancer, prognosis
Introduction
The role of DNA mismatch repair (MMR) proteins in sporadic and hereditary colorectal carcinoma has been extensively investigated,1 but the role of these proteins in the molecular pathogenesis of non-small-cell lung cancer (NSCLC) is poorly understood. Alterations in DNA produced during replication and recombination are repaired by the MMR system in an effort to maintain genomic stability, and tumours lacking MMR function exhibit a mutator phenotype.2 In addition, the cytotoxic effects of a number of alkylating agents used in the treatment of cancer are dependent on a functional MMR system.2 Hereditary non-polyposis colorectal cancer (HNPCC/Lynch syndrome) is an autosomal dominant condition, which comprises 2–5% of all colorectal cancers. It results from germ-line mutations in MMR genes, with alterations of hMLH1 and hMSH2 accounting for the vast majority of cases.1 This syndrome is characterized by genetic instability and the propensity to develop a number of neoplasms, particularly colorectal cancer and, to a lesser extent, malignancies of the endometrium, stomach, pancreas, ureters, ovaries, brain and skin.3 Pulmonary neoplasms are not a characteristic feature of this syndrome, suggesting that defective MMR gene function may not play a major role in the pathogenesis of NSCLC. Interestingly, alterations in expression of MMR proteins MLH1 and MSH2 have been reported in a variable proportion of NSCLC ranging from 18%4 to 61%,5 but no studies have investigated the role of reduced protein expression in precursor lesions of NSCLC and very few have investigated their potential prognostic significance in invasive carcinomas.
Methyl guanine DNA methyltransferase (MGMT) is a DNA repair enzyme involved in removal of abnormal adducts from the O6 position of guanine, providing protection from mutagenic agents and conferring resistance to alkylating chemotherapeutic drugs.2 MGMT expression is thought to be induced by a number of toxic agents, including cigarette smoke.6 Promoter region methylation resulting in reduced expression of MGMT occurs commonly in a variety of tumours such as colorectal cancer and melanoma7,8 and has been reported as an unfavourable prognostic factor in NSCLC.9 However, very few studies have investigated the role of altered MGMT protein expression in NSCLC.
In this study, expression of DNA repair proteins MLH1, MSH2 and MGMT were investigated in early-stage NSCLC and precursor lesions using tissue microarrays (TMAs) and the results have been correlated with clinicopathological parameters and patient survival.
Materials and methods
Patient cohort
Tumour samples and clinical follow-up data were obtained from a cohort of 108 stage I–II NSCLC patients treated at the Royal Prince Alfred Hospital, Sydney, Australia between 1997 and 1999. The cohort included 70 (64.8%) men and 38 women (35.2%) with a median age at diagnosis of 67 years (range 41–81 years) and median survival time of 72 months (range 3.3–97.5 months), excluding patients with survival <60 days. Histological tumour subtypes were assessed using the World Health Organization classification,10 and there were 48 (44.4%) adenocarcinomas (ADCs) [including seven bronchioloalveolar carcinomas (BACs)], 19 (17.6%) large cell carcinomas (LCCs), 40 (37.0%) squamous cell carcinomas (SCCs) and one (0.9%) mixed adenosquamous carcinoma. For survival analyses, invasive ADCs and (non-invasive) BACs were assessed both separately and as a single group. Tumours were staged using the American Joint Committee on Cancer Tumor-Node-Metastasis classification11 and consisted of 88 (81.5%) stage Ι and 20 (18.5%) stage ΙΙ tumours. Regional lymph node metastases were available from nine patients. Precursor lesions were also assessed when available, and there were up to 13 cases of bronchial squamous epithelial metaplasia, two with low-grade dysplasia, and eight cases of bronchial SCC in situ; however, there was insufficient material for adequate assessment of all of these cases with all three markers. Follow-up information of at least 5 years was available for this study.
Tumour samples
TMAs were constructed using three to four donor cores of tumour, 1 mm in diameter, from appropriate areas in formalin-fixed paraffin-embedded tissue blocks. These tissue cores were arrayed into a recipient paraffin block using a tissue arraying instrument (Beecher Instruments, Silver Springs, MD, USA). Serial sections were cut from the TMA blocks at 4 μm thickness and mounted on glass slides. The use of TMAs to investigate immunohistochemical expression of MLH1 and MSH2 as opposed to whole sections of tumours has been validated in a study of colorectal carcinomas.12
Immunohistochemistry
Sections were deparaffinized with xylene and rehydrated through graded alcohols to water. Immunohistochemical analysis for protein expression of the MGMT, MLH1 and MSH2 genes was undertaken using the following antibodies. MLH1 (clone G168-15; BD Pharmingen, San Diego, CA, USA; diluted 1:100), MSH2 (clone FE11; Oncogene, San Diego, CA, USA; diluted 1:1000) and MGMT (clone MT23.2; Zymed, Carlsbad, CA, USA; diluted 1:600). Immunohistochemistry was performed using Goat Anti-Mouse IgG, Polymer-Horseradish Peroxidase IHC amplification reagent (Chemicon, Temecula, CA, USA) as the detection system and 3,3′-diaminobenzidine as the substrate chromogen (ICN Biomedicals, Aurora, OH, USA). Heat-induced antigen retrieval was performed by heating in a pressure cooker (Decloaking Chamber; Biocare Medical, Concord, CA, USA) in preheated citrate buffer (10 mmol/l, pH 6.0) for 5 min. In some cases, the tissue sections were microwave treated in preheated ethylenediamine tetraaceticacid buffer for 20–40 min. All immunohistochemistry was performed on a Sequenza rack with Coverplate (ThermoShandon, Pittsburgh, PA, USA). The slides were treated with 1% goat serum and then incubated with primary antibody overnight at room temperature. Upon completion of a Tris buffer wash, the slides were incubated with enzyme-conjugated polymer with goat antimouse IgG antibody for 30 min at room temperature and then washed in buffer. After reaction with diaminobenzidene/hydrogen peroxide for 5 min, slides were rinsed in distilled water and immersed in 1% copper sulphate solution for 1 min. After washing, the sections were counterstained in Gill's Haematoxylin 2 (Australian Biostain, Traralgon, Australia) solution for 30 s, followed by blueing solution for 15 s.
The positive controls were matched samples of normal bronchial mucosa and peripheral lung parenchyma incorporated into the tissue arrays. Samples of normal bronchial mucosa showed positive staining for MMR proteins in an average of 95–100% of cells in all cases except for two cases stained with MLH1, which were excluded from the analysis. Samples from normal spleen were also incorporated into the arrays to use as both external positive controls and also as reference points within the arrays. Nuclear expression of MLH1 was also seen in all splenic samples, but the reactivity was not scored. A negative control slide was incubated with non-immune serum instead of the primary antibody.
Scoring
Two pathologists (W.A.C. and C.C.) independently scored each case without knowledge of the patient's clinical details and an average of the two scores obtained was used. Immunohistochemical expression of MLH1 was scored semiquantitatively by multiplying the percentage of cells showing nuclear expression and the intensity of immunoreactivity using a three-tier grading system (1 = weak, 2 = moderate and 3 = strong). An average score was obtained from the multiple samples of each case and reduced protein expression was taken as cases with a score of <200. Where markedly discrepant, the case was reviewed before deciding on an appropriate score. There was good correlation between the scores obtained from each pathologist (MLH1 correlation coefficient R = 0.94, P < 0.01; MSH2 R = 0.79, P < 0.001 and MGMT R = 0.96, P < 0.001).
Statistical analyses
The Pearson χ2 test and Fisher's exact test (two-sided) were used to compare associations between protein expression and various clinicopathological characteristics. The Kaplan–Meier log rank and Cox proportional regression model were used for survival analyses. SPSS statistical software package version 13.0 was used for all analyses (SPSS Inc., Chicago, IL, USA). P-values of <0.05 were regarded as statistically significant.
Results
MMR protein expression
Nuclear expression of MLH1 was seen extensively in normal tissues, but was reduced in 62 out of the 106 cases of NSCLC (58.5%) (Figure 1). In SCC, MLH1 expression was reduced in 27/39 (69.2%), in LCCs 5/18 (27.8%) and in ADCs 30/48 (62.5%), including 5/7 (71.4%) BACs. Of eight cases of bronchial epithelial squamous carcinoma in situ, only one case (12.5%) showed reduced MLH1 expression. Thirteen cases of bronchial epithelial squamous metaplasia were assessed and none had reduced expression of MLH1, including two cases with low-grade dysplasia. Of nine cases with lymph node metastases, only two (22.2%) showed reduced MLH1 expression.
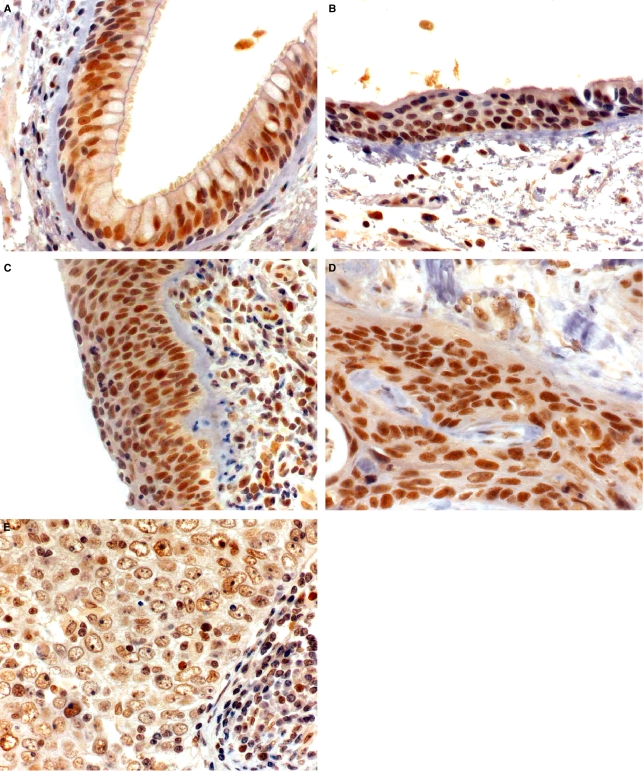
Figure 1.
Immunohistochemistry for MLH1. Strong expression of MLH1 is seen in (A) normal bronchial epithelium, (B) bronchial epithelium with squamous metaplasia, (C) bronchial epithelium with carcinoma in situ (most cases) and most invasive squamous cell carcinomas (SCCs) (D). Some cases of invasive SCC show reduced expression of MLH1 (E).
There was reduced MSH2 expression in 19 of 105 cases (18.1%) (Figure 2). In different histological subtypes, MSH2 was reduced in 4/39 SCCs (10.3%), 3/16 (18.8%) LCCs and 11 of 48 (22.9%) ADCs, including 2/7 (28.6%) BACs. MSH2 was not reduced in any of the seven cases of bronchial epithelial squamous carcinoma in situ or any of the 11 cases of squamous metaplasia with or without dysplasia in bronchial epithelium. There was a total of nine lymph node metastases, with none showing reduced MSH2 expression.
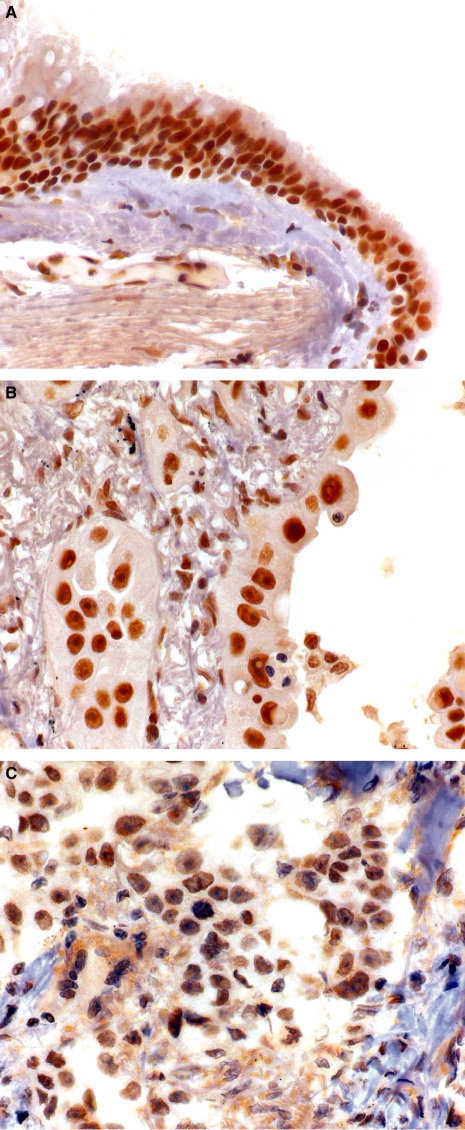
Figure 2.
Immunohistochemistry of MSH2 in (A) normal bronchial epithelium, (B) adenocarcinoma with normal expression of MSH2, and (C) adenocarcinoma with reduced expression of MSH2.
Reduced MGMT scores were seen in 84 of 108 cases (77.8%) of NSCLC (Figure 3). In SCC, 32 (80%) showed reduced expression of MGMT, LCC 13 (68.4%) and in ADCs 38 (79.2%). In eight cases of bronchial epithelial squamous carcinoma in situ, there was reduced MGMT expression in two (25%) cases. Samples of bronchial epithelial squamous metaplasia did not show any reduction in MGMT reactivity in the eight cases (0%) without dysplasia or the two cases (0%) with low-grade dysplasia. Of nine lymph node metastases, four cases (44.4%) had reduced expression of MGMT.
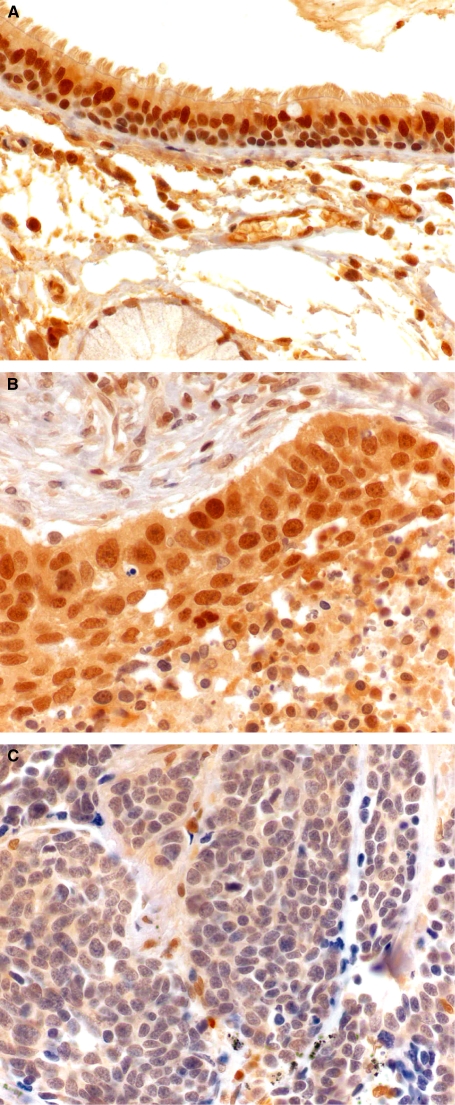
Figure 3.
Immunohistochemistry of methyl guanine DNA methyltransferase (MGMT) in (A) normal bronchial epithelium, (B) large cell carcinoma with normal expression of MGMT, and (C) large cell carcinoma with reduced expression of MGMT.
Correlations between expression of different proteins
Reduction of either MLH1 or MSH2 was found in 64/104 cases (61.5%) and co-reduction of both MLH1 and MSH2 proteins was found in 16 cases (15.4%). There was an inverse relationship between expression of the two proteins that was statistically significant (P = 0.012). Ninety-two of 106 (86.8%) showed reduction of either MLH1 or MGMT, and 52 cases (49.1%) showed reduction of both MLH1 and MGMT. Eighty-three of 105 (79.0%) showed reduction of either MSH2 and MGMT, and 17 (16.2%) showed reduction of both of these proteins.
Correlations with pathological and clinical variables
Reduced expression of MLH1 was found to be less common in LCCs (P = 0.004, Pearson's χ2; P = 0.007, Fisher's exact test two-sided) and poorly differentiated tumours (P = 0.044, Pearson's χ2) (Table 1). Other pathological or patient factors such as gender, age, tumour size, stage, lymphatic invasion, blood vessel invasion, perineural invasion and involvement of the bronchial surgical margin were evenly distributed between those with and without reduced MLH1 expression. MSH2 and MGMT expression did not correlate with any of the measured clinicopathological parameters (Table 1).
Table 1.
Relationship between protein expression and clinicopathological characteristics of patients
| MLH1 expression | MSH2 expression | MGMT expression | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Normal | Reduced | χ2, P-value | Normal | Reduced | χ2, P-value | Normal | Reduced | χ2, P-value | |
| Tumour type | 0.028* | 0.29 | 0.64 | ||||||
| ADC | 16 | 24 | 0.81 | 37 | 11 | 0.24 | 10 | 37 | 0.84 |
| BAC | 2 | 5 | 0.47 | 5 | 2 | 0.46 | 2 | 5 | 0.68 |
| SCC | 12 | 27 | 0.087 | 35 | 4 | 0.11 | 8 | 32 | 0.67 |
| LCC | 13 | 5 | 0.004* | 13 | 3 | 0.94 | 6 | 13 | 0.28 |
| Mixed | 1 | 1 | † | 1 | 1 | † | 0 | 2 | † |
| Differentiation | 0.11 | 0.84 | 0.61 | ||||||
| Well | 2 | 6 | 0.32 | 6 | 2 | 0.60 | 3 | 6 | 0.40 |
| Mod | 20 | 37 | 0.15 | 50 | 10 | 0.66 | 11 | 46 | 0.44 |
| Poor | 22 | 19 | 0.044* | 30 | 7 | 0.87 | 10 | 32 | 0.75 |
| Size, mm | 0.79 | 0.39 | 0.33 | ||||||
| ≤30 | 23 | 34 | 45 | 12 | 15 | 43 | |||
| >30 | 21 | 28 | 41 | 7 | 9 | 41 | |||
| Sex | 0.17 | 0.95 | 0.24 | ||||||
| Male | 32 | 37 | 55 | 12 | 18 | 52 | |||
| Female | 12 | 25 | 31 | 7 | 6 | 32 | |||
| Age, years | 0.74 | 0.87 | 0.61 | ||||||
| <67 | 22 | 33 | 39 | 9 | 10 | 40 | |||
| ≥67 | 22 | 29 | 47 | 20 | 14 | 44 | |||
| Stage | 0.76 | 0.84 | 0.47 | ||||||
| 1A | 11 | 19 | 0.52 | 24 | 7 | 0.44 | 9 | 22 | 0.28 |
| 1B | 23 | 33 | 0.92 | 46 | 8 | 0.37 | 10 | 47 | 0.22 |
| 2A | 3 | 2 | 0.39 | 4 | 1 | 0.91 | 2 | 3 | 0.33 |
| 2B | 7 | 8 | 0.66 | 12 | 3 | 0.84 | 3 | 12 | 0.82 |
| BVI | 0.81 | 0.14 | 0.28 | ||||||
| Absent | 41 | 57 | 81 | 16 | 21 | 79 | |||
| Present | 3 | 5 | 5 | 3 | 3 | 5 | |||
| LVI | 0.32 | 0.60 | 0.84 | ||||||
| Absent | 42 | 56 | 80 | 17 | 22 | 78 | |||
| Present | 2 | 6 | 6 | 2 | 2 | 6 | |||
| PNI | 0.23 | 0.24 | 0.45 | ||||||
| Absent | 44 | 60 | 5 | 18 | 24 | 82 | |||
| Present | 0 | 2 | 1 | 1 | 0 | 2 | |||
| Margin | 0.39 | 0.20 | 0.60 | ||||||
| Not involved | 40 | 59 | 79 | 19 | 23 | 78 | |||
| Involved | 4 | 3 | 7 | 0 | 1 | 6 | |||
Statistically significant (P < 0.05).
Too few cases to assess.
ADC, Adenocarcinoma; BAC, bronchioloalveolar carcinoma; SCC, squamous cell carcinoma; LCC, large cell carcinoma; BVI, blood vessel invasion; LVI, lymphovascular invasion; PNI, perineural invasion.
MMR protein expression and patient survival
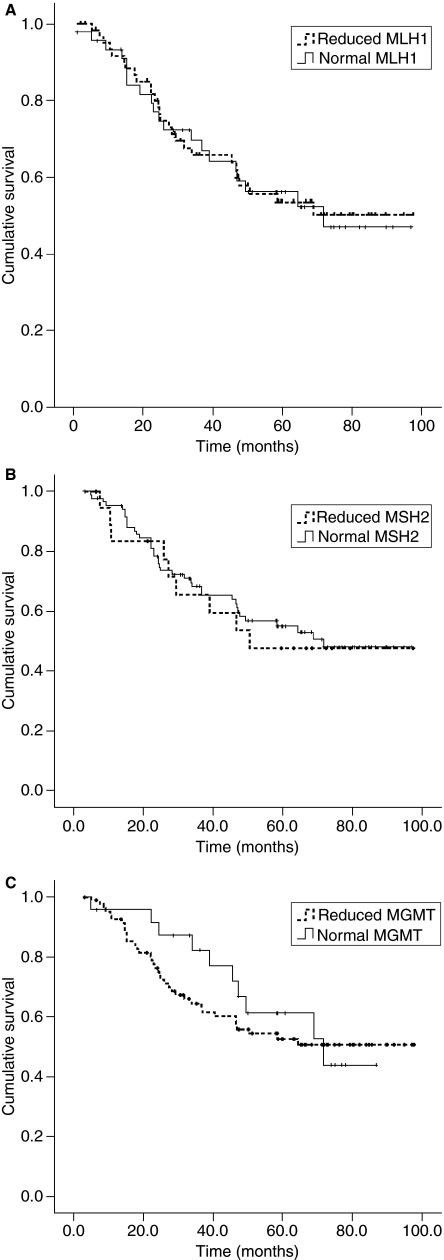
Expression of MMR proteins was compared with overall patient survival using Kaplan–Meier survival analysis. No significant correlation was found between survival and expression of MLH1 (P = 0.92), MSH2 (P = 0.78) or MGMT (P = 0.57) (Figure 4). Tumours that showed a reduction of either MLH1 or MSH2 (or both) were not associated with survival (P = 0.83). Similarly, other combinations of DNA repair protein expression (MLH1 and/or MGMT reduced, MSH2 and/or MGMT reduced) did not correlate with survival (data not shown). Analysis of survival based on reduced MMR protein expression using the Kaplan–Meier method was not significant when the data were subanalysed based on tumour grade, histological type, stage or patient gender (data not shown). Similarly, using Cox regression analysis, no significant correlation was found between survival and expression of MLH1 [P = 0.94, hazard ratio (HR) 0.98, 95% confidence interval (CI) 0.54, 1.76], MSH2 (P = 0.78, HR 0.90, 95% CI 0.44, 1.87) or MGMT (P = 0.57, HR 0.82, 95% CI 0.41, 1.6).
Figure 4.
A, Probability of survival according to MLH1 expression [Kaplan–Meier survival curve, P = 0.92, log rank (Mantel–Cox)]. Reduced MLH1 expression n = 62 (dotted line), normal MLH1 expression n = 44 (solid line). There were 60 censored cases. B, Probability of survival according to MSH2 expression using immunohistochemical score (Kaplan–Meier survival, P = 0.78, log rank test). Reduced MSH2 expression n = 19 (dotted line), normal MSH21 expression n = 86 (solid line). There were 58 censored cases. C, Probability of survival according to MGMT expression [(Kaplan–Meier survival curve, P = 0.57, log rank (Mantel–Cox); P = 0.25, Breslow; P = 0.36, Tarone–Ware]. Reduced MGMT expression n = 83 (dotted line), normal MGMT expression n = 24 (solid line). There were 60 censored cases.
Discussion
In this study, expression of DNA repair proteins MLH1, MSH2 and MGMT have been investigated in early-stage NSCLC and precursor lesions using TMAs. Although the role of MMR proteins in sporadic and hereditary colorectal carcinoma has been extensively investigated,1 the role of these proteins in the molecular pathogenesis of lung cancer is poorly understood. Molecular alterations of MMR genes have been found in a significant number of NSCLCs, and promoter methylation is thought to be the predominant mechanism of silencing hMLH1 and hMSH2 genes in these cases.4,13,14 Some studies have found that loss of heterozygosity at loci for DNA MMR genes is relatively frequent in NSCLC,15,16 whereas others have not been able to demonstrate hMLH1 promoter methylation.17 Homozygous deletions or rearrangements have not been demonstrated in the hMLH1 gene in NSCLC.15,16 In this study, we have demonstrated that decreased DNA repair protein expression is relatively common in NSCLC with MLH1, MSH2 and MGMT reduced in 58.5%, 18.1% and 77.8% of NSCLC cases, respectively. Reduced expression of MLH1 in NSCLC has been reported at frequencies of 20–61%,5,18 and in some studies has been associated with microsatellite instability.5,18 Decreased MSH2 expression has been reported at similar rates ranging from 18%4 to 58%,16 and in our study population altered MSH2 was less frequent than MLH1. The differences between studies possibly relate to different patient populations and different criteria being applied to determine reduction of protein expression.
In a study of 150 cases of NSCLC, Xinarianos et al.16 have found reduced expression of MLH1 protein in 58.6% of cases, MSH2 in 57.8% and either MLH1 or MSH2 in 82%. Although we have found reduced expression of MSH2 in considerably fewer cases, our results for MLH1 and reduction of either protein were similar. They did not find any correlation between reduced MLH1 expression and age, gender, tumour differentiation or TNM T stage, but they did demonstrate an association with heavy smoking and nodal metastases in SCC.16 In contrast, in our study reduced MLH1 was less common in poorly differentiated tumours and large cell-type carcinomas. Interestingly, colorectal carcinomas associated with altered MMR protein function are associated with a variety of clinicopathological characteristics, including a propensity to be poorly differentiated.19,20 As in our study, Xinarianos et al.16 have not found hMSH2 expression to be correlated with any of the clinicopathological parameters assessed.
Whereas others have reported no loss of expression of MMR proteins MLH1, MSH2 or MSH6 in the non-invasive bronchioloalveolar subtype of ADC,21 we were able to demonstrate reduction of MLH1 in 71.4% and MSH2 in 28.6% of this type of carcinoma. Our results for this subset of ADC are very similar to our findings in all ADCs, suggesting the role of altered DNA MMR function in both types of tumour, if any, is likely to be similar. The investigation reported by Aubry et al.21 did not include invasive ADCs, criteria used for assessing immunohistochemical expression were not clearly defined, and it is possible that methodological differences could account for the discrepant findings.
MGMT promoter region methylation has been demonstrated in 7–55% of NSCLC14,22,23 and is an independent predictor of poor prognosis in one study,9 whereas others have not been able to demonstrate a significant association with survival.24 Immunohistochemical expression of MGMT has been studied in a group of 83 stage I–III NSCLC and reduced expression was found in only 25% of cases,6 compared with 77.8% in our study. There was a significant difference in MGMT expression between smokers and non-smokers (in whom the protein was more frequently reduced)6 and between ADC and LCC, with none of the LCCs showing loss of expression, whereas in our study we did not demonstrate any association with tumour type and found loss of expression in a considerable number of LCCs (68.4%). As in our study, they found no association between MGMT expression and age, gender, stage or histological tumour type.
The development of pulmonary SCC is known to occur through a stepwise progression of bronchial epithelial abnormalities starting from squamous metaplasia through to dysplasia, carcinoma in situ and, ultimately, invasive carcinoma. Although the histomorphological changes in this process are recognized, the underlying molecular alterations are only poorly understood. Early alterations include overexpression of cyclin D1, cyclin E and p53, whereas loss of retinoblastoma expression occurs late in the development of invasive carcinoma.25 We found that although reduced expression of MLH1, MSH2 and MGMT was not uncommon in invasive NSCLC, it was relatively rare in precursor lesions, with no reductions seen in the earliest histological abnormalities of metaplasia and dysplasia. Only very few cases of carcinoma in situ showed reduced expression of the DNA repair proteins, suggesting such alterations occur relatively late in the pathogenesis of malignancy. Alternatively, the precursor abnormalities studied could represent genetically distinct lesions that had not given rise to the corresponding invasive carcinoma present in the resected specimen. However, the number of cases with precursor squamous lesions was too low in our study to draw a meaningful conclusion. Interestingly, reduced protein expression was relatively uncommon in lymph node metastasis, suggesting these alterations are unlikely to play an important role in the development of metastatic potential; however, the sample numbers with regional node spread were small and a larger study would be required to validate this finding.
Although reduced expression of DNA repair proteins has been demonstrated in a variable proportion of NSCLCs, the prognostic significance of these alterations has only very rarely been reported. In our study, we have been unable to demonstrate any association between alterations in expression of the DNA repair proteins MLH1, MSH2 and MGMT and survival. Similarly, in a study of non-smoking Taiwanese female patients with NSCLC, neither MLH1 expression nor promoter hypermethylation were significantly associated with prognosis.13 Although the authors were able to demonstrate an association between MSH2 promoter methylation and shorter overall survival, results of the relationship between MSH2 protein expression and survival were not reported.13 Other studies examining the methylation status of MLH1 and MGMT gene promoters have not shown any correlation with overall survival.14 This is in contrast to colorectal carcinomas, where defective MMR protein function is a beneficial prognostic feature.1,26,27
Acknowledgments
M.R.J.K.-C. is a Cancer Institute NSW Fellow.
The authors thank the Cancer Institute NSW, National Health and Medical Research Council and the RT Hall Trust for their grant support.
Glossary
Abbreviations
- ADC
adenocarcinoma
- BAC
bronchioloalveolar carcinoma
- CI
confidence interval
- HR
hazard ratio
- LCC
large cell carcinoma
- MGMT
methyl guanine DNA methyltransferase
- MMR
mismatch repair
- NSCLC
non-small-cell lung cancer
- SCC
squamous cell carcinoma
- TMA
tissue microarray
Competing interests
None to declare.
References
- 1.Haydon AMM, Jass JA. Emerging pathways in colorectal cancer development. Lancet Oncol. 2002;3:83–88. doi: 10.1016/s1470-2045(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 2.Bellacosa A. Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ. 2001;8:1076–1092. doi: 10.1038/sj.cdd.4400948. [DOI] [PubMed] [Google Scholar]
- 3.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J. Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y-C, Lu Y-P, Tseng R-C, et al. Inactivation of hMLH1 and hMSH2 by promotor methylation in primary non-small cell lung tumors and matched sputum samples. J. Clin. Invest. 2003;111:887–895. doi: 10.1172/JCI15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang J-W, Chen Y-C, Chen C-Y, Chen J-T, Chen S-K, Wang Y-C. Correlation of genetic instability with mismatch repair protein expression and p53 mutations in non-small cell lung cancer. Clin. Cancer. Res. 2000;6:1639–1646. [PubMed] [Google Scholar]
- 6.Mattern J, Koomagi R, Volm M. Smoking-related increase of O6-methylguanine-DNA methyltransferase expression in human lung carcinomas. Carcinogenesis. 1998;19:1247–1250. doi: 10.1093/carcin/19.7.1247. [DOI] [PubMed] [Google Scholar]
- 7.Kohonen-Corish MRJ, Cooper WA, Saab J, Thompson JF, Trent RJA, Millward MJ. Promoter hypermethylation of the O6-methylguanine DNA methyltransferase gene and microsatellite instability in metastatic melanoma. J. Invest. Dermatol. 2006;126:167–171. doi: 10.1038/sj.jid.5700005. [DOI] [PubMed] [Google Scholar]
- 8.Kohonen-Corish MRJ, Daniel JJ, Chan C, et al. Low microsatellite instability is associated with poor prognosis is stage C colon cancer. J. Clin. Oncol. 2005;23:2318–2324. doi: 10.1200/JCO.2005.00.109. [DOI] [PubMed] [Google Scholar]
- 9.Brabender J, Usadel H, Metzger R, et al. Quantitative O6-methylguanine DNA methyltransferase methylation analysis in curatively resected non-small.cell lung cancer: associations with clinical outcome. Clin. Cancer Res. 2003;9:223–227. [PubMed] [Google Scholar]
- 10.Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC. World Health Organisation classification of tumours. Pathology and genetics of tumours of the lung, pleura, thymus and heart. Lyon: IARC Press; 2004. [Google Scholar]
- 11.Grondin SC, Liptay MJ. Current concepts in the staging of non-small cell lung cancer. Surg. Oncol. 2002;11:181–190. doi: 10.1016/s0960-7404(02)00050-6. [DOI] [PubMed] [Google Scholar]
- 12.Hendriks Y, Franken P, Dierssen JW, et al. Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am. J. Pathol. 2003;162:469–477. doi: 10.1016/S0002-9440(10)63841-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu H-S, Wen C-K, Tang Y-A, et al. Promoter hypermethylation is the predominant mechanism in hMLH1 and hMSH2 deregulation and is a poor prognostic factor in nonsmoking lung cancer. Clin. Cancer Res. 2005;11:5410–5416. doi: 10.1158/1078-0432.CCR-05-0601. [DOI] [PubMed] [Google Scholar]
- 14.Safar AM, Spencer H, Su X, et al. Methylation profiling of archived non-small cell lung cancer: a promising prognostic system. Clin. Cancer Res. 2005;11:4400–4405. doi: 10.1158/1078-0432.CCR-04-2378. [DOI] [PubMed] [Google Scholar]
- 15.Benachenhou N, Guiral S, Gorska-Flipot I, Labuda D, Sinnett D. High resolution deletion mapping reveals frequent allelic losses at the DNA mismatch repair loci hMLH1 and hMSH3 in non-small cell lung cancer. Int. J. Cancer. 1998;77:173–180. doi: 10.1002/(sici)1097-0215(19980717)77:2<173::aid-ijc1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 16.Xinarianos G, Liloglou T, Prime W, et al. hMLH1 hMSH2 expression correlates with allelic imbalance on chromosome 3p in non-small cell lung carcinomas. Cancer Res. 2000;60:4216–4221. [PubMed] [Google Scholar]
- 17.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 18.Takahashi Y, Kondo K, Hirose T, et al. Microsatellite instability and protein expression of the DNA mismatch repair gene, hMLH1, of lung cancer in chromate-exposed workers. Mol. Carcinog. 2005;42:150–158. doi: 10.1002/mc.20073. [DOI] [PubMed] [Google Scholar]
- 19.Bocker T, Schlegel J, Kullmann F, et al. Genomic instability in colorectal carcinomas: comparison of different evaluation methods and their biological significance. J. Pathol. 1996;179:15–19. doi: 10.1002/(SICI)1096-9896(199605)179:1<15::AID-PATH553>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 20.Chaves P, Cruz C, Lage P, et al. Immunohistochemical detection of mismatch repair gene proteins as a useful tool for the identification of colorectal carcinoma with the mutator phenotype. J. Pathol. 2000;191:355–360. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH644>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 21.Aubry M-C, Halling KC, Myers JL, Tazelaar HD, Yang P, Thibodeau SN. DNA mismatch repair genes hMLH1, hMSH2, and hMSH6 are not inactivated in bronchioloalveolar carcinomas of the lung. Cancer. 2001;92:2898–2901. doi: 10.1002/1097-0142(20011201)92:11<2898::aid-cncr10104>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 22.Russo AL, Thiagalingam A, Pan H, et al. Differential DNA hypermethylation of critical genes mediates the stage-specific tobacco smoke-induced neoplastic progression of lung cancer. Clin. Cancer Res. 2005;11:2466–2470. doi: 10.1158/1078-0432.CCR-04-1962. [DOI] [PubMed] [Google Scholar]
- 23.Esteller M, Hamiltion SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 24.Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–255. [PubMed] [Google Scholar]
- 25.Lonardo F, Rusch V, Langenfeld J, Dmitrovsky E, Klimstra D. Overexpression of cyclins D1 and E is frequent in bronchial preneoplasia and precedes squamous cell carcinoma development. Cancer Res. 1999;59:2470–2476. [PubMed] [Google Scholar]
- 26.Cawkwell L, Gray S, Murgatroyd H, et al. Choice of management strategy for colorectal cancer based on a diagnostic immunohistochemical test for defective mismatch repair. Gut. 1999;45:409–415. doi: 10.1136/gut.45.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright CM, Dent OF, Barker MA. Prognostic significance of extensive microsatellite instability in sporadic clinicopathological stage C colorectal cancer. Br. J. Surg. 2000;87:1197–1202. doi: 10.1046/j.1365-2168.2000.01508.x. [DOI] [PubMed] [Google Scholar]