Abstract
In an attempt to quantify the rates of protein sequence divergence in Drosophila, we have devised a screen to differentiate between slow and fast evolving genes. We find that over one-third of randomly drawn cDNAs from a Drosophila melanogaster library do not cross-hybridize with Drosophila virilis DNA, indicating that they evolve with a very high rate. To determine the evolutionary characteristics of such protein sequences, we sequenced their homologs from a more closely related species (Drosophila yakuba). The amino acid substitution rates among these cDNAs are among the fastest known and several are only about 2-fold lower than the corresponding values for silent substitutions. An analysis of within-species polymorphisms for one of these sequences reveals an exceptionally high number of polymorphic amino acid positions, indicating that the protein is not under strong negative selection. We conclude that the Drosophila genome harbors a substantial proportion of genes with a very high divergence rate.
Keywords: protein evolution, neutral evolution/expressed sequence tag/genome evolution
Evolutionary novelties are usually thought to be brought about either by changes in the regulatory interactions of genes or by gene duplication with subsequent diversification (1–4). The latter assumption in particular has lead to the notion that the large number of genes that can be found in highly evolved organisms can potentially be derived from a limited number of functional protein modules. Estimates of the number of functional modules range between 1,000 and 7,000 (5, 6). However, in the current eukaryotic genome projects, a relatively high number of ORFs are identified that do not show a match with other known protein motifs in databases (7–9). Usually it is assumed that this means that their homologs and their respective modules are yet to be discovered. However, an alternative interpretation would be that some proteins evolve so fast that their homologs cannot be discovered over larger evolutionary distances. There are some indications that such fast evolving sequences do indeed exist. First, even in proteins harboring functionally highly conserved motifs, such as transcription factors, a substantial proportion of the protein can diverge so quickly that the alignment between homologs from different species may become impossible outside of the conserved DNA-binding domain. Second, for some of the genes known to be involved in early embryogenesis in Drosophila, it has proven to be difficult or even impossible to obtain homologs from outside the insects (10). Finally, a few genes from Drosophila have been identified that diverge so fast that they cannot even be cloned from distantly related Drosophilids (11–13). Such examples show that it may be worthwhile to systematically analyze how many fast evolving genes exist in a given organism. The results of such a screen would not only bear on the question of the number of ancient conserved protein modules, but also on the question of the origin and material basis of evolutionary novelties.
We have devised such a screen in Drosophila with the following rationale. Randomly picked clones from a Drosophila melanogaster cDNA library are hybridized against a panel of genomic DNA from different insect species with increasing evolutionary distance. The most closely related species in this panel is Drosophila virilis, which has split from D. melanogaster between 40 and 60 million years ago. As it is known that the neutral evolution rate in Drosophilids lies between 1 and 2% per million years (14, 15), one should assume that sequences evolving with a neutral or close to neutral rate should not cross-hybridize between these two species under moderately stringent conditions. On the other hand, conserved protein sequences should readily cross-hybridize with D. virilis and also with some of the other species in the panel. After this initial screen, the non-cross-hybridizing genes are analyzed more closely. Their homologs are isolated from Drosophila yakuba, a species that has split from D. melanogaster only about 10–15 million years ago, which should allow recovery of homologs of sequences evolving with a neutral rate. The respective cDNAs from both species are then fully sequenced and compared with each other to verify that they have a homologous ORF, which should be indicative of functional genes. Moreover, the sequence comparison of the coding region between these two species allows an estimate of the evolutionary rates of the respective protein. As a final test of whether truly fast evolving genes are recovered, one can determine the level of within species polymorphism for a certain gene. It is expected under a neutral model of molecular evolution that a gene that shows a high divergence rate between species should also be polymorphic within a species.
We show that this strategy does indeed identify a large number of genes with very high evolutionary divergence rates. Only a few of them include a known protein module, whereas for most of them, homologs cannot be detected in the databases. These results support the notion that a whole class of genes exists in a typical eukaryotic genome that has not been systematically taken account of so far.
MATERIALS AND METHODS
Screening of Clones.
An oligo(dT) primed and directionally cloned λ ZAPII cDNA library encompassing the first 2–14 hr of embryonic D. melanogaster development (Stratagene) was plated at low density. Five hundred and seventy-six single clones were picked and checked via cross hybridization for duplicates. One hundred and five nonduplicate cDNA clones were used for the further experiments. The plasmids were in vivo excised with the ExAssist helper phage (Stratagene). The 5′ end of each plasmid was cycle sequenced and run on an Applied Biosystems model 377 automated sequencer (Perkin–Elmer) resulting in 300–600 bases of sequence. The sequences were then used to search nonredundant databases with the blastx program via E-mail (matrix: BLOSUM 62). Scores were considered significant for P < 10−3, though most P values were much smaller (i.e., P < 10−7). The expressed sequence tag (EST) sequences were submitted to the EST division of GenBank with accession numbers AA433202–AA433290. For the cross-hybridization experiments, genomic DNA of the respective species was purified by CsCl centrifugation. The DNA was digested with EcoRI, and 2 μg of D. melanogaster, 4 μg of D. virilis, 8 μg of Musca domestica (housefly), and 10 μg of Tenebrio molitor (large flour beetle) were loaded per lane on an 0.8% agarose gel. The different amounts of DNA roughly reflect the different genome sizes. The gel was blotted and hybridized with the cDNA inserts (obtained by PCR with flanking primers and labeled with 32P by random priming) at 65°C in 5× standard saline citrate (SSC). The filters were washed three times for 15 min at 65°C with 2× SSC and exposed to x-ray film. These conditions represent a hybridization stringency, which should allow up to 35% mismatch in a probe with balanced GC-content.
D. yakuba cDNA Clones.
To construct the D. yakuba embryonic cDNA library, poly(A+) RNA was isolated from total RNA of 0–14 hr of embryonic development with the PolyATract System (Promega), and the library was constructed with the λ ZAPII cDNA synthesis and library construction kit (Stratagene). The phages were plated and transferred onto nylon membranes. Filters were hybridized with a 32P-labeled cDNA fragment of D. melanogaster under the same conditions as above. cDNAs were completely sequenced using a shotgun procedure in combination with custom designed primers. Alignment was usually unequivocal, and the number of synonymous and nonsynonymous substitutions per site was estimated according to Comeron (16).
Polymorphism Analysis.
Genomic DNA from single flies of a collection of isofemale lines from throughout the world (kindly provided by M. Kidwell, University of Arizona) was prepared using a proteinase K/SDS and phenol/chloroform purification procedure. An 868-bp segment of the reading frame of clone 1G5 was amplified with primers 1G5-PR3 [5′-AAGTATCTAGCCGA(CT)GAGGAC-3′] and 1G5-PR4 (5′-TACCCAGCTCTCATTCATCTC-3′) in 20 μl reaction volume with 1 pmol of each primer, 200 pmol dNTP, 1× Taq buffer, and 0.5 unit of AmpliTaq DNA polymerase (Perkin–Elmer). The DNA was gel-purified and directly sequenced with the amplification primers and internal primers on an Applied Biosystems model 377 sequencer. Each base was sequenced from both directions. Accession numbers are AF005865–AF005881.
In Situ Hybridization.
Digoxigenin-labeled DNA or RNA probes were produced by random priming or in vitro transcription from plasmids containing the cDNA insert according to manufacturer’s protocols (Boehringer Mannheim). In situ hybridization was done essentially as described (17).
RESULTS
Evolutionary Conservation of Random cDNA Clones.
One hundred and five nonredundant cDNA clones were randomly isolated from an embryonic D. melanogaster cDNA library and analyzed in two ways. First, 300–600 bp were sequenced from the 5′ end and the sequences were compared with sequence databases to allow the identification of already known genes and genes with clear homologs. Second, low-stringency hybridization to genomic DNA from a panel of insect species was employed to analyze the degree of conservation of the genes. The most closely related species in this panel was D. virilis (split around 40–60 million years ago), the next one the housefly M. domestica (split around 100–120 million years ago) and finally the beetle T. molitor (split around 250–270 Mya). The results of these experiments are summarized in Table 1.
Table 1.
Summary statistics for the characterization of the random cDNA clones from D. melanogaster (n = 105)
| Character | No. (%) |
|---|---|
| Database matches of partial cDNA sequences | |
| Exact match with D. melanogaster | 26 (24%) |
| Match with other species | 17 (16%) |
| No match | 62 (60%) |
| Conservation in genomic cross-hybridization | |
| D. melanogaster only | 56 (53%) |
| Drosophilids | 33 (31%) |
| Drosophilids and Musca | 6 (6%) |
| All four species | 10 (10%) |
The upper part of the table shows results of the database search with the partial sequences. Note that most of the genes (23 of 26) showing an exact match in D. melanogaster also showed matches in other species. The lower part shows results of the genomic cross-hybridization analysis between a panel of different species.
The database searches with the partial sequences revealed that a fraction of 60% showed no matches with known genes, which is similar to the published results of large scale expressed sequence tag sequencing projects with Caenorhabditis elegans, Arabidopsis, and humans (7–9). The cross-hybridization study showed that more than half of the clones (53%) hybridize with D. melanogaster DNA only, 31% hybridize in both Drosophilids, and only 10% in all four species. Among the latter are well-known conserved genes that also had matches in the databases, thus confirming the utility of the cross-hybridization approach.
The proportion of sequences that hybridize to D. melanogaster DNA, but not to D. virilis DNA, is surprisingly large. There are in fact potential sources of error in this approach, most notably the possibility of including incomplete cDNA clones containing only untranslated 3′ ends. We found, however, that only 4 of the 26 already known sequences from D. melanogaster represented 3′ ends only and more than half of them were full-length cDNAs. Thus, though some correction is necessary, we estimate that well over one-third of the randomly chosen cDNAs are derived from fast evolving genes according to this assay.
We have also analyzed the spatio-temporal expression patterns of all sequences in this study by whole-mount in situ hybridization. The results show that the fast evolving genes do not significantly differ in their expression characteristics from the slow evolving ones (Table 2), indicating that they constitute a representative sample of all genes.
Table 2.
Comparison of expression patterns
| Expression | Conserved | Rapidly evolving |
|---|---|---|
| Spatially restricted | 15 | 14 |
| Homogenous | 34 | 42 |
Whole-mount in situ hybridizations of D. melanogaster embryos were done for all clones used in the study. The expression patterns of clones with exact D. melanogaster database matches were obtained from the literature. The differences in numbers found in the different classes are not significant (G = 0.40, P > 0.5).
Comparison of Rapidly Evolving cDNA Clones Between D. melanogaster and D. yakuba.
To analyze the fraction of fast evolving sequences in detail and to estimate their amino acid substitution rates, we compared them to their homologs from the closely related species, D. yakuba. The cDNAs of 10 fast evolving and 1 highly conserved clone were recovered from a D. yakuba library, and the clones from both species were sequenced completely. Nine pairs of clones, including the highly conserved one, contained a homologous ORF in both species and are thus likely to encode functional proteins. This shows also that the above described hybridization results are not simply due to the inclusion of a high proportion of noncoding cDNA fragments. However, in two pairs of clones no homologous ORF could be identified and they were omitted from the further analysis. In subsequent database searches with the complete cDNA sequences, the highly conserved clone and three of the fast evolving clones gave significant matches. The conserved clone 2A12 matches with kinesin-like proteins (best match with CHO1; GenBank accession no. X83575; blast score, 472; P = 2 × 10−65), clone 1E9 with zinc-finger proteins (best match with Xenopus Znc6; Protein Identification Resource accession no. PC1144; blast score, 72; P = 8.8 × 10−9), and clone 2D9 with LIM domain proteins (best match with sunflower LIM domain protein SF3, Protein Identification Resource accession no. S37656; blast score, 110; P = 1.4 × 10−7), and 2A5 shows a weak similarity to the yeast ATP11 precursor protein (SWISS-PROT accession no. P32453; blast score, 89; P = 0.00015). The matches of 1E9 and 2D9 concern only the respective protein modules, but not sequences outside of them. The other six clones did not yield any significant matches in the database.
Table 3 lists the number of synonymous (Ks) and nonsynonymous (Ka) nucleotide substitutions per site for these cDNA clones and other genes of which complete sequences from D. melanogaster and D. yakuba are available. The highest proportion of replacement substitutions are found among four genes that were identified in our screen. In each case, the values are only about twofold lower than the corresponding numbers of synonymous substitutions of the same genes. Moreover, they are only about 3-fold lower than the value of 0.32 substitutions per site that was found for the neutrally evolving parts of the rDNA internal transcribed spacer regions from this species pair (15).
Table 3.
Comparison of nonsynonymous (Ka) and synonymous (Ks) substitutions per site of different genes from D. melanogaster and D. yakuba
| Gene | Accession | Codons | Ka | 95% CI | Ks | 95% CI |
|---|---|---|---|---|---|---|
| 1G5 | MEL: AF005851; | 347 | 0.168 | 0.252–0.441 | 0.346 | 0.134–0.199 |
| YAK: AF005852 | ||||||
| 1E9 | MEL: AF005847 | 393 | 0.116 | 0.092–0.139 | 0.279 | 0.205–0.356 |
| YAK: AF005848 | ||||||
| 2D9 | MEL: AF005861; | 280 | 0.108 | 0.070–0.143 | 0.233 | 0.157–0.303 |
| YAK: AF005862 | ||||||
| 1A3 | MEL: AF005843; | 227 | 0.092 | 0.063–0.120 | 0.196 | 0.119–0.279 |
| YAK: AF005844 | ||||||
| Anon-3B1.2 | MEL: X76206; | 260 | 0.079 | 0.056–0.104 | 0.284 | 0.191–0.367 |
| YAK: X84413 | ||||||
| 1A4 | MEL: AF005846; | 185 | 0.048 | 0.022–0.079 | 0.213 | 0.132–0.302 |
| YAK: AF005845 | ||||||
| Period | MEL: X76206; | 1233 | 0.032 | 0.026–0.040 | 0.309 | 0.257–0.340 |
| YAK: S53298 | ||||||
| 2D7 | MEL: AF005859; | 283 | 0.023 | 0.010–0.032 | 0.234 | 0.159–0.311 |
| YAK: AF005860 | ||||||
| 2A5 | MEL: AF005855; | 274 | 0.020 | 0.008–0.030 | 0.218 | 0.149–0.281 |
| YAK: AF005856 | ||||||
| Amylase | MEL: L22717; | 494 | 0.020 | 0.012–0.028 | 0.110 | 0.072–0.142 |
| YAK: X04569 | ||||||
| 2C9 | MEL: AF005857; | 265 | 0.018 | 0.005–0.028 | 0.112 | 0.057–0.155 |
| YAK: AF005858 | ||||||
| G-S-T | MEL: X14233; | 208 | 0.017 | 0.002–0.030 | 0.114 | 0.058–0.168 |
| YAK: M84580 | ||||||
| Adh | MEL: J01066; | 256 | 0.015 | 0.005–0.026 | 0.156 | 0.094–0.211 |
| YAK: X54120 | ||||||
| G-6-P | MEL: L27540; | 558 | 0.011 | 0.005–0.017 | 0.198 | 0.151–0.240 |
| YAK: L27673 | ||||||
| hb | MEL: Y00274; | 829 | 0.011 | 0.005–0.015 | 0.183 | 0.143–0.220 |
| YAK: unpubl. | ||||||
| CO I | MEL: U37541; | 498 | 0.007 | 0.002–0.0012 | 0.315 | 0.251–0.396 |
| YAK: X03240 | ||||||
| 2A12 | MEL: AF005853; | 498 | 0.006 | 0.001–0.011 | 0.325 | 0.260–0.401 |
| YAK: AF005854 |
The sequences of eight additional genes were retrieved from the Genbank database. G-S-T, glutathion S-transferase; Adh, alcohol dehydrogenase; G-6-P, glucose-6-phosphatase; COI, cytochrome oxidase I; anon-3B1.2, anonymous ORF close to period. CI is confidence interval, calculation according to Comeron (16).
To ensure that the fastest evolving genes are not paralogs of duplicated gene pairs, we have done an additional Southern hybridization analysis with D. yakuba DNA. Only one hybridizing band was found in most cases, suggesting that the genes are indeed unique. Additional evidence that paralogs are not a problem comes from the fact that even though multiple cDNA clones were recovered for each of the genes from D. yakuba, they could all be allocated to the same gene by sequence comparison with the longest clone. Finally, the fact that the synonymous rates are nearly the same across all the genes compared between D. melanogaster and D. yakuba suggests also that these are not paralogs with a substantially longer evolutionary divergence time than the split of the two species.
Population Polymorphism.
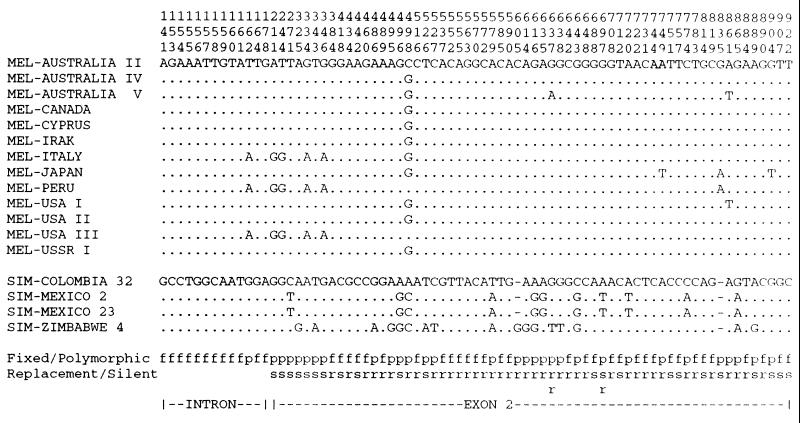
Differences in the amino acid sequence of homologous proteins from different species may be due to fixation of neutral substitutions by random drift or to fixation of adaptive substitutions by natural selection. The McDonald–Kreitman approach (18) can be used to differentiate between the two processes. It is based on the prediction of the neutral evolutionary theory that genes with a high number of substitutions between species should also show a high degree of within-species polymorphism. We have tested this for the fastest evolving clone in our sample. Most of the coding region of clone 1G5 was amplified and sequenced from 13 strains of D. melanogaster and 4 strains of the sibling species Drosophila simulans. A large number of polymorphic sites were found in both species (Fig. 1), many more than in comparable studies for other genes (18–20). Two lines of evidence suggest that the polymorphisms found reflect a mutation-drift equilibrium in a neutrally or near neutrally evolving sequence. First, there are more polymorphic replacement sites than synonymous sites in agreement with the fact that there are also more potential replacement sites in the region. Second, the fraction of polymorphic sites in the short intron is comparable to the fraction of polymorphic sites in the coding region (1.6% vs. 3.9%; G = 0.85, P > 0.3). This suggests that almost none of the amino acid positions may be under strong selective constraint. A comparison between fixed and polymorphic sites between the two species shows also no significant deviation from the assumption of a neutral evolution in this region (Table 4). Thus, both the within- and between-species parameters indicate that this coding region evolves with a near neutral rate and under apparently neutral conditions.
Figure 1.
Polymorphic sites of 1G5 in different populations of D. melanogaster and D. simulans. Dots represent identical nucleotides with respect to the top sequences. The region surveyed includes bases 58–922 of clone 1G5 (total length of coding region is 1,081 bp and includes a small intron of 61 bp). The “fixed/polymorphic” line indicates whether a substitution is fixed (f) or polymorphic (p), and the “replacement/silent” line indicates replacement (r) and silent (s) substitutions. At position 605 is a 6-bp indel polymorphism and at position 835 a fixed indel difference of 9 bp which are disregarded for the calculation shown in Table 4. Positions 637 and 697 show two replacement polymorphisms each.
Table 4.
Polymorphisms found for sequence 1G5 in D. melanogaster and D. simulans
| Fixed | Polymorphic | |
|---|---|---|
| Replacement | 23 | 20 |
| Synonymous | 10 | 12 |
Fixed and polymorphic sites were assessed as described in ref. 10. A G test of independence did not reject the null hypothesis that the proportion of replacement substitutions is independent of whether the substitutions are fixed or polymorphic (G = 0.37, P > 0.5).
DISCUSSION
Our results have general implications on how one can envisage the evolution of genes and genomes. Some scenarios have suggested that there may only be a limited number of stable protein folds or domains that were already present early in the evolution of life and that were used over and over again in different combinations to make up most of the genes existing in todays species (4–6). Our results would suggest that there is a substantial fraction of coding sequences that does not follow such a pattern. The high amino acid replacement rates found in these sequences raises the question of whether these proteins might be able to assume many different stable protein folds that can easily be changed during evolution. On the other hand, it is well known that conservation of folding structure does not necessarily require the conservation of the primary sequence (21). These fast-evolving genes will therefore be a test case of whether fold structures are generally more conserved than primary sequences, or whether evolution is indeed able to explore many different new folds. In any case, our results suggest that the failure of finding homologs for a substantial proportion of genes in genome comparisons may not imply that these code for novel proteins, but might simply indicate that these are fast evolving.
A previous study investigating the general conservation of cDNAs between Drosophilids by DNA–DNA hybridization has suggested that cDNAs are not evolving as fast as single-copy noncoding DNA and that there might be even differences in embryonic messages versus adult messages in this respect (22). Furthermore, we note that most genes analyzed so far from Drosophila species tend to be more conserved than the fast-evolving genes characterized here. However, there are exceptions such as transformer (11), period (23) and Acp-26Aa (24), all three of which may be directly involved in sexual selection or species specific differentiations. Another interesting case is the gene spalt adjacent. It shows a high evolutionary rate between closely related species (12) and does not seem to have a function, at least not under laboratory conditions. Neither an allele with a stop codon at the beginning of the reading frame nor artificial over-expression lead to any recognizable phenotype (13). Still, an ORF is retained between the different species analyzed (12), indicating that it is not a pseudogene. It remains to be shown whether the apparent lack of an overt function is typical for most fast evolving genes, but this would at least explain why this class of genes has not been found more often so far. Lack of a phenotype under laboratory conditions does not imply that a gene lacks a function in the wild. Genes providing partially redundant functions (25) or genes involved in local adaptation, parasite defense or speciation might have subtle adult phenotypic effects when mutant and would thus have escaped the genetic screening regimes that have been employed so far.
Acknowledgments
We thank Margaret Kidwell for providing wild-type strians, Charles N. David for enthusiastic encouragement, and Charles N. David, Michael Ashburner, Svante Pääbo, Loredana Nigro, and Markus Friedrich for comments on the manuscript. This work was supported by a Gerhard Hess Preis from the Deutsche Forschungsgemeinschaft to D.T. and by a Ph.D. studentship from the Graduiertenkolleg “Zelluläre und molekulare Aspekte der Entwicklung” to K.S.
Footnotes
References
- 1.King M C, Wilson A. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 2.Carroll S B. Nature (London) 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 3.Palopoli M F, Patel N. Curr Opin Genet Dev. 1996;6:502–508. doi: 10.1016/s0959-437x(96)80074-8. [DOI] [PubMed] [Google Scholar]
- 4.Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- 5.Dorit R L, Schoenbach L, Gilbert W. Science. 1990;250:1377–1382. doi: 10.1126/science.2255907. [DOI] [PubMed] [Google Scholar]
- 6.Chotia C. Nature (London) 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- 7.Waterston R, Martin C, Craxton M, Huynh C, Coulson A, Hillier L, Durbin R, Green P, Shownkeen R, Halloran N, Metzstein M, Hawkins T, Wilson R, Berks M, Du Z, Thomas K, Thierry-Mieg J, Sulston J. Nat Genet. 1992;1:114–123. doi: 10.1038/ng0592-114. [DOI] [PubMed] [Google Scholar]
- 8.Adams, M. D., Kerlavage, A. R., Fleischmann, R. D., Fuldner, R. A., Bult, C. J., et al. (1995) Nature (London) 377, Suppl., 3–174. [PubMed]
- 9.Dujon P. Trends Genet. 1996;12:263–270. doi: 10.1016/0168-9525(96)10027-5. [DOI] [PubMed] [Google Scholar]
- 10.Akam M, Averof M, Castelli-Gair J, Dawes R, Falciani F, Ferrier D. Development (Cambridge, UK) Suppl. 1994;1994:209–215. [PubMed] [Google Scholar]
- 11.O’Neil M T, Belote J M. Genetics. 1992;131:113–128. doi: 10.1093/genetics/131.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reuter D, Schuh R, Jäckle H. Proc Natl Acad Sci USA. 1989;86:5483–5486. doi: 10.1073/pnas.86.14.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reuter D, Kühnlein R P, Frommer G, Barrio R, Kafatos F C, Jäckle H. Chromosoma. 1996;104:445–454. doi: 10.1007/BF00352268. [DOI] [PubMed] [Google Scholar]
- 14.Sharp P M, Li W-H. J Mol Evol. 1989;28:398–402. doi: 10.1007/BF02603075. [DOI] [PubMed] [Google Scholar]
- 15.Schlötterer C, Hauser M-T, von Haeseler A, Tautz D. Mol Biol Evol. 1994;11:513–522. doi: 10.1093/oxfordjournals.molbev.a040131. [DOI] [PubMed] [Google Scholar]
- 16.Comeron J M. J Mol Evol. 1995;41:1152–1159. doi: 10.1007/BF00173196. [DOI] [PubMed] [Google Scholar]
- 17.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 18.McDonald J H, Kreitman M. Nature (London) 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 19.Eanes W F, Kirchner M, Yoon J. Proc Natl Acad Sci USA. 1993;90:7475–7479. doi: 10.1073/pnas.90.16.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hey J, Kliman R M. Mol Biol Evol. 1993;10:804–822. doi: 10.1093/oxfordjournals.molbev.a040044. [DOI] [PubMed] [Google Scholar]
- 21.Sander C, Schneider R. Proteins. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- 22.Powell J R, Caccone A, Gleason J M, Nigro L. Genetics. 1993;133:291–298. doi: 10.1093/genetics/133.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thackeray J R, Kyriacou C. J Mol Evol. 1990;31:389–401. doi: 10.1007/BF02106054. [DOI] [PubMed] [Google Scholar]
- 24.Tsaur S-C, Wu C-I. Mol Biol Evol. 1997;14:544–549. doi: 10.1093/oxfordjournals.molbev.a025791. [DOI] [PubMed] [Google Scholar]
- 25.Tautz D. BioEssays. 1992;14:263–266. doi: 10.1002/bies.950140410. [DOI] [PubMed] [Google Scholar]