Abstract
Surface-enhanced Raman scattering (SERS) nanoparticles are emerging as a new approach for optical detection of biomolecules. In a model assay in formalin-fixed paraffin-embedded (FFPE) prostate tissue sections, we detect prostate-specific antigen (PSA) using antibody (Ab) conjugated to composite organic–inorganic nanoparticles (COINs), and we use identical staining protocols to compare COIN-Ab and Alexa–Ab conjugates in adjacent tissue sections. Spectral analysis illustrates the fundamental difference between fluorescence and Raman signatures and accurately extracts COIN probe signals from background autofluorescence. Probe signals are used to generate images of PSA expression on the tissue, and quality measures are presented to characterize the performance of the COIN assay in comparison to Alexa. Staining accuracy (ability to correctly identify PSA expression in epithelial cells) is somewhat less for COIN than Alexa, which is attributed to an elevated false negative rate of the COIN. However, COIN provided signal intensities comparable to Alexa, and good intra-, inter-, and lot-to-lot consistencies. Overall, COIN and Alexa detection reagents possess similar performance with FFPE tissues, supporting the further development of Raman probes for this application. This manuscript contains online supplemental material at http://www.jhc.org. Please visit this article online to view these materials. (J Histochem Cytochem 56:371–379, 2008)
Keywords: formalin-fixed paraffin-embedded tissue, multiplex detection, surface-enhanced Raman scattering, autofluorescence, spectral analysis
Immunohistochemistry (IHC) and immunofluorescence (IF) have been developed to visualize protein expression in the context of tissue morphology. Recently, both methods have been applied to simultaneously detect multiple proteins in a single sample, which reduces demands on tissue specimens. This “multiplex” detection requires availability of different color probes as well as instrumentation capable of separating the signals from individual probes (Tsurui et al. 2000; Camp et al. 2002; Fountaine et al. 2006; Levenson and Mansfield 2006; Byers et al. 2007). The desire for greater multiplexing potential has motivated development of fluorescent quantum dots and, more recently, development of nanoparticle probes based on Raman emission. Here we directly compare staining by antibody (Ab)-conjugated Raman probes and Alexa fluorescent probes for the case of a single antigen on adjacent sections of formalin-fixed paraffin-embedded (FFPE) prostate tissue. Detailed quality measures are developed and presented to allow detailed evaluation of the staining quality to provide quantitative benchmarks for further probe development.
Raman and fluorescence emission result from very different mechanisms, but both involve excitation of a molecule with light and emission of light at longer wavelengths. Unlike the single broad peaks of molecular fluorophores (50–70 nm) and quantum dots (30–40 nm), Raman emission is characterized by a series of very narrow peaks (∼2 nm). Raman signatures can be detected using the same spectral instrumentation that is increasingly applied for fluorescence analysis, but Raman emission intensity is normally much too weak to serve as an optical label. Raman probes overcome this limitation by exploiting an effect known as surface-enhanced Raman scattering (SERS), which occurs when molecules bind to certain metal surfaces (Fleischmann et al. 1974; Albrecht and Creighton 1977; Jeanmaire and Vanduyne 1977) including silver and gold nanoparticles (Nie and Emery 1997; Kneipp et al. 1998; Ni et al. 1999; Doering and Nie 2002). Raman probes can be fabricated by a variety of methods (Ni et al. 1999; Cao et al. 2002; Doering and Nie 2003; Grubisha et al. 2003; Mulvaney et al. 2003; Nithipatikom et al. 2003; Kneipp et al. 2005; Su et al. 2005; Vo-Dinh et al. 2005; Xu et al. 2005; Kim et al. 2006; McCabe et al. 2006; Schlucker et al. 2006; Chen et al. 2007; Gong et al. 2007; Jun et al. 2007), and the degree of Raman signal enhancement depends on the approach. Composite organic–inorganic nanoparticles (COIN) are one type of Raman probe made by aggregating silver nanoparticles in the presence of a chosen organic label molecule with a distinct Raman signature (Su et al. 2005; Sun et al. 2007). Each COIN retains the distinct signature of the chosen Raman label(s), but the enhancement provides an extremely bright probe emission suitable for direct conjugation to biomolecule detection reagents.
Raman probes are a relatively new approach for biomolecule detection. The most advanced applications have focused on analysis of proteins or nucleic acids in solution, including quantitative detection of single proteins in sandwich-binding assays (Ni et al. 1999; Grubisha et al. 2003; Driskell et al. 2005; Su et al. 2005; Xu et al. 2005; Cui et al. 2006; Gong et al. 2007; Kim et al. 2007; Sun et al. 2007) and for multiplex DNA hybridization assays both in solution (Faulds et al. 2005,2007) and in plate format (Cao et al. 2002). A small number of applications have demonstrated detection of Raman probes in living or fixed cell samples (Nithipatikom et al. 2003; Kneipp et al. 2005; Vo-Dinh et al. 2005; Kim et al. 2006; Hu et al. 2007; Lee et al. 2007). Relatively little work has been done thus far to develop Raman probes for tissue-based analysis (Schlucker et al. 2006; Sun et al. 2007), despite strong potential for multiplexing and to overcome interference from tissue autofluorescence. As a first step to address the potential of multiplex protein analysis in human tissue sections, we recently demonstrated the ability to separate signals of two colocalized COIN probes from tissue autofluorescence in FFPE tissue sections (Sun et al. 2007).
We set out to describe in more detail the features and quality of COIN–Ab direct conjugates for protein detection in FFPE tissue samples. We designed parallel experiments with COIN and Alexa in FFPE prostate tissue. As a model assay, we targeted detection of prostate-specific antigen (PSA), a highly abundant protein specifically expressed in prostate epithelium. This approach highlights the features of Raman-based probes by comparison with a familiar and commonly used Alexa Fluor dye and thereby provides a reference to clearly characterize the assay performance of COIN–Ab conjugates.
Materials and Methods
Preparation of COIN–Ab and Alexa–Ab Conjugates
COIN nanoparticle probes were fabricated, stabilized by encapsulation, and conjugated to Abs as described previously (Su et al. 2005; Sun et al. 2007). Briefly, fabricated silver nanoparticles were aggregated under controlled conditions in the presence of a chosen Raman label molecule with a distinct optical signature. Raman labels captured within the silver aggregates show an enormous enhancement of Raman signal intensity. Unlike fluorescence emission from molecular fluorophores, Raman emission is insensitive to photobleaching, and we see little effect from illumination for the laser powers used here. COIN aggregates were stabilized by encapsulation with cross-linked BSA, resulting in COIN with an average diameter of 60 nm. Chemical groups on the BSA coating were used to directly conjugate the COIN to polyclonal anti-PSA Ab (AF1344; R&D Systems, Minneapolis, MN). The resulting anti-PSA–COIN conjugate provided a ready-to-use reagent that was stable for months (Sun et al. 2007).
The same anti-PSA Ab was conjugated to Alexa Fluor 568 following manufacturer's protocols (Alexa Fluor 568 Monoclonal Antibody Labeling Kit, A-20184; Invitrogen, Carlsbad, CA). Reactions yielded ∼8 mol of dye molecule per Ab for the anti-PSA–Alexa conjugates.
Prior to application to tissues, COIN and Alexa conjugates were routinely tested in a plate-binding assay as described previously (Sun et al. 2007) using immobilized PSA protein as a target (purified PSA from human semen, P3338; Sigma, St Louis, MO).
Tissue Preparation and Staining Procedures
Prostate tissues were collected at the University of Washington under an Institutional Review Board-approved protocol. FFPE human prostate tissue was cut into 5-μm sections, transferred to Fisher Probe-on Plus Capillary gap slides (Fisher Scientific; Pittsburgh, PA), baked, and stored at 4C until use. Sections were deparaffinized in three changes of xylene, rehydrated through graded ethanol (three changes in 100% ethanol for 2 min each, two changes in 95% ethanol for 2 min each, and one change of 70% ethanol for 1 min), and washed in PBS (5 min). Enzymatic antigen retrieval was performed by Proteinase-K (S3020; Dako, Carpinteria, CA) treatment for 5 min at room temperature, followed by washing in PBST (PBS buffer with 0.1% Tween). Tissue sections were incubated with 3% BSA/PBS for 30 min at room temperature to provide general protein blocking, followed by blotting to remove the blocking buffer.
Working solutions of the anti-PSA–COIN or anti-PSA–Alexa conjugates were prepared fresh each day by diluting stock solutions in 3% BSA/PBS. Prepared slides were incubated with 150 μl of anti-PSA–COIN or anti-PSA–Alexa in a humidified chamber for 30 min at room temperature. After incubation, slides were washed (two changes of PBST for 5 min each, one change of PBS for 5 min), rinsed with 0.1 M NaCl, and coverslipped.
Tissue Data Acquisition and Analysis
An epi-illumination microscope was modified for laser excitation and spectral acquisition by using a mirror in the filter cube head to direct a laser through the objective and collect emitted light through the same objective (Su et al. 2005; Sun et al. 2007). The laser (532 nm) was focused to a single spot on the sample to excite the probe, and emitted light from the sample spot was directed to a spectrograph (Acton 2300i; Princeton Instruments, Acton, MA) and CCD detector (256 × 1024 pixels; Princeton Instruments 7500) to record the spectrally resolved emission (spectral range 530–660 nm across 1024 camera pixels). Each spectrum was acquired with a ×20 objective using 0.l-sec acquisition time and 650-μW laser power at the sample surface. The same microscope provided conventional brightfield imaging and filter-based imaging normally used for fluorescence analysis.
The imaging system used a two-axis computer-controlled sample stage (Prior ProScan CS152KB; Prior Scientific, Rockland, MA) allowing automated spectral acquisition from each point in a raster pattern across a chosen prostate gland. The number of spots and the spacing were chosen based on the desired level of spatial resolution (e.g., 30 × 30 spot array, 5-μm spacing), and a full spectrum was recorded at each point (0.1 sec per spot, 90 sec typical for a full image). Probe intensity was determined by linear least squares regression of the measured spectra using the appropriate reference spectra for the probe (COIN or Alexa), representative tissue autofluorescence, and a freely varying polynomial to account for unknown variations in the autofluorescence background (“regress” function in MATLAB; The MathWorks, Natick, MA). Error in the spectral fitting was calculated as the average percent error across the spectral range, and typical values were <5%. Images for the COIN and Alexa stains were reported either as intensity maps or as binary maps in which each point is classified as positive or negative based on a single intensity threshold. To generate a binary image of a gland, the brightfield tissue image was manually traced in ImageJ to create a mask that identified points overlying the epithelium and stroma. Points in the lumen were rejected from further analysis due to the occasional presence of stained tissue debris in the lumen. Binary images were generated by classifying each pixel as positive or negative for the target based on a threshold intensity value. The threshold for each probe was automatically set at the value that minimized false positive and false negative pixel classifications in that image. Custom MATLAB code was used for the spectral fitting procedure, image analysis, and image generation. The procedure and analysis for imaging are illustrated in Results.
Alexa-stained slides were also imaged on the same microscope using conventional mercury lamp excitation and the appropriate filter set for the dye (Calcium Crimson 41,027; Chroma Technology, Rockingham, VT). The image was flat-field corrected to remove variations in the illumination, and the resulting intensity image was analyzed in parallel with the spectral data.
We summarized the quality of staining from images by reporting three overall quality measures as described in Results. Quality measures were calculated for each gland, and reported values are the average and SD for a set of glands. Comparisons between measurements (e.g., sets of identical glands measured in adjacent slides) were performed using p values from the Student's t-test for two-tailed distributions and paired-data sets (MS Excel).
Results
PSA was chosen as a model protein for characterization of a COIN-based detection assay in tissue sections due to its ubiquitous and robust expression in normal and cancerous prostate epithelium, its high expression level, availability of quality Abs, and the opportunity to assess the specificity of staining independently based on tissue morphology. In each experiment, adjacent tissue sections were stained with anti-PSA–COIN and anti-PSA–Alexa, and staining signals were quantified in matching gland pairs. Glands for COIN intensity measurements were selected based on the tissue morphology or on Alexa–anti-PSA staining. Spectral data sets were acquired for both the COIN and Alexa stains, and the Alexa slide was also imaged using conventional filter cubes.
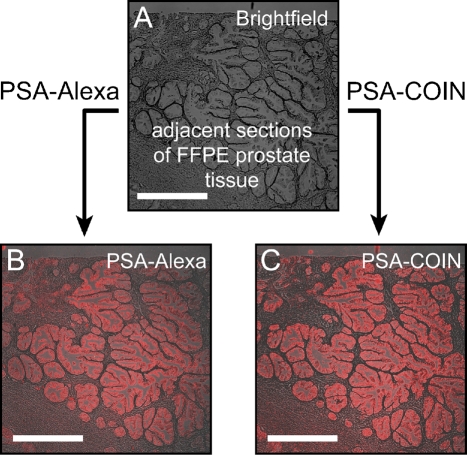
Conventional fluorescence analysis uses filter sets to provide a broad window of excitation matched to the dye absorption and to collect a broad window of emission around the fluorescence peak of a specific dye. Comparable to a conventional fluorescence microscope, our Raman microscope was equipped with filter cubes to allow measurements of emitted light from tissue sections stained with COIN-conjugated or Alexa dye-conjugated Abs. Filter cube imaging is ideally suited for the initial evaluation of the staining quality of the COIN-conjugated PSA Ab and its specificity for epithelial cells. Figure 1 shows the fluorescence emission from anti-PSA–Alexa 568 and the Raman emission from anti-PSA–COIN imaged using a conventional filter set and identical measurement conditions. The filter set was chosen to provide optimal excitation and fluorescence collection for the Alexa dye, and the emission filter also captured the majority of Raman peaks from the COIN signature. The opportunity to obtain measurements from precisely the same microscopic field with Alexa- or COIN-conjugated anti-PSA Abs permitted side-by-side comparison of the same gland stained by two reagents. We observed similar qualitative results for Alexa- or COIN-conjugated anti-PSA Abs and obtained similar values of staining intensities and specificities across multiple glands.
Figure 1.
Comparison of filter cube images of anti-PSA–COIN and anti-PSA–Alexa on adjacent sections of formalin-fixed paraffin-embedded (FFPE) prostate tissue. Images are acquired using conventional filter set imaging (×5 objective, Calcium Crimson filter set, exposure 5 sec). (A) Brightfield image of a prostate tissue section. (B) Fluorescence emission from anti-PSA–Alexa 568 on FFPE prostate tissue. (C) Raman emission from anti-PSA–COIN for the same region as B in the adjacent slide. Both stains report PSA expression in the epithelium of prostate glands but not in the stroma, and the COIN and Alexa show comparable brightness (true intensities shown). Bar = 500 μm.
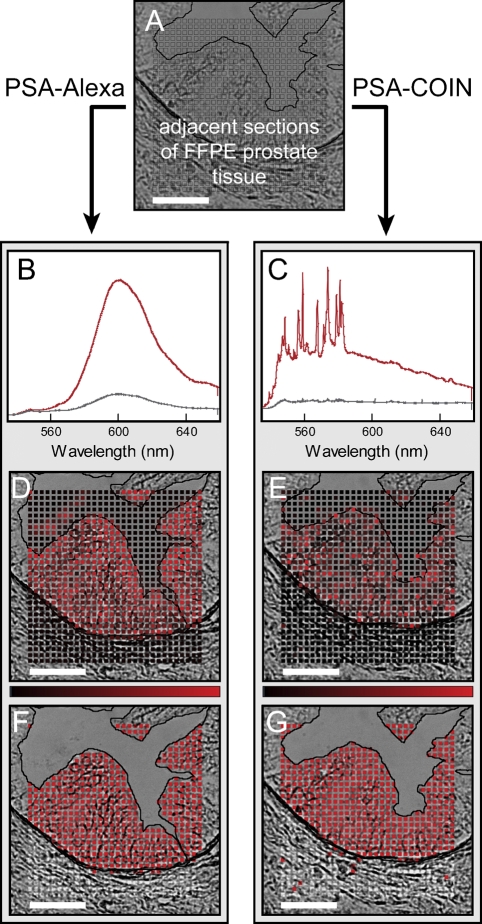
Filter cube imaging rejects important spectral information by reporting a single intensity integrated over a wide wavelength range (e.g., 60 nm for the emission filter in Figure 1). Spectral analysis significantly improves the ability to identify signals from multiple probes in the presence of background autofluorescence (Levenson and Mansfield 2006). Figure 2 illustrates the spectral acquisition and analysis used to extract and report probe intensities for Alexa and COIN. We located identical glands on adjacent sections stained by COIN and Alexa and measured spectra for each point (boxes in Figure 2A) in a raster pattern spanning the stroma, epithelium, and lumen. Spectral acquisition for protein detection with COIN- or Alexa-conjugated Abs was performed at ×20 magnification (Figure 2), but similar results were obtained at ×40. Figure 2B shows the characteristic broad peak of Alexa in representative spectra recorded from the epithelium (red) and stroma (gray). High signal levels are seen in the epithelium as expected for the PSA stain, but nonspecific Alexa background was uniformly present throughout the stroma. The red spectrum in Figure 2C shows the distinct signature of COIN, including a broad underlying signal typical of SERS enhancement. Unlike the Alexa stain, background signal from COIN shown in Figure 2B (gray) results from a small number of particles, whereas the majority of points in the stroma have essentially zero signal. We extracted the pure signals by fitting the measured spectrum using reference spectra for the probe (Alexa or COIN) and representative tissue autofluorescence, as well as a freely varying polynomial to account for unknown differences in the autofluorescence background. Figures 2D and 2E show the extracted probe intensities for the Alexa and COIN mapped onto their respective brightfield images where the brightness of the red color is proportional to the signal intensity of the spot (see Materials and Methods). Figures 2G and 2H simplify the visual representation using binary reporting in which each point is classified as positive (red) or negative (gray), based on the optimum intensity threshold for each gland.
Figure 2.
Illustration of spectral analysis for COIN and Alexa stains on adjacent tissue sections. (A) Adjacent sections of FFPE prostate tissue were stained either with anti-PSA–COIN or with anti-PSA–Alexa. Spectra were acquired at each point (boxes) in a raster pattern spanning the stroma, epithelium, and lumen of a single gland. (B) Representative spectra from anti-PSA–Alexa Fluor 568 measured in the epithelium (red) and stroma (gray). The upper spectrum (red) was also used as the reference spectrum for subsequent Alexa spectral fitting. Nonspecific Alexa staining (gray spectrum) occurred uniformly across the stroma at ∼20% of positive signal. (C) Representative spectra from anti-PSA–COIN measured in the epithelium (red) and stroma (gray). The upper spectrum (red) was also used as the reference spectrum for subsequent COIN spectral fitting. (D,E) Intensities use a linear scale from black (zero) to red, as shown by the intensity bar; some high-intensity values are truncated. (F,G) Binary results for Alexa and COIN where each pixel is classified as positive (red) or negative (gray). The lumen region is rejected from the binary analysis due to the difficulty of accurately identifying this boundary in brightfield images as well as the occasional presence of stained debris in the lumen. Note the presence of stained tissue debris in D. Bar = 50 μm.
Biological variability in the amount of PSA expression across glands in the same tissue section is significant due to varied levels of differentiation and atrophy of the epithelium. This variability is sufficient to compare COIN and Alexa stains over a range of expression levels. By analyzing multiple glands within the same prostate, we avoid variability due to tissue collection, fixation, and processing that we cannot easily determine. Therefore, we designed experiments to measure the same gland in adjacent tissue sections that were stained with COIN- or Alexa-conjugated Abs. To compare the assay characteristics with COIN–Ab conjugates to Alexa–Ab conjugates, we constructed three overall quality measures: signal-to-background (S/B) ratio, spot-to-spot variability, and staining accuracy. S/B ratio is the average signal in the epithelium, where PSA is expressed, divided by the average signal in the stroma where PSA is not expressed. These measures did not include contributions from the autofluorescence, which is specifically removed by the spectral deconvolution algorithm before intensities are reported. Therefore, the background is defined here as nonspecific binding of the Ab-conjugated probe to the stroma. Spot-to-spot variability for a gland and raster is calculated from the SD of spot intensities within the epithelium divided by the mean signal intensity (i.e., %CV). In a visual representation, spot-to-spot variability is recognized as the overall non-uniformity of staining and depicted qualitatively in Figures 2D and 2E. Accuracy is the fraction of points correctly classified as positive in the epithelium or negative in the stroma using the optimum intensity threshold for each gland. Accuracy is demonstrated qualitatively in Figures 2F and 2G.
The three quality measures were calculated for each individual gland, and Table 1 shows summary statistics for 10 glands stained by anti-PSA–COIN or anti-PSA–Alexa in adjacent slides. For reference, results are shown for conventional analysis using filter cube images of the same glands. Accuracy of the COIN assay is comparable to typical fluorescence analysis using filter imaging but less than the reference Alexa assay with spectral imaging (p<0.01). S/B ratio is larger for the COIN assay (p<0.01) than for the Alexa gold standard assay. As noted above, background staining by Alexa was distributed uniformly throughout the stroma, whereas the COIN background resulted from a few errant particles. We expected the larger S/B ratio of COIN to yield greater accuracy by improving the ability to differentiate positive and negative signals, but found the opposite. This difference may result from two contributions: (1) a distribution of COIN intensities in the epithelium as seen in Table 1 by the larger spot-to-spot variability for COIN and/or (2) a background signal arising from a small number of COIN bound to the stroma.
Table 1.
Summary statistics for anti-PSA–COIN and anti-PSA–Alexa on adjacent slides
| Detection | Analysis method | Signal/background (mean ± SD) | Spot-to-spot variation (mean ± SD) | Accuracya (mean ± SD) |
|---|---|---|---|---|
| PSA–Alexa | Image analysisb | 3 ± 1 | 43 ± 9 | 0.91 ± 0.03 |
| PSA–Alexa | Spectral analysis | 5 ± 1 | 47 ± 7 | 0.97 ± 0.01 |
| PSA–COIN | Spectral analysis | 8 ± 2 | 97 ± 19 | 0.90 ± 0.04 |
Fraction of correctly classified spots in the epithelium and stroma, i.e., (true positive pixels + true negative pixels)/total pixels.
See Materials and Methods.
We reasoned that the increased spot-to-spot variability of the signal from COIN–Ab conjugates is caused either by aggregation of COIN–Ab particles, which generates high intensity spots, or by Abs attached to small, poorly functional COIN, which results in a dim signal. To further explore these causes of spot-to-spot variability, we detected bound primary COIN-conjugated Abs with a secondary Ab–Alexa conjugate (Figure SF1). If Ab is bound, but no COIN signal is detectable, the primary COIN–Ab conjugate can be visualized after binding to a secondary Alexa-conjugated Ab. Staining results revealed a significant decrease in spot-to-spot variability of the secondary Alexa stain compared with the primary COIN stain (Table ST1), suggesting that the low-intensity spots were occupied by primary Abs that emit little or no COIN signal. As expected, because several secondary Abs bind a single primary Ab, the signal intensity under conditions using a secondary Alexa-conjugated Ab was greater than with the directly conjugated primary Alexa Ab (p=0.01). Spot-to-spot variability and accuracy of the secondary Alexa Ab were similar to conditions using direct Alexa–Ab conjugates (p=0.32, p=0.74, respectively). As a next step, we tested whether spot-to-spot variability can be improved by size purification of the COIN, which eliminates aggregates. Although we observed a reduction in particle diameter after size purification (Figure SF2), we did not observe a corresponding decline in spot-to-spot variability (p=0.15, Table ST1). We conclude from these data that COIN–Ab aggregates do not reduce the quality of the COIN reagent, and that an improvement of staining characteristics should be achievable by eliminating the low-intensity COIN particles.
Table 2 presents results to demonstrate the reproducibility of the COIN assay, and Alexa results from parallel experiments are included for comparison. Intra-assay variability is calculated from glands measured within the same slide, and inter-assay variability includes glands measured on different days using the same reagents. Reported values are the %CV among individual glands, and values in parentheses are the average and SD that were used for the calculation. Spot-to-spot variabilities and accuracies were of high consistency for both COIN and Alexa stains, whereas S/B ratios were considerably more variable. We attribute this variability (%CVs >40) to the significant component of biological variability in PSA expression between glands. Inter-lot variability includes glands measured in four independent experiments using different COIN preparations. In all cases, COIN assays were reproducible (p>0.19 for comparison of %CVs in the intra- and inter-assay analysis). Thus, despite variation between lots due to multiple steps of the COIN fabrication process, COIN–Ab conjugates from different lots provide consistent results in repeated assays.
Table 2.
Reproducibility of PSA measurements with COIN- or Alexa-conjugated antibody reagents
| Detection method | Signal/background %CV (mean ± SD) | Spot-to-spot variation %CV (mean ± SD) | Accuracya %CV (mean ± SD) |
|---|---|---|---|
| Intra-assay variabilityb | |||
| PSA–COIN | 43% (13 ± 6) | 14% (111 ± 16) | 3% (0.91 ± 0.03) |
| PSA–Alexa | 45% (6 ± 3) | 11% (34 ± 4) | 2% (0.97 ± 0.02) |
| Inter-assay variabilityc | |||
| PSA–COIN | 54% (10 ± 5) | 10% (109 ± 11) | 4% (0.89 ± 0.04) |
| PSA–Alexa | 41% (5 ± 2) | 13% (33 ± 4) | 2% (0.97 ± 0.02) |
| Inter-lot variabilityd | |||
| PSA–COIN | 48% (9 ± 4) | 19% (100 ± 19) | 3% (0.89 ± 0.03) |
Fraction of correctly classified spots in the epithelium and stroma, i.e., (true positive pixels + true negative pixels)/total pixels.
Variability between individual glands on the same slide. Data include nine gland pairs measured in parallel for COIN and Alexa in one experiment. This data set is representative of individual experiments, four of which are summarized by the inter-lot variability below.
Variability between glands measured on different days using the same reagents. Data include a total of 13 gland pairs measured in parallel for COIN and Alexa in two experiments.
Variability between glands measured on different days using different reagents. Data include independent experiments from four different COIN.
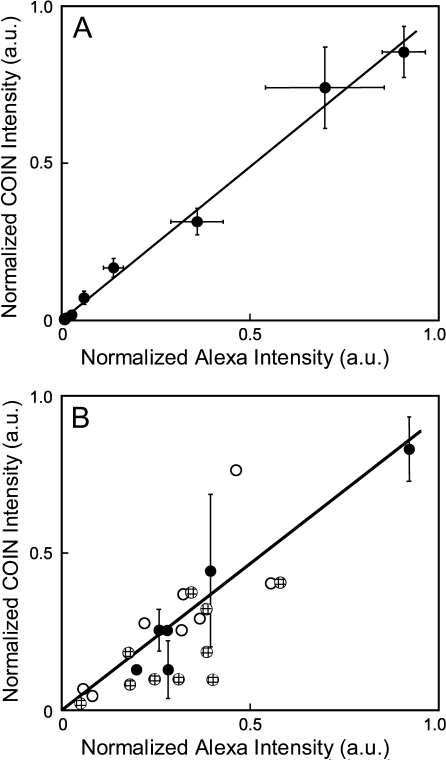
To determine whether COIN–Ab conjugates provide an accurate quantitative measurement of PSA concentration, we determined the correlation between quantitative measurements by COIN and Alexa stains. Figure 3A shows a direct comparison of COIN- and Alexa-conjugated Abs in the simple format of a plate-binding assay. A Pearson correlation coefficient of 0.996 was obtained for parallel Alexa and COIN stains on 3 separate days. Next we determined the correlation between COIN and Alexa stains in tissues by measuring the same gland on adjacent slides that were either stained with COIN- or Alexa-conjugated Abs. Figure 3B directly compares the COIN and Alexa signals for 24 individual glands, including measurements performed on different days and with different COIN preparations. One set of data points includes multiple measurements of the same gland on adjacent tissue sections (black circles), and error bars represent the SD. A similar comparison has been reported for immunofluorescence of the c-Met Ab, and correlation coefficients of 0.17–0.8 were observed in the datasets of sequential sections (Pozner-Moulis et al. 2007). Despite the biological variation in PSA expression between adjacent slides, results show that COIN and Alexa report highly correlated measurements of signal intensities (Pearson correlation coefficient 0.85).
Figure 3.
Direct quantitative comparison between anti-PSA–Alexa and anti-PSA–COIN in parallel plate assays and tissue assays. (A) Direct comparison of intensities reported by anti-PSA–Alexa and anti-PSA–COIN for parallel wells in a plate-binding assay. Each data point is the average of three experiments conducted using the same COIN and Alexa reagents on different days, with corresponding error bars. Pearson correlation coefficient for the entire set is 0.996. (B) Direct comparison of intensities reported by anti-PSA–Alexa or anti-PSA–COIN from the same glands in adjacent tissue sections. COIN intensities are normalized by the intensity of the corresponding direct binding assay for each COIN preparation. COIN and Alexa intensities are reported as the signal-to-background ratio for each gland. Data include experiments performed the same day on adjacent slides (black circles), experiments using the same COIN on different days (black circles and cross-hatched circles), and experiments using completely different COIN preparations (black circles and white circles). Black symbols represent measurements using three different COIN preparations applied to adjacent slides and measured on the same day, and error bars represent the precision of replicate measurements. Pearson correlation coefficient for the entire set is 0.85.
Discussion
Protein detection in tissues with directly conjugated Abs requires exceptionally bright labels, and a major challenge for Raman probe development has been fabrication of probes with sufficient brightness. COIN takes advantage of the relatively large enhancement provided by clustering silver nanoparticles (Michaels et al. 2000; Doering and Nie 2002; Jiang et al. 2003). A small number of applications have used Raman probes directly conjugated to Abs to perform tissue (Schlucker et al. 2006; Sun et al. 2007) or cell assays (Nithipatikom et al. 2003; Kim et al. 2006). We showed here that COIN–Ab direct conjugates have intensities comparable to the bright Alexa Fluor 568 for identical assays and measurement conditions (Figure 1 and Figures 2B and 2C). Further, the bright signals from COIN allowed very short spectral acquisition times (0.1 sec per point, 650-μW laser power), which permitted us to use raster imaging containing up to 900 spots for routine measurements.
As with other nanoparticle probes including quantum dots (Medintz et al. 2005), probes and assay conditions must be developed to deal with nanoparticle-specific effects, such as the potential for Ab inactivation, steric interference, and nonspecific binding. BSA encapsulation of COIN was developed to provide robust conjugation of COIN nanoparticles to Abs and to reduce nonspecific binding to tissue (Sun et al. 2007). In addition, antigen retrieval conditions, blocking buffers, and staining conditions required minor adjustments for COIN–Ab conjugates, but the same protocol was used in our experiments for both anti-PSA–COIN and anti-PSA–Alexa. To explore potential steric effects, we have performed duplex dose-titration experiments in a plate-binding format and on tissue in which the concentration of one COIN was varied, whereas the concentration of the other was held constant. We found no significant interference of COIN signals over a 4-fold variation in each COIN concentration (unpublished data), although more work is needed to identify steric hindrances that may occur with other COIN–Ab conjugates.
In this study the Alexa stain served as a well-established and frequently used reference method for comparison to the COIN assay. We observe that in archival sections of radical prostatectomies, which are often poorly fixed, Alexa reports nearly perfect accuracy for binary identification of PSA expression when data are acquired by spectral analysis (Table 1). Results demonstrate that spectral detection and analysis improve specificity compared with the conventional filter cube method and validates our approach for signal quantitation.
Each spot in the raster provides a specific signal intensity, and we use the variability of spot intensity as a parameter in the evaluation of the COIN staining. Typically, spot-to-spot variability of Alexa stain is approximately half as large as the variability of the COIN stain. We clearly demonstrated that the increased spot-to-spot differences in the COIN staining are due to COIN-free Ab or poorly functional COIN because a secondary Alexa Ab interacted in a uniform fashion with epithelium that was presaturated with COIN Ab (Figure SF1, Table ST1). The protocol we developed with primary COIN and secondary Alexa Ab staining can be used as an assay to follow the removal of poorly functional COIN–Ab conjugates, which we expect will enhance the quality of COIN staining. Improved uniformity may be particularly valuable for high-resolution imaging (e.g., Figure 2), whereas low-resolution imaging (e.g., Figure 1) and reporting of average gland intensities (e.g., Figure 3) do not suffer in quality. We note that imaging with this detail has not been reported for Raman probes nor have quality measures of this detail been reported. The thorough and systematic evaluation of COIN–Ab conjugates in comparison to Alexa dyes clearly demonstrates the excellent performance and reproducibility of COIN–Ab reagents (Table 1 and Table 2) and encourages future COIN reagent and assay development for analysis of human tissue sections.
Quantitative abilities of Raman probes including COIN (e.g., Figure 3A) have been clearly demonstrated in solution (Faulds et al. 2005,2007) and plate-binding assays (Grubisha et al. 2003; Driskell et al. 2005; Su et al. 2005; Xu et al. 2005; Gong et al. 2007; Sun et al. 2007). We showed that Alexa and COIN report highly correlated intensities for matched glands on adjacent slides (Figure 3B), which suggests the potential to extend quantitative analysis with COIN–Ab conjugates to complex samples. The true potential of Raman probes for quantitative signal detection in tissues lies in the ability to accurately separate probe signals from background autofluroescence, which is particularly advantageous for low-abundant analytes and low-intensity signals. For the anti-PSA–COIN assay used here, COIN intensities were many times greater than background autofluorescence (Figure 2C), but we have shown in a previous study that weaker COIN signals can be accurately separated from autofluorescence of comparable intensity (Sun et al. 2007). In our experiments with FFPE prostate tissues, we routinely find large variations in magnitude and shape of autofluorescence across a single sample, and variations between samples are dependent on fixation and antigen retrieval (not shown). The combination of COIN probes and spectral analysis allows us to specifically remove autofluorescence at each point due to the dramatic difference between the sharp Raman peaks and the broad autofluorescence signature. Specifically, we account for unknown variations in background by using a freely varying polynomial in the spectral deconvolution algorithm. The polynomial is able to fit unknown, smoothly varying background such as autofluorescence, without interfering with the fit of Raman probes. In contrast, measurements from fluorophores are difficult to distinguish from unknown autofluorescence because probe signals share similar spectral features with autofluorescence.
Quantitation and multiplex detection of proteins require probes that can be differentiated from one another and from confounding background signals. Advances in immunofluorescence detection have been realized both by adoption of spectral instrumentation and by development of probes with improved optical properties (Levenson and Mansfield 2006). For example, quantum dots provide a significant improvement over conventional fluorophores due to features such as reduced peak width, reduced photobleaching, single-laser excitation, and brightness sufficient for direct conjugation to detection molecules (Medintz et al. 2005; True and Gao 2007). Similar benefits have motivated the recent development of Raman probes, with the added advantage of greatly reduced peak width (∼2 nm, Figure 2C) compared with quantum dots (30–40 nm) or molecular fluorophores (50–70 nm). The unique fingerprint of narrow peaks emitted by Raman probes increases the multiplexing potential and the ability to quantitatively separate signals from background autofluorescence. We have shown simultaneous detection and deconvolution of two colocalized COIN in FFPE prostate tissues (Sun et al. 2007), and we are currently working on triplex detection using two COIN–Ab direct conjugates and a conventional fluorophore.
In this study we illustrated the features of COIN assay in comparison to a conventional fluorophore, determined the assay characteristics of COIN for protein detection in FFPE tissue sections, and demonstrated initial steps toward quantitative antigen detection using COIN–Ab conjugates.
Supplementary Material
Acknowledgments
The authors thank Intel Digital Health Group's Biomedical and Life Sciences research team for development and synthesis of COIN reagents. We thank Dr. Kung-bin Sung for his role in development and initial application of COIN for tissue analysis. The authors thank Kim Adolphson at the Fred Hutchinson Cancer Research Center for significant contributions to development of COIN assay conditions and Dr. Mark Roth at the Fred Hutchinson Cancer Research Center for facilitating the collaboration and inspiring project discussion.
References
- Albrecht MG, Creighton JA (1977) Anomalously intense Raman-spectra of pyridine at a silver electrode. J Am Chem Soc 99:5215–5217 [Google Scholar]
- Byers RJ, Di Vizio D, O'Connell F, Tholouli E, Levenson RM, Gossard K, Twomey D, et al. (2007) Semiautomated multiplexed quantum dot-based in situ hybridization and spectral deconvolution. J Mol Diagn 9:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp RL, Chung GG, Rimm DL (2002) Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med 8:1323–1328 [DOI] [PubMed] [Google Scholar]
- Cao YWC, Jin RC, Mirkin CA (2002) Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science 297:1536–1540 [DOI] [PubMed] [Google Scholar]
- Chen JW, Jiang JH, Gao X, Gong JL, Shen GL, Yu RQ (2007) Gold-aggregated, dye-embedded, polymer-protected nanoparticles (GDPNs): a new type of tags for detection with SERS. Colloids and Surfaces A: Physicochem Eng Aspects 294:80–85 [Google Scholar]
- Cui Y, Ren B, Yao JL, Gu RA, Tian ZQ (2006) Synthesis of AgcoreAushell bimetallic nanoparticles for immunoassay based on surface-enhanced Raman spectroscopy. J Phys Chem B 110:4002–4006 [DOI] [PubMed] [Google Scholar]
- Doering WE, Nie SM (2002) Single-molecule and single-nanoparticle SERS: examining the roles of surface active sites and chemical enhancement. J Phys Chem B 106:311–317 [Google Scholar]
- Doering WE, Nie SM (2003) Spectroscopic tags using dye-embedded nanoparticles and surface-enhanced Raman scattering. Anal Chem 75:6171–6176 [DOI] [PubMed] [Google Scholar]
- Driskell JD, Kwarta KM, Lipert RJ, Porter MD, Neill JD, Ridpath JF (2005) Low-level detection of viral pathogens by a surface-enhanced Raman scattering based immunoassay. Anal Chem 77:6147–6154 [DOI] [PubMed] [Google Scholar]
- Faulds K, McKenzie F, Smith WE, Graham D (2007) Quantitative simultaneous multianalyte detection of DNA by dual-wavelength surface-enhanced resonance Raman scattering. Angew Chem Int Ed 46:1829–1831 [DOI] [PubMed] [Google Scholar]
- Faulds K, Stewart L, Smith WE, Graham D (2005) Quantitative detection of dye labelled DNA using surface enhanced resonance Raman scattering (SERRS) from silver nanoparticles. Talanta 67:667–671 [DOI] [PubMed] [Google Scholar]
- Fleischmann M, Hendra PJ, McQuillan A (1974) Raman-spectra of pyridine adsorbed at a silver electrode. Chem Phys Lett 26:163–166 [Google Scholar]
- Fountaine TJ, Wincovitch SM, Geho DH, Garfield SH, Pittaluga S (2006) Multispectral imaging of clinically relevant cellular targets in tonsil and lymphoid tissue using semiconductor quantum dots. Mod Pathol 19:1181–1191 [DOI] [PubMed] [Google Scholar]
- Gong J-L, Liang Y, Huang Y, Chen J-W, Jiang J-H, Shen G-L, Yu R-Q (2007) Ag/SiO2 core-shell nanoparticle-based surface-enhanced Raman probes for immunoassay of cancer marker using silica-coated magnetic nanoparticles as separation tools. Biosens Bioelectron 22:1501–1507 [DOI] [PubMed] [Google Scholar]
- Grubisha DS, Lipert RJ, Park HY, Driskell J, Porter MD (2003) Femtomolar detection of prostate-specific antigen: an immunoassay based on surface-enhanced Raman scattering and immunogold labels. Anal Chem 75:5936–5943 [DOI] [PubMed] [Google Scholar]
- Hu QY, Tay LL, Noestheden M, Pezacki JP (2007) Mammalian cell surface imaging with nitrile-functionalized nanoprobes: biophysical characterization of aggregation and polarization anisotropy in SERS imaging. J Am Chem Soc 129:14–15 [DOI] [PubMed] [Google Scholar]
- Jeanmaire DL, Vanduyne RP (1977) Surface Raman spectroelectrochemistry. 1. Heterocyclic, aromatic, and aliphatic-amines adsorbed on anodized silver electrode. J Electroanal Chem 84:1–20 [Google Scholar]
- Jiang J, Bosnick K, Maillard M, Brus L (2003) Single molecule Raman spectroscopy at the junctions of large Ag nanocrystals. J Phys Chem B 107:9964–9972 [Google Scholar]
- Jun BH, Kim JH, Park H, Kim JS, Yu KN, Lee SM, Choi H, et al. (2007) Surface-enhanced Raman spectroscopic-encoded beads for multiplex immunoassay. J Comb Chem 9:237–244 [DOI] [PubMed] [Google Scholar]
- Kim JH, Kim JS, Choi H, Lee SM, Jun BH, Yu KN, Kuk E, et al. (2006) Nanoparticle probes with surface enhanced Raman spectroscopic tags for cellular cancer targeting. Anal Chem 78:6967–6973 [DOI] [PubMed] [Google Scholar]
- Kim K, Lee HS, Kim NH (2007) Silver-particle-based surface-enhanced resonance Raman scattering spectroscopy for biomolecular sensing and recognition. Anal Bioanal Chem 388:81–88 [DOI] [PubMed] [Google Scholar]
- Kneipp J, Kneipp H, Rice WL, Kneipp K (2005) Optical probes for biological applications based on surface-enhanced Raman scattering from indocyanine green on gold nanoparticles. Anal Chem 77:2381–2385 [DOI] [PubMed] [Google Scholar]
- Kneipp K, Kneipp H, Manoharan R, Hanlon EB, Itzkan I, Dasari RR, Feld MS (1998) Extremely large enhancement factors in surface-enhanced Raman scattering for molecules on colloidal gold clusters. Appl Spectrosc 52:1493–1497 [Google Scholar]
- Lee S, Kim S, Choo J, Shin SY, Lee YH, Choi HY, Ha S, et al. (2007) Biological imaging of HEK293 cells expressing PLCγ1 using surface-enhanced Raman microscopy. Anal Chem 79:916–922 [DOI] [PubMed] [Google Scholar]
- Levenson RM, Mansfield JR (2006) Multispectral imaging in biology and medicine: slices of life. Cytometry A 69A:748–758 [DOI] [PubMed] [Google Scholar]
- McCabe AF, Eliasson C, Prasath RA, Hernandez-Santana A, Stevenson L, Apple I, Cormack PAG, et al. (2006) SERRS labelled beads for multiplex detection. Faraday Discuss 132:303–308 [DOI] [PubMed] [Google Scholar]
- Medintz IL, Uyeda HT, Goldman ER, Mattoussi H (2005) Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater 4:435–446 [DOI] [PubMed] [Google Scholar]
- Michaels AM, Jiang J, Brus L (2000) Ag nanocrystal junctions as the site for surface-enhanced Raman scattering of single rhodamine 6G molecules. J Phys Chem B 104:11965–11971 [Google Scholar]
- Mulvaney SP, Musick MD, Keating CD, Natan MJ (2003) Glass-coated, analyte-tagged nanoparticles: a new tagging system based on detection with surface-enhanced Raman scattering. Langmuir 19:4784–4790 [Google Scholar]
- Ni J, Lipert RJ, Dawson GB, Porter MD (1999) Immunoassay readout method using extrinsic Raman labels adsorbed on immunogold colloids. Anal Chem 71:4903–4908 [DOI] [PubMed] [Google Scholar]
- Nie SM, Emery SR (1997) Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science 275:1102–1106 [DOI] [PubMed] [Google Scholar]
- Nithipatikom K, McCoy MJ, Hawi SR, Nakamoto K, Adar F, Campbell WB (2003) Characterization and application of Raman labels for confocal Raman microspectroscopic detection of cellular proteins in single cells. Anal Biochem 322:198–207 [DOI] [PubMed] [Google Scholar]
- Pozner-Moulis S, Cregger M, Camp RL, Rimm DL (2007) Antibody validation by quantitative analysis of protein expression using expression of Met in breast cancer as a model. Lab Invest 87:251–260 [DOI] [PubMed] [Google Scholar]
- Schlucker S, Kustner B, Punge A, Bonfig R, Marx A, Strobel P (2006) Immuno-Raman microspectroscopy: in situ detection of antigens in tissue specimens by surface-enhanced Raman scattering. J Raman Spectrosc 37:719–721 [Google Scholar]
- Su X, Zhang J, Sun L, Koo TW, Chan S, Sundararajan N, Yamakawa M, et al. (2005) Composite organic-inorganic nanoparticles (COINs) with chemically encoded optical signatures. Nano Lett 5:49–54 [DOI] [PubMed] [Google Scholar]
- Sun L, Sung KB, Dentinger C, Lutz B, Nguyen L, Zhang JW, Qin HY, et al. (2007) Composite organic-inorganic nanoparticles as Raman labels for tissue analysis. Nano Lett 7:351–356 [DOI] [PubMed] [Google Scholar]
- True LD, Gao XH (2007) Quantum dots for molecular pathology—their time has arrived. J Mol Diagn 9:7–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurui H, Nishimura H, Hattori S, Hirose S, Okumura K, Shirai T (2000) Seven-color fluorescence imaging of tissue samples based on Fourier spectroscopy and singular value decomposition. J Histochem Cytochem 48:653–662 [DOI] [PubMed] [Google Scholar]
- Vo-Dinh T, Yan F, Wabuyele MB (2005) Surface-enhanced Raman scattering for medical diagnostics and biological imaging. J Raman Spectrosc 36:640–647 [Google Scholar]
- Xu S, Ji X, Xu W, Zhao B, Dou X, Bai Y, Ozaki Y (2005) Surface-enhanced Raman scattering studies on immunoassay. J Biomed Opt 10:031112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.