Abstract
The genomes of most eukaryotes are composed of genes arranged on the chromosomes without regard to function, with each gene transcribed from a promoter at its 5′ end. However, the genome of the free-living nematode Caenorhabditis elegans contains numerous polycistronic clusters similar to bacterial operons in which the genes are transcribed sequentially from a single promoter at the 5′ end of the cluster. The resulting polycistronic pre-mRNAs are processed into monocistronic mRNAs by conventional 3′ end formation, cleavage, and polyadenylation, accompanied by trans-splicing with a specialized spliced leader (SL), SL2. To determine whether this mode of gene organization and expression, apparently unique among the animals, occurs in other species, we have investigated genes in a distantly related free-living rhabditid nematode in the genus Dolichorhabditis (strain CEW1). We have identified both SL1 and SL2 RNAs in this species. In addition, we have sequenced a Dolichorhabditis genomic region containing a gene cluster with all of the characteristics of the C. elegans operons. We show that the downstream gene is trans-spliced to SL2. We also present evidence that suggests that these two genes are also clustered in the C. elegans and Caenorhabditis briggsae genomes. Thus, it appears that the arrangement of genes in operons pre-dates the divergence of the genus Caenorhabditis from the other genera in the family Rhabditidae, and may be more widespread than is currently appreciated.
Keywords: polycistronic transcription, Caenorhabditis elegans, splice leader, SL RNA
In bacteria and archaea, the genomes are primarily organized in arrays of genes whose products have related functions. These gene clusters, called operons, are cotranscribed from an upstream promoter and the resulting polycistronic mRNA is translated by ribosomes initiating at or near the 5′ end of the RNA. These operons serve to efficiently coregulate proteins that function together. In contrast, eukaryotes have genomes composed of genes arranged apparently at random, with each transcribed by a promoter at its 5′ end. However, in a group of primitive eukaryotic protozoa, the trypanosomes, genes are transcribed polycistronically (1–3). In this case, the polycistronic pre-mRNA is processed by 3′ end formation and trans-splicing to create conventional eukaryotic monocistronic mRNAs. The trans-splicing reaction that creates the 5′ ends of the mRNAs is related to the cis-splicing of higher eukaryotes; it proceeds through a 2′–5′ branched intermediate, the splice sites have the same consensus sequences, and it is catalyzed by some of the same small nuclear ribonucleoprotein particles (4, 5).
Trans-splicing was first discovered in trypanosomatids (4, 6), and later shown to occur also in Caenorhabditis elegans and other nematodes (ref. 7; reviewed in refs. 8 and 9), in Euglena (10), and in flatworms (11, 12). In contrast to trypanosomes, in which only trans-splicing is present, the genes in the other organisms also contain cis-spliced introns. It was presumed that these genes were monocistronic and arranged randomly on the chromosomes as in other eukaryotes. However, it was recently discovered that the free-living nematode C. elegans does have polycistronic transcription units, as in trypanosomes, and it uses a special small nuclear ribonucleoprotein, the SL2 small nuclear ribonucleoprotein, to process the downstream genes in the polycistronic pre-mRNAs into monocistronic mRNAs (13, 14).
Trans-splicing in nematodes involves the donation of a 22 nucleotide spliced leader (SL), including a trimethylguanosine cap, from a small nuclear RNA to the 5′ ends of pre-mRNAs (15–20). While the exact function of the SL is unknown, it has recently been shown to increase translation efficiency of those mRNAs containing it at their 5′ ends in Ascaris lumbricoides in vitro extracts (21).
C. elegans has two different SLs, each 22 nt long. The first to be discovered, SL1 (7), is trans-spliced onto the 5′ ends of about half of C. elegans mRNAs (14). The signal for SL1 trans-splicing is the presence of a short sequence, called an outron, at the 5′ end of the pre-mRNA (22). The outron is simply an AU-rich sequence lacking a functional 5′ splice site, followed by a 3′ splice site (23, 24).
The second SL to be discovered, SL2 (25), was shown to be reserved for splicing to downstream genes in polycistronic pre-mRNAs derived from cotranscribed gene clusters, or operons (13). The resulting polycistronic pre-mRNAs are processed into monocistronic units by cleavage and polyadenylation and by trans-splicing as they are synthesized. Approximately 25% of C. elegans genes are arranged in operons (14). The genes in these clusters are oriented in the same 5′ to 3′ direction and are separated by ≈100 bp of intercistronic DNA, or in rarer cases 300–400 bp (26). In every case investigated, the first gene in the cluster is either trans-spliced to SL1 or is not trans-spliced, whereas the downstream genes are trans-spliced, either exclusively to SL2 or to a mixture of SL2 and SL1 (13, 14). Since SL2 has only been found trans-spliced to downstream genes in operons, we interpret the presence of SL2 on the 5′ end of an mRNA as evidence of an operon (27).
Although C. elegans operons may not always serve to coregulate genes whose products function together, as in bacteria, there are a few clear examples of operons that serve this purpose. For example the deg-3 operon contains two subunits of the same acetylcholine receptor (ref. 28; M. Treinin and M. Chalfie, personal communication), and the lin-15 operon encodes two unrelated proteins whose products collaborate in the cell signaling pathway resulting in vulva formation (29, 30).
It is not yet known how widespread SL2 and operons are in the nematode phylum. SL1 has been detected in all nematode species studied with virtually no sequence variation (reviewed in ref. 9). In contrast, SL2 has been found only in Caenorhabditis species (25). Comparison of C. elegans with Caenorhabditis briggsae, two species that have been estimated to have been separated for 25–50 million years (31), shows perfect conservation of SL2 and in several cases, the same operons found in the C. elegans genome are present in the genome of C. briggsae (32–34).
In this paper we report, to the best of our knowledge, the first example of an operon and of SL2 trans-splicing in a nematode outside of the genus Caenorhabditis: the rhabditid Dolichorhabditis (CEW1). The isolate CEW1 is an undescribed sibling species of Rhabditis (Oscheius) tipulae, a member of the Dolichura Group (35). Placement of CEW1 and R. tipulae in a genus is actively under investigation (L. Carta, personal communication), but CEW1 is provisionally called Dolichorhabditis. Although Dolichorhabditis is a small free-living nematode, it is much more evolutionarily distant from C. elegans than is C. briggsae. This conclusion is based on the sequence of SL1 and SL2 RNAs, as well as a comparison of the vit-6 genes from the two species, where most introns interrupt the gene at different locations and even the preferred codon usage is somewhat different (36). Molecular phylogenetic analysis of full-length small subunit ribosomal RNA sequences (M. Blaxter, P. DeLey, L. Liu, and J. Garey, personal communication) places Dolichorhabditis in a clade of rhabditid nematodes that includes the genera Rhabditis (free-living bacteriovores) and Heterorhabditis (entomopathogenic nematodes), as well as the order Strongylida (encompassing vertebrate–parasitic species such as Haemonchus contortus and Ostertagia circumcincta). Our results demonstrate that operons are more widespread, at least among the nematodes, than has been appreciated.
MATERIALS AND METHODS
Standard molecular biological procedures were performed as described (37, 38).
Cloning the Dolichorhabditis rpl-29/rpp-1 Operon.
Construction of the Dolichorhabditis genomic library has been described (36). A 204-bp BamHI–SalI digested C. elegans rpp-1 cDNA PCR product (oligos: rp21–6Sal, 5′-CCCGGTCGACTTACGATTGAAGAATGGC-3′ and rp21–7Bam, 5′-CCCGGGATCCGAGACAGAAGTGATGAGG-3′) was labeled using the Prime-It Random Primer Labeling Kit (Stratagene). This probe was used to screen the Dolichorhabditis genomic library. DNA from a single hybridizing plaque was digested with multiple restriction endonucleases and a 2.4-kb HindIII fragment, which hybridized to the probe, was subcloned into pTZ18U, and sequenced (39) using Sequenase (United States Biochemical).
Determination of SL Specificity of Dolichorhabditis rpl-29 and rpp-1.
This experiment was performed as previously described (14). Reverse transcription (RT) of Dolichorhabditis rpl-29 and rpp-1 mRNAs was performed using gene-specific oligonucleotides (L27B65′-4, 5′-GCCTTCTCAGCGCTGACC-3′ and RP21B65′-1, 5′-CCGGTGATAGCGACCT-3′) followed by PCR using the L27B65′-4 and RP21B65′-1 oligos as upstream primers and the SL1–20 and Dolichorhabditis SL2–20 oligos as upstream primers. The SL oligonucleotides consist of the 5′ 20 nt of the C. elegans SL1 and Dolichorhabditis SL2β and γ spliced leader sequences, respectively. The products were separated on 10% denaturing polyacrylamide gels, transferred by electroblotting to Hybond-N (Amersham), and hybridized with either a Dolichorhabditis rpl-29 RT-PCR product or a C. elegans rpp-1 cDNA PCR product.
Southern Blots.
Genomic DNA isolated from Dolichorhabditis, C. elegans, and C. briggsae was digested with restriction endonucleases, separated electrophoretically on a 1% agarose gel, and transferred to Hybond-N. This blot was then probed with a 283-bp Dolichorhabditis rpl-29 PCR product (oligos: L27B65′-1, 5′-GCACCCTGGAGGACGTG-3′ and L27B65′-4, 5′-GCCTTCTCAGCGCTGACC-3′), which was labeled as above. The low stringency hybridization conditions were 5× SSC, 2× Denhardt’s solution, 0.05 M NaPO4 (pH 6.5), 0.1% SDS, and 0.2 mg/ml salmon sperm DNA, incubated at 48°C. The wash conditions were 2× SSC at room temperature and 1× SSC at 50°C. The annealed probe was removed from the blot (confirmed by autoradiographic exposure) and then the blot was hybridized with the C. elegans rpp-1 cDNA PCR product used to select the Dolichorhabditis clone. Conditions were the same, except that the rpp-1 probe was hybridized at 50°C.
Cloning Dolichorhabditis SL1 Genes.
The Dolichorhabditis genomic library was screened with a 32P-kinased oligonucleotide composed of the first 20 nt of the C. elegans SL1 spliced leader (SL1–20: 5′-GGTTTAATTACCCAAGTTTG-3′). Plaques that hybridized to this probe were isolated. At least two different clones were isolated based on digestion with multiple restriction endonucleases. Fragments that hybridized to the probe were subcloned and sequenced as above.
Cloning Dolichorhabditis SL2 Genes.
To identify the 5′ end of the rpp-1 mRNA, primer extension sequencing was performed essentially as described (18) using an oligodeoxynucleotide, PESEQB6RP21, which hybridized to the Dolichorhabditis rpp-1 RNA and ended 8 nt from the predicted trans-splice site (PESEQB6RP21: 5′-GTAGCCTTAGCTCAAAGAG-3′). The results of this experiment allowed the design of a degenerate oligonucleotide (B6D6 SL2–20: 5′-TTTTVCCCAGNNTCTCAAG-3′), which was used in RT-PCR employing total Dolichorhabditis RNA and an oligonucleotide complementary to nt 90–110 of C. elegans SL2 RNA (SKSL2–3′: 5′-TTTGCTCTACCGGATGACCCC-3′). This PCR product was labeled and used to probe the Dolichorhabditis genomic library. Plaques hybridizing to the probe were isolated as above. Five different clones were obtained based on digestion with multiple restriction endonucleases. Fragments from these digests that hybridized to the probe were subcloned into pTZ18U and sequenced as above.
RESULTS
Cloning of the Dolichorhabditis Genomic Sequence Containing the rpp-1 Gene.
In C. elegans, the gene that encodes an acidic, large-subunit ribosomal protein homologous to P1 in a wide variety of organisms and to RP21c of Drosophila, is trans-spliced to SL2 (RP21, A. Fire and S. Harrison, personal communication). It is thus presumed to be a downstream gene in an operon. Although the region of the C. elegans genome containing the RP21/P1-encoding gene has not yet been cloned or found in the sequenced cosmids of the Genome Sequencing Consortium, we examined the genomic structure of this gene in Dolichorhabditis because ribosomal protein genes tend to be highly conserved and therefore potentially easier to obtain by cross-hybridization than other known SL2-accepting genes. To clone the genomic region containing the Dolichorhabditis homolog of RP21, a Dolichorhabditis genomic library was probed with a 200-bp fragment of C. elegans RP21 cDNA at moderate stringency. A single hybridizing phage was selected; a 2.4-kb HindIII fragment that annealed to the probe was cloned into pTZ18U and its sequence was determined. This genomic fragment contained two entire genes arranged in the same orientation, as expected for an operon (Fig. 1). The rpl-29 gene, encoding another large subunit ribosomal protein (the L29 homolog), was present upstream of the gene encoding a P1/RP21 homolog, rpp-1.
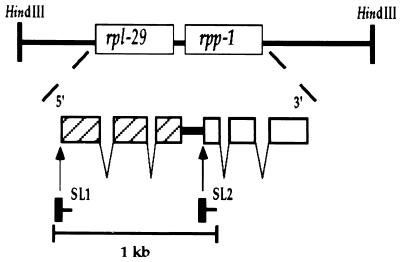
Figure 1.
Schematic diagram of the Dolichorhabditis rpl-29/rpp-1 operon. Exons are denoted by boxes (rpl-29 shaded and rpp-1 open) and introns by angled lines. The intercistronic sequence (87 nt) is represented by a thick line. The trans-splicing reaction in which the spliced leader, represented by solid boxes, is added to the 5′ ends of pre-mRNAs, is represented by arrows from the SL RNAs to the trans-splice site.
The genes in most C. elegans operons are separated by ≈100 bp (26). To determine the location of the 3′ end of the rpl-29 gene, RT-PCR was performed with an oligonucleotide near the end of the coding region and oligo(dT) (data not shown). The size of the fragment indicated that the 3′ end of the mRNA was located 11 bp downstream of an AAUAAA cleavage and polyadenylation signal. The 5′ end of the downstream gene, rpp-1, defined as the trans-splice site, was determined by primer extension sequencing from a primer near the 5′ end of the gene (see below). The results demonstrated that there are only 87 bp between the 3′ end of rpl-29 and the 5′ end of rpp-1. This distance is similar to, but shorter than, the intercistronic distance found in the known C. elegans operons.
Determination of Trans-Splicing Specificity.
If these two ribosomal protein genes constitute an operon, then rpp-1 should be trans-spliced to SL2, and rpl-29 would be trans-spliced to SL1 if it is trans-spliced at all. As a first test, RT-PCR experiments were performed with gene-specific oligonucleotides and oligonucleotides consisting of the first 20 bp of C. elegans SL1 and SL2 sequences (not shown). By this test, rpl-29, the upstream gene was trans-spliced exclusively to SL1, but no trans-spliced product was detectable for rpp-1. This suggested the possibility that Dolichorhabditis SL2, if it existed, was too diverged from C. elegans SL2 to allow cross-hybridization at the stringency used. To determine the sequence of the 5′ end of rpp-1 mRNA, we performed primer extension sequencing with an rpp-1-specific oligonucleotide (not shown). The determined sequence was identical to the genomic sequence of Dolichorhabditis rpp-1 up to the trans-splice site, preceded by a sequence similar to, but slightly different from, C. elegans SL2. The sequence was ambiguous at several sites, which may indicate that rpp-1 in Dolichorhabditis receives a mixture of SL2-related spliced leaders (as do SL2-accepting mRNAs in C. elegans). This suggests that Dolichorhabditis has operons and, that like C. elegans, it trans-splices downstream genes using SL2. Once the Dolichorhabditis SL2 sequence had been determined (see below) it was possible to test the SL specificity of Dolichorhabditis rpl-29 and rpp-1 by RT-PCR (Fig. 2). This demonstrated that rpl-29 is predominantly SL1 trans-spliced, whereas the majority of the rpp-1 mRNA is SL2 trans-spliced. The surprising low-level SL2 trans-splicing to rpl-29 has not been further investigated, but it should be noted that we do not in general see low-level SL2 trans-splicing to C. elegans mRNAs whose genes immediately follow promoters. In contrast, many SL2-accepting mRNAs have been shown to also accept SL1 at detectable levels in C. elegans.
Figure 2.
SL specificity of Dolichorhabditis rpl-29 and rpp-1 mRNAs. Dolichorhabditis RNA was reverse transcribed with gene-specific oligonucleotides, and PCR was performed with the same oligonucleotides and SL1–20 or Dolichorhabditis SL2–20. The products were electrophoresed on 10% polyacrylamide gels, blotted, and probed with either a Dolichorhabditis rpl-29 probe (Left) or a C. elegans rpp-1 probe (Right).
The rpl-29/rpp-1 Clustering Is Conserved.
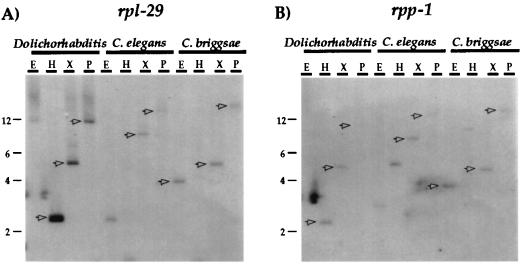
To determine whether the operon arrangement of rpl-29/rpp-1 found in Dolichorhabditis may be conserved in Caenorhabditis, genomic Southern blot analysis of DNA from Dolichorhabditis, C. elegans, and C. briggsae was performed. Each genomic DNA preparation was digested with several different restriction endonucleases, and blots were hybridized at low stringency sequentially with a Dolichorhabditis rpl-29 probe (Fig. 3A), and then with a C. elegans rpp-1 probe (Fig. 3B). If the rpl-29 and rpp-1 genes are clustered in C. elegans and C. briggsae, then the probes made from the two genes should hybridize to at least some of the same genomic fragments (marked with arrows in Fig. 3). C. elegans genomic DNA digested with XbaI or PstI resulted in fragments of ≈8 kb and >12 kb, respectively, that hybridized to both probes. C. briggsae genomic DNA digested with EcoRI, XbaI, or PstI resulted in fragments of ≈3.8 kb, 4.8 kb, and >12 kb, respectively, that hybridized to both probes. These data show that the C. elegans and C. briggsae genomes also contain the rpl-29 and rpp-1 genes in close proximity to one another and indicates that the same two genes may be contained in an operon, as in Dolichorhabditis.
Figure 3.
The rpl-29/rpp-1 cluster is present in C. elegans and C. briggsae. Genomic DNA from Dolichorhabditis, C. elegans, and C. briggsae was cut with the restriction endonucleases EcoRI (E), HindIII (H), XbaI (X), and PstI (P), separated by electrophoresis, transferred to Hybond, and hybridized to an rpl-29 32P-labeled probe (A). The blot was then stripped and rehybridized with an rpp-1 probe (B). Fragments hybridizing to both probes are indicated by arrowheads. Molecular weight markers (in kb) are indicated at the left of each panel.
Cloning of Dolichorhabditis SL1 and SL2 RNA Genes.
In C. elegans SL1 and SL2 are donated by small nuclear RNAs, called SL RNAs, 95 and 110 nt in length, respectively. SL1 RNA is encoded by about 110 genes contained on the same 1-kb tandem repeat that specifies 5S rRNA, but on the opposite strand (7). In contrast, SL2 RNAs are encoded by over 30 genes that are not closely linked to one another (refs. 25 and 40, and see below). However, Southern blot analysis of Dolichorhabditis SL1 RNA genes indicates that they are dispersed throughout the genome and are not associated with the 5S rRNA genes (data not shown).
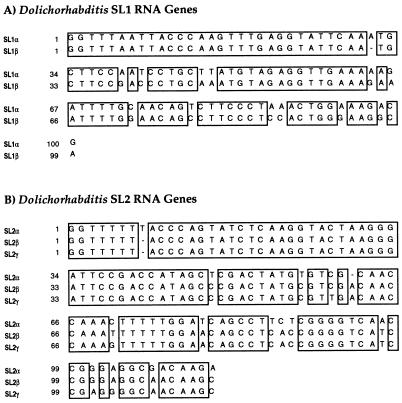
To clone Dolichorhabditis SL1 RNA genes, a Dolichorhabditis genomic library was probed with a labeled oligonucleotide complementary to the first 20 nt of the C. elegans SL1 spliced leader. Plaques that hybridized to this probe were purified, and restriction endonuclease fragments hybridizing to the probe were subcloned and sequenced (Fig. 4A). Each clone contained a single SL1 RNA gene, based on a sequence identical to the SL1 sequence, followed by ≈80 bp that could be folded into the canonical SL RNA secondary structure, three stem-loops with a typical Sm-binding site located in the single-stranded region between the second and third stem-loops (Fig. 5).
Figure 4.
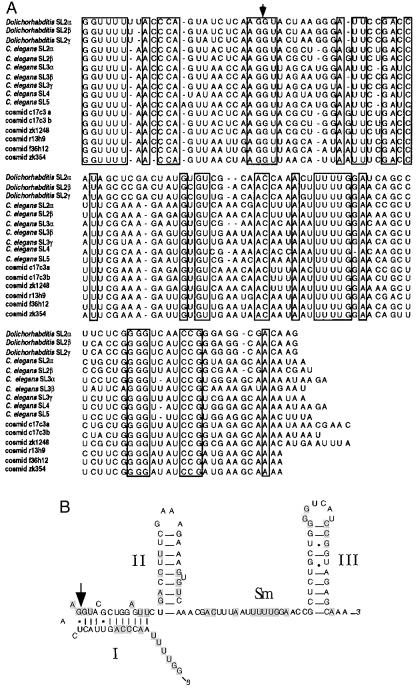
Sequence of Dolichorhabditis SL1 and SL2 RNA genes. Sequences of Dolichorhabditis SL1 and SL2 RNA genes were aligned manually. (A) Nucleotide positions that are conserved in both Dolichorhabditis SL1 genes are boxed. (B) Nucleotide positions conserved in all three sequenced Dolichorhabditis SL2 genes are boxed.
Figure 5.
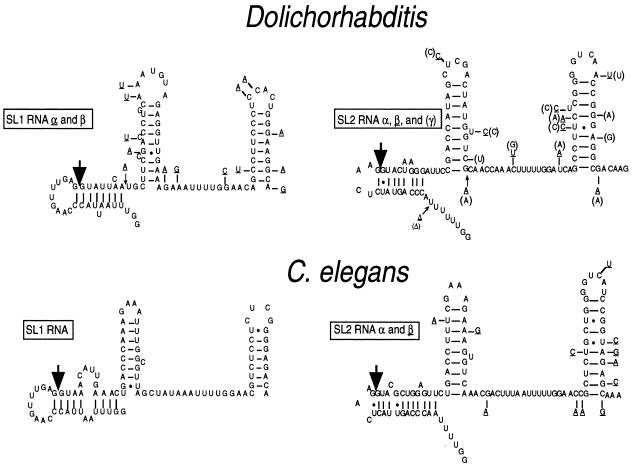
Comparison of proposed secondary structures of SL1 and SL2 RNAs from C. elegans and Dolichorhabditis. Large arrows indicate trans-splice donor sites. Small arrows denote insertions in gene variants. Dashes specify nucleotide changes in gene variants at that position. A Δ represents a gap in the alignment. Dolichorhabditis SL1β is shown with Dolichorhabditis SL1α changes underlined. Dolichorhabditis SL2α is shown with Dolichorhabditis SL2β changes underlined and Dolichorhabditis SL2γ changes in parentheses. C. elegans SL2α is shown with C. elegans SL2β changes underlined.
To clone the Dolichorhabditis SL2 RNA genes, a degenerate oligonucleotide based on the primer extension sequence from the 5′ end of the rpp-1 mRNA was used in combination with an oligonucleotide complementary to nt 90 to 110 of C. elegans SL2 RNA to perform RT-PCR with Dolichorhabditis total RNA. The resulting PCR product was used to probe a Dolichorhabditis genomic library. Twelve clones representing five regions of the genome, were obtained and three SL2 RNA genes were subcloned and sequenced (Fig. 4B). These clones all encode slightly different SL2 RNAs, but each can adopt the 3-stem-loop secondary structure characteristic of nematode SL RNA genes and each contains an appropriately located Sm-binding site (Fig. 5) (17). Whereas Dolichorhabditis SL2 β and γ genes specify typical 22-nt leaders, Dolichorhabditis SL2 α specifies a 23-nt leader (as does one of the C. elegans SL2 variants) containing a string of six uridines instead of five near the 5′ end. There is no other variation in the SL or within the first stem of these three Dolichorhabditis genes. Similarly, there is very little variability in the second stem: only one base of the loop and the bulged pyrimidine vary between the three genes. The proposed single-stranded region containing the Sm-binding site varies at three positions, including one variable position within the Sm-binding site. The third stem is the most variable, with the base of the proposed stem varying at several positions.
These genes represent the first non-C. elegans SL2 RNA genes to be identified. An alignment of their sequences with the homologous C. elegans SL2 RNA sequences (Fig. 6A), reveals several conserved features that may be important for SL2 RNA function. First, the SL2 sequence itself is not as highly conserved as is SL1, which is perfectly conserved throughout the nematodes. The 5′-most sequences of SL2 are however perfectly conserved, as is the trans-splice donor site. It is interesting to note that the splice site on SL2 of both species differs at several positions from the consensus 5′ splice site (G/GURAGU). In C. elegans, the SL2 RNA trans-splice donor site consensus sequence is G/GUWMRH, where W is A or U, M is C or A, R is G or A, and H is C, A, or U. In Dolichorhabditis, the trans-splice donor site sequence is G/GUACUA. Second, the intron portion of SL2 RNA has diverged substantially, but there are a few elements that appear to be conserved, including part of the Sm-binding site (UUUUG), part of stem II, and the top of stem-loop III (Fig. 6B).
Figure 6.
Comparative phylogenetic analysis of SL2 RNAs. (A) All known SL2 genes were aligned manually. The trans-splice site is indicated by an arrow. Nucleotide positions that are conserved in 100% of the sequences are boxed. Cosmids c17c3, zk1248, r13h9, f36h12, and zk354 were identified by performing blast analysis of the output of the C. elegans genome sequencing project using the complete sequence of the C. elegans SL2α gene. (B) Proposed secondary structure of the C. elegans SL2α RNA. Those nucleotide positions that are conserved in 100% of known SL2 genes (from above) are shaded. The stems are numbered as they are referred to in the text. The Sm-binding site is labeled. Note major areas of conservation: spliced leader/trans-splice donor site, Sm-binding region, and third stem-loop.
DISCUSSION
The existence of operons in eukaryotes is evolutionarily intriguing. While operons are a common form of genome organization in both the archaea and the eubacteria, they have in general not been found in eukaryotes. Just how widespread are eukaryotic operons? We have begun to answer this question by identifying both a conserved operon and SL2 trans-splicing in another free-living nematode, Dolichorhabditis. Dolichorhabditis is a free-living rhabditid nematode belonging to the same family as C. elegans (36, 41). Although the two species are morphologically similar, C. elegans 28S rRNA differs from Dolichorhabditis 28S rRNA at 48 of 241 positions (K. Thomas, personal communication). Comparison of the C. elegans and Dolichorhabditis vit-6 genes demonstrates that whereas the primary structure of the encoded protein is related, it is so distant that even its codon usage and intron positions are quite different (36). Recently, an analysis of the 18S rRNA from Dolichorhabditis showed that it is a rhabditid species, most closely related to the parasitic strongylids (M. Blaxter, P. DeLey, L. Liu, and J. Garey, personal communication).
The rpl-29/rpp-1 Operon May Predate the Separation of the Genus Caenorhabditis and the Rest of the Family Rhabditidae.
Although this is the first report of the rpl-29/rpp-1 operon, it was already known that the C. elegans product encoding RP21/P1 is SL2 trans-spliced (A. Fire and S. Harrison, personal communication). However, the gene(s) in the operon upstream of rpp-1 in C. elegans had not previously been identified. Upon discovering that rpl-29 is upstream of rpp-1 in Dolichorhabditis, we asked if this operon could also be present in Caenorhabditis. We showed by genomic Southern blot analysis that the rpl-29/rpp-1 gene cluster is present in both C. elegans and C. briggsae (Fig. 3). Thus, since the two genes are clustered in Caenorhabditis and since one of the genes does receive SL2, this is likely to represent an operon in Caenorhabditis. Hence, it seems probable that this operon was present in the last common ancestor of Caenorhabditis and the Dolichorhabditis/Heterorhabditis/Strongylida clade.
SL2 Trans-Splicing of Downstream Operon mRNAs Is Conserved.
Using primer extension sequencing, we showed that rpp-1 is trans-spliced to an SL2-like spliced leader in Dolichorhabditis. Ambiguity in the primer extension sequencing data may mean that rpp-1 in Dolichorhabditis receives a mixture of SL2-related spliced leaders. Similar observations have been made in C. elegans, where several novel SL2-related spliced leaders were identified on protein kinase C1A, TRA-2, and casein kinase IIb mRNAs (40, 42–44). Using RT-PCR we have subsequently demonstrated that rpl-29 mRNA is predominantly SL1 trans-spliced, whereas rpp-1 is primarily SL2 trans-spliced (Fig. 2). These findings strongly suggest that other nematodes outside the genus Caenorhabditis also use SL2 trans-splicing to process polycistronic pre-mRNAs encoded by operons in a manner similar to that found in C. elegans.
Comparison of SL2 RNA Genes Reveals Conservation of Regions Potentially Important for SL2 Function.
Three genes encoding Dolichorhabditis SL2-like RNAs were cloned and sequenced (Fig. 4). These RNAs are all slightly different. The RNAs encoded by these genes can fold into a secondary structure similar to that proposed for C. elegans SL2 RNA (Fig. 5). Dolichorhabditis SL2α specifies a 23-nt spliced leader. A 23-nt SL2-related spliced leader, known as SL5, has also been identified in C. elegans (40).
By searching the output from the C. elegans sequencing project we identified several other novel C. elegans SL2 RNA genes (Fig. 6A). With this growing number of SL2 RNA gene sequences available, it is possible to identify elements of the SL2 RNA necessary for function using phylogenetic comparative analysis. Alignment of these sequences reveals elements that have been conserved, including the 5′ end of the SL, the trans-splice donor site, part of the second stem, the Sm-binding region, and the top of the third stem-loop (Fig. 6B). The conservation of these elements can be explained in the context of previous experiments.
The spliced leader of the SL1 RNA of Ascaris lumbricoides is necessary for SL RNA transcription in vitro (45). However, conservation of the spliced leader sequence, length, or structure is not required for trans-splicing in vitro (46). Similarly, it is possible that the SL2 sequence itself may not be required for the trans-splicing reaction, but for the transcription of the SL2 RNA gene. Alternatively, it may be conserved because the SL2 sequence is important for some other function such as translation.
The importance of the 3′ half of the second stem and the Sm-binding site was also demonstrated in A. lumbricoides in vitro experiments showing that mutations that prevented formation of an SL ribonucleoprotein prevented efficient trans-splicing (47). Mutations in the Sm-binding site of A. lumbricoides SL RNA that prevent the binding of SL RNP-specific proteins also result in failure to trans-splice in vitro (48).
The last region that is conserved in SL2 RNA is the top of the third stem-loop. Interestingly, the SL1 RNAs of Dolichorhabditis and C. elegans do not show conservation of this region of the third stem-loop (Fig. 5). Therefore, the fact that it is conserved among all SL2 RNAs in Dolichorhabditis and C. elegans is potentially revealing. It is possible that this region of SL2 RNA contributes to the determination of SL2 specificity, perhaps by binding an SL2 RNA-specific polypeptide. SL2 trans-splicing is mechanistically linked to 3′ end formation of the upstream gene (13, 49). We therefore hypothesize that this third stem-loop is a binding site for a protein that is either involved in 3′ end formation of the upsteam gene or can interact with proteins that are. We are currently addressing this hypothesis experimentally.
In conclusion, we report here the discovery of an operon and SL2 trans-splicing in Dolichorhabditis. Furthermore, the gene cluster containing this operon is conserved between C. elegans, C. briggsae, and Dolichorhabditis. This discovery strongly suggests that SL2 trans-splicing was found in the common ancestor of Caenorhabditis and Dolichorhabditis, which implies that eukaryotic operons are evolutionarily older than was previously appreciated and may still be discovered in other eukaryotes.
Acknowledgments
We thank Mark Blaxter and Kelly Thomas for sharing unpublished results on the phylogenetic identification of Dolichorhabditis. This work was supported by Grant GM42432 from the National Institute of General Medical Sciences.
ABBREVIATIONS
- SL
spliced leader
- RT
reverse transcription
Footnotes
References
- 1.Johnson P J, Kooter J M, Borst P. Cell. 1987;51:273–281. doi: 10.1016/0092-8674(87)90154-1. [DOI] [PubMed] [Google Scholar]
- 2.Muhich M L, Boothroyd J C. Mol Cell Biol. 1988;8:3837–3846. doi: 10.1128/mcb.8.9.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tschudi C, Ullu E. EMBO J. 1988;7:455–463. doi: 10.1002/j.1460-2075.1988.tb02833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutton R E, Boothroyd J C. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tschudi C, Ullu E. Cell. 1990;61:459–466. doi: 10.1016/0092-8674(90)90527-l. [DOI] [PubMed] [Google Scholar]
- 6.Murphy W J, Watkins K P, Agabian N. Cell. 1986;47:517–525. doi: 10.1016/0092-8674(86)90616-1. [DOI] [PubMed] [Google Scholar]
- 7.Krause M, Hirsh D. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilsen T W. Annu Rev Microbiol. 1993;47:413–440. doi: 10.1146/annurev.mi.47.100193.002213. [DOI] [PubMed] [Google Scholar]
- 9.Davis R E. Parasitol Today. 1995;12:33–40. doi: 10.1016/0169-4758(96)80643-0. [DOI] [PubMed] [Google Scholar]
- 10.Tessier L H, Keller M, Chan R L, Fournier R, Weil J H, Imbault P. EMBO J. 1991;10:2621–2625. doi: 10.1002/j.1460-2075.1991.tb07804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajkovic A, Davis R E, Simonsen J N, Rottman F M. Proc Natl Acad Sci USA. 1990;87:8879–8883. doi: 10.1073/pnas.87.22.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis R E, Singh H, Botka C, Hardwick C, Ashraf el Meanawy M, Villanueva J. J Biol Chem. 1994;269:20026–20030. [PubMed] [Google Scholar]
- 13.Spieth J, Brooke G, Kuersten S, Lea K, Blumenthal T. Cell. 1993;73:521–532. doi: 10.1016/0092-8674(93)90139-h. [DOI] [PubMed] [Google Scholar]
- 14.Zorio D A R, Cheng N N, Blumenthal T, Spieth J. Nature (London) 1994;372:270–272. doi: 10.1038/372270a0. [DOI] [PubMed] [Google Scholar]
- 15.Thomas J D, Conrad R C, Blumenthal T. Cell. 1988;54:533–539. doi: 10.1016/0092-8674(88)90075-x. [DOI] [PubMed] [Google Scholar]
- 16.Van Doren K, Hirsh D. Nature (London) 1988;335:556–559. doi: 10.1038/335556a0. [DOI] [PubMed] [Google Scholar]
- 17.Bruzik J P, Van Doren K, Hirsh D, Steitz J A. Nature (London) 1988;335:559–562. doi: 10.1038/335559a0. [DOI] [PubMed] [Google Scholar]
- 18.Bektesh S, Van Doren K, Hirsh D. Genes Dev. 1988;2:1277–1283. doi: 10.1101/gad.2.10.1277. [DOI] [PubMed] [Google Scholar]
- 19.Liou R, Blumenthal T. Mol Cell Biol. 1990;10:1764–1768. doi: 10.1128/mcb.10.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Doren K, Hirsh D. Mol Cell Biol. 1990;10:1769–1772. doi: 10.1128/mcb.10.4.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maroney P A, Denker J A, Darzynkiewicz E, Laneve R, Nilsen T W. RNA. 1995;1:714–723. [PMC free article] [PubMed] [Google Scholar]
- 22.Conrad R, Thomas J, Spieth J, Blumenthal T. Mol Cell Biol. 1991;11:1921–1926. doi: 10.1128/mcb.11.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conrad R, Liou R F, Blumenthal T. EMBO J. 1993;12:1249–1255. doi: 10.1002/j.1460-2075.1993.tb05766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conrad R, Lea K, Blumenthal T. RNA. 1995;1:164–170. [PMC free article] [PubMed] [Google Scholar]
- 25.Huang X, Hirsh D. Proc Natl Acad Sci USA. 1989;86:8640–8644. doi: 10.1073/pnas.86.22.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blumenthal T, Steward K. In: Caenorhabditis elegans II. Riddle D, Blumenthal T, Meyer B, Priess J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 117–145. [Google Scholar]
- 27.Blumenthal T. Trends Genet. 1995;11:132–136. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- 28.Treinin M, Chalfie M. Neuron. 1995;14:871–877. doi: 10.1016/0896-6273(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 29.Clark G S, Xiaowei L, Horvitz H R. Genetics. 1994;137:987–997. doi: 10.1093/genetics/137.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang L S, Tzou P, Sternberg P W. Mol Biol Cell. 1994;5:395–411. doi: 10.1091/mbc.5.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy B P, Aamodt E J, Allen F L, Chung M A, Heschl M F P, McGhee J D. J Mol Biol. 1993;229:890–908. doi: 10.1006/jmbi.1993.1094. [DOI] [PubMed] [Google Scholar]
- 32.Lee Y H, Huang X, Hirsh D, Fox G E, Hecht R M. Gene. 1992;121:227–235. doi: 10.1016/0378-1119(92)90126-a. [DOI] [PubMed] [Google Scholar]
- 33.Kuwabara P E, Shah S. Nucleic Acids Res. 1994;22:4414–4418. doi: 10.1093/nar/22.21.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hengartner M O, Horvitz H R. Cell. 1994;76:665–676. doi: 10.1016/0092-8674(94)90506-1. [DOI] [PubMed] [Google Scholar]
- 35.Sudhaus W, Hooper D J. Nematologica. 1994;40:508–533. [Google Scholar]
- 36.Winter C E, Penha C, Blumenthal T. Mol Biol Evol. 1996;13:674–684. doi: 10.1093/oxfordjournals.molbev.a025628. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 38.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross L H, Freedman J H, Rubin C S. J Biol Chem. 1995;270:22066–22075. doi: 10.1074/jbc.270.37.22066. [DOI] [PubMed] [Google Scholar]
- 41.Winter C E. Comp Biochem Physiol B. 1992;103:189–196. [Google Scholar]
- 42.Land M, Islas-Trejo A, Rubin C S. J Biol Chem. 1994;269:14820–14827. [PubMed] [Google Scholar]
- 43.Kuwabara P E, Okkema P G, Kimble J. Mol Biol Cell. 1992;3:461–473. doi: 10.1091/mbc.3.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu E, Rubin C S. J Biol Chem. 1991;266:19796–19802. [PubMed] [Google Scholar]
- 45.Hannon G J, Maroney P A, Denker J A, Nilsen T W. Cell. 1990;61:1247–1255. doi: 10.1016/0092-8674(90)90689-c. [DOI] [PubMed] [Google Scholar]
- 46.Maroney P A, Hannon G J, Shambaugh J D, Nilsen T W. EMBO J. 1991;10:3869–3875. doi: 10.1002/j.1460-2075.1991.tb04956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maroney P A, Hannon G J, Denker J A, Nilsen T W. EMBO J. 1990;9:3667–3673. doi: 10.1002/j.1460-2075.1990.tb07578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denker J A, Maroney P A, Yu Y T, Kanost R A, Nilsen T W. RNA. 1996;2:746–755. [PMC free article] [PubMed] [Google Scholar]
- 49.Kuersten S, Lea K, MacMorris M, Spieth J, Blumenthal T. RNA. 1997;3:269–278. [PMC free article] [PubMed] [Google Scholar]