Abstract
Sequence divergence acts as a potent barrier to homologous recombination; much of this barrier derives from an antirecombination activity exerted by mismatch repair proteins. An inverted repeat assay system with recombination substrates ranging in identity from 74% to 100% has been used to define the relationship between sequence divergence and the rate of mitotic crossing-over in yeast. To elucidate the role of the mismatch repair machinery in regulating recombination between mismatched substrates, we performed experiments in both wild-type and mismatch repair defective strains. We find that a single mismatch is sufficient to inhibit recombination between otherwise identical sequences, and that this inhibition is dependent on the mismatch repair system. Additional mismatches have a cumulative negative effect on the recombination rate. With sequence divergence of up to approximately 10%, the inhibitory effect of mismatches results mainly from antirecombination activity of the mismatch repair system. With greater levels of divergence, recombination is inefficient even in the absence of mismatch repair activity. In both wild-type and mismatch repair defective strains, an approximate log-linear relationship is observed between the recombination rate and the level of sequence divergence.
Recombination between sequences at identical (allelic) positions on homologous chromosomes is important for mitotic DNA repair and for meiotic chromosome disjunction. In addition to the normal allelic interactions, eukaryotic genomes contain large numbers of repeated sequences that can potentially serve as substrates for nonallelic (ectopic) recombination events. Ectopic recombination can be either reciprocal or nonreciprocal in nature and can lead to alterations in genome structure. Nonreciprocal recombination events (gene conversions), which involve the unidirectional transfer of information from one repeat to another, result in either the homogenization of repeats or the generation of novel sequences. Reciprocal recombination events (crossovers) involve the physical exchange of information between repeats and give rise to various types of genome rearrangements (e.g., duplications, deletions, inversions, and translocations). Given the genome-destabilizing effects of ectopic recombination, it is important that such events occur at low rates relative to allelic recombination.
Two physical features of repeated sequences may be important for limiting potentially deleterious interactions (1): the total length of the repeats (substrate size) and the degree of sequence identity between the repeats (substrate homology). Studies in both prokaryotic systems (2, 3) and eukaryotic systems (4–8) have shown that substrate size does influence the efficiency of recombination. Similarly, studies done in a variety of organisms have shown that sequence divergence can be a potent barrier to homologous recombination (9–23). The molecular basis of this barrier is not entirely understood, but it is presumably related to the formation and/or removal of mismatches in recombination intermediates (24, 25). Prokaryotic studies have demonstrated that the recombination barrier afforded by sequence divergence derives largely from antirecombination activity of the replication-associated mismatch repair machinery (10, 11, 13, 14).
The best characterized mismatch repair system is the methyl-directed mismatch repair system of Escherichia coli, which contains three key components (MutS, MutL, and MutH). MutS binds to mismatched bases; MutH incises the newly replicated unmethylated DNA strand at hemi-methylated dam sites; and MutL functions as a molecular matchmaker to bring MutS and MutH together (26). Eukaryotes possess multiple MutS and MutL homologs (no MutH homologs have been identified), which have attracted attention because of their role in preventing the accumulation of cancer-promoting mutations (27). In the yeast Saccharomyces cerevisiae six MutS homologs (Msh1p-6p) and four MutL homologs (Pms1p, Mlh1p-3p) have been identified. Msh2p, Msh3p, Msh6p, Pms1p, and Mlh1p are involved in correcting DNA replication errors; strains defective for any of these proteins have a mutator phenotype (28, 29). In addition to editing the products of DNA replication, there is evidence that these proteins also regulate recombination between mismatched DNA substrates in yeast (17–19, 22). Although elimination of MutS and MutL homologs can yield comparable mutator phenotypes, MutS homologs appear to play a greater role than MutL homologs in regulating recombination between diverged sequences (17, 18).
We described previously an intron-based recombination system that can be used to examine reciprocal recombination between diverged sequences in yeast (17). This system was used to measure mitotic crossing-over between identical (100%) sequences, 91%-identical sequences, and 77%-identical sequences in both wild-type and mismatch repair (MMR) defective strains. Recombination between the 91%- and the 77%-identical sequences was very inefficient relative to recombination between 100% substrates in wild-type strains. In MMR-defective strains, however, recombination between the 91% substrates improved dramatically and was almost as efficient as that between 100% substrates; recombination between the 77% substrates remained inefficient in MMR-defective strains. These results indicate that the MMR machinery imposes the major barrier to recombination between sequences of low divergence; at higher levels of divergence, the sequences themselves may impose a barrier to the recombination machinery. In the work reported here, a large number of recombination substrates exhibiting a wide range of sequence divergence have been used to determine the number of mismatches necessary to trigger the antirecombination activity of the yeast MMR machinery and to define more precisely the distinct roles of DNA sequence divergence and mismatch repair activity in limiting genomic rearrangements.
MATERIALS AND METHODS
Media and Growth Conditions.
S. cerevisiae strains were grown at 30°C. Yeast extract/peptone (YEP) medium (1% yeast extract/2% Bacto-peptone; 2.5% agar for plates) supplemented with 2% glycerol and 4% galactose (YEPGG) or 2% dextrose (YEPD) was used for nonselective growth. Synthetic complete medium (30) supplemented with 2% glycerol and 4% galactose but deficient in histidine (SGG-his) was used to select for prototrophs in the His+ rate measurement experiments.
Plasmid Constructions.
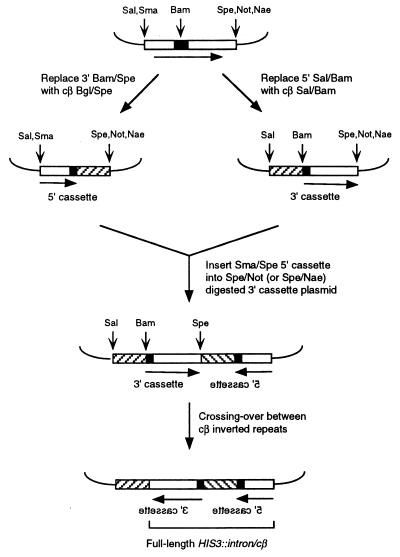
Plasmid pSR266 contains the pGAL-HIS3::intron construct (for details, see ref. 17) and was used as the starting point for constructing chicken β-tubulin (cβ) inverted repeat substrates according to the general scheme shown in Fig. 1. Appropriate 350 bp segments of cDNAs encoding cβ isoforms were amplified by PCR using as template plasmids obtained from the laboratory of D. Cleveland (31–34). PCR was performed either in the presence of 1.5–3.0 mM MgCl2/0 mM MnCl2 (high fidelity conditions) or 0.5–1.5 mM MgCl2/0.25–0.75 mM MnCl2 (low fidelity conditions; see ref. 35). Amplification primers contained restriction endonuclease sites near the 5′ ends; the forward primer added SalI and BglII sites to the amplified DNA while the reverse primer contained a BamHI site. The amplified product was digested with SalI and BamHI and was inserted into SalI/BamHI-digested pBluescript SK, generating pBluescript-cβ plasmids. The 1-bp changes in the cβ2 substrates were introduced by site-directed mutagenesis of the cβ2a 3′ cassette plasmid using the Chameleon kit (Stratagene). Other sequence changes were introduced as a result of PCR errors. The cβ inserts were sequenced as appropriate.
Figure 1.
Construction of inverted repeat substrates. The pGAL-HIS3::intron construct contained on plasmid pSR266 is shown at the top. Open boxes correspond to HIS3 sequences, solid boxes to artificial intron sequences, and cross-hatched boxes to cβ sequences; boxes are not to scale. Only those restriction sites relevant to the constructions are shown: Sal, SalI; Sma, SmaI; Bam, BamHI; Spe, SpeI; Not, NotI; Nae, NaeI; Bgl, BglII.
Yeast Strain Constructions.
All strains used in this study were derived by LiAc transformation (36) of isogenic strains SJR231 (MATα ade2–101oc his3Δ200 ura3-Nhe) and GCY121 (MATα ade2–101oc his3Δ200 ura3-Nhe msh2Δ msh3Δ::hisG; see ref. 17). Plasmids containing the inverted repeat constructs were targeted to integrate at the URA3 locus by digesting DNA with StuI before transformation. Ura+ transformants were analyzed by Southern blot analysis to identify those containing only a single copy of the plasmid integrated at URA3.
Measuring Recombination Rates.
Recombination rates were determined by the method of the median (37) as follows. Two-day-old colonies were excised from YEPD plates, inoculated into 5 ml of YEPGG medium, and grown for 2 days on a roller drum. Cells were harvested, washed once with sterile H2O, and resuspended in 1 ml H2O. Aliquots (100 μl) of appropriately diluted (or undiluted) cells were plated on SGG-his selective medium to determine the number of His+ recombinants per culture or on YEPD to assess the total number of viable cells per culture. His+ colonies were counted on day 4 after selective plating. The wild-type and MMR-defective strains containing a given pair of substrates were grown and plated at the same time. Generally 4–6 independent cultures were plated on a given day, and a minimum of 10 independent cultures (representing at least two independent platings) was used for each rate determination. Recombination rates between different strains were statistically compared using the χ2 method described by Wierdl et al. (38).
Estimation of Minimal Efficient Processing Segment (MEPS).
With a knowledge of the sequence divergence between pairs of substrates, one can estimate the length of a MEPS (3) for recombination as being approximately equal to the slope of the regression line of ln (recombination rate) versus sequence divergence (39), where the sequence divergence in the regression equation is expressed on a scale from zero (perfect identity) to one (no identity). This method is applicable to a variety of data sets as it does not require knowledge of the exact number of mismatches in any given segment. If the exact number of mismatches is known, a better approximation of the MEPS is given by L(1 − ea), where L is the length of the substrates (350 bp in the experiments reported here) and a is the slope of the regression of ln(recombination rate) versus the number of mismatches.
RESULTS
Intron-Based Recombination Assay System.
The essential features of the intron-based recombination system used in this study are illustrated in Fig. 1; a detailed description of the system can be found in Datta et al. (17). The starting plasmid in all manipulations was an integrative URA3 vector containing a HIS gene into which an artificial intron had been inserted (pGAL-HIS3::intron). A plasmid containing a 5′ recombination cassette (the 5′ end of HIS3, the 5′ portion of the intron, and a recombination substrate) was constructed by replacing sequences downstream of the 5′ intron splice consensus element with a 350-bp fragment derived from one of several cβ cDNAs (cβ2, cβ3, cβ6, or cβ7). A plasmid containing a 3′ recombination cassette (a recombination substrate, the 3′ portion of the intron, and the 3′ end of HIS3) was constructed by replacing sequences upstream of the intron TACTAAC element with a 350-bp fragment derived from a cβ cDNA. To combine 5′ and 3′ cassettes into a single plasmid, a fragment containing the 5′ cassette was inserted in inverted orientation at the 3′ end of the 3′ cassette. Finally, a single copy of this plasmid was integrated at the URA3 locus on chromosome V. Crossing-over between the 5′ cassette cβ sequences and the 3′ cassette cβ sequences inverts the region between them, placing the 5′ and 3′ parts of the HIS3 gene in the same orientation and reconstituting a functional cβ-containing intron. Because the recombinant cβ sequences are spliced out of the primary HIS3 transcript, there are no functional constraints on the recombination products and the degree of sequence identity can be varied over a very broad range. It should be noted that this system detects only crossover events and will not detect simple gene conversions.
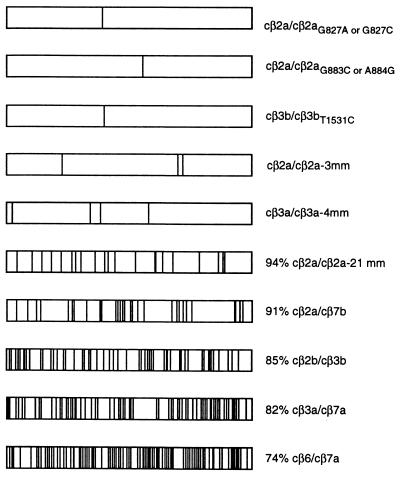
Combinations of 5′ and 3′ cassettes containing cβ sequences derived from cDNAs encoding different β-tubulin isoforms were used to achieve variable amounts of sequence divergence. For example, a cβ2 5′ cassette and a cβ7 3′ cassette were combined to generate 91%-identical substrates. In addition to directly using cβ cDNA sequences, random mismatches were introduced by mutagenic PCR or specific mismatches were introduced by site-directed mutagenesis. As controls for the diverged sequences, 100%-identical substrates were constructed by combining 5′ and 3′ cassettes containing the same cβ sequences (e.g., cβ2a/cβ2a). The distributions of nonidentical bases in the recombination substrates are shown in Fig. 2.
Figure 2.
Alignments of recombination substrates derived from cβ cDNA sequences. All sequences are approximately 350 bp. Each potential mismatch between a given pair of sequences is indicated by a vertical line.
Recombination Rates Between Mismatch-Containing Substrates.
Our previous work (17) demonstrated that recombination between sequences containing many mismatches is regulated by the MMR machinery, but provided no information concerning the number of mismatches that are necessary to trigger this antirecombination role. To specifically address this issue, substrates containing one or a few mismatches were introduced into the genomes of both wild-type (MMR+) and msh2Δmsh3Δ (MMR−) strains. Given what is known about yeast MMR, a msh2Δmsh3Δ double mutant should be devoid of all mismatch repair activity (28). Recombination rates between the substrates were inferred by measuring the rates of His+ prototroph formation (Table 1). Five different substrate pairs of 100% identity were analyzed as controls: cβ2a/cβ2a, cβ3a/cβ3a, cβ3b/cβ3b, cβ6/cβ6, and cβ7a/cβ7a. Each pair of 100%-identical substrates recombined at a rate of approximately 1 × 10−6 in the MMR+ background. For each substrate pair, the rate of recombination was elevated 2- to 4-fold in the MMR− strain relative to the MMR+ strain.
Table 1.
Rates of His+ recombinants
| % identity | Substrates | MMR+ rate, ×10−8 | MMR− rate, ×10−8 | MMR−/MMR+ |
|---|---|---|---|---|
| 100% | cβ2a/cβ2a | 92 | 350 | 3.8 |
| cβ3a/cβ3a | 86 | 190 | 2.2 | |
| cβ3b/cβ3b | 110 | 230 | 2.1 | |
| cβ6/cβ6 | 71 | 110 | 1.6 | |
| cβ7a/cβ7a | 170 | 480 | 2.8 | |
| 99.7% | cβ2a/cβ2aG827A | 21 | 290 | 14 |
| (1 mismatch) | cβ2a/cβ2aG827C | 30 | 390 | 13 |
| cβ2a/cβ2aA884G | 23 | 410 | 18 | |
| cβ2a/cβ2aG883C | 31 | 310 | 10 | |
| cβ3b/cβ3bT1531C | 29 | 280 | 9.7 | |
| 99% | cβ2a/cβ2a-3mm | 11 | 320 | 29 |
| cβ3a/cβ3a-4mm | 12 | 290 | 24 | |
| 94% | cβ2a/cβ2a-21 mm | 1.6 | 220 | 140 |
| 91% | cβ2a/cβ7b | 2.9 | 130 | 45 |
| 85% | cβ2b/cβ3b | 0.45 | 35 | 77 |
| 82% | cβ3a/cβ7a | 0.22 | 18 | 82 |
| 74% | cβ6/cβ7a | 0.024 | 1.32 | 55 |
cβ sequences were numbered as in the GenBank files; accession nos. for cβ2, cβ3, cβ6, and cβ7 are M11443, M14228, J02828, and X07011, respectively. cβ2a sequences consisted of nt 690–1038, cβ2b sequences of nt 657–1007, cβ3a of nt 1093–1441, cβ3b of nt 1391–1741, cβ6 of nt 429–779, cβ7a of nt 450–800, and cβ7b of nt 781–1129. For substrates containing a single mismatch, the nature and position of the mismatch are indicated as a subscript. The cβ2 sequence with three mismatches (cβ2-3mm) contains the following changes: A769T, A934G, and T942C. The cβ3a sequence with four mismatches (cβ3a-4mm) contains the following changes: A1216G, A1228G, C1297T, and deletion of G1101. Two types of statistical analyses indicate that a 2-fold difference between recombination rates is significant. First, the cβ2a/cβ2a substrates were used as an internal control in most fluctuation experiments, which yielded 15 different, but very similar, rate measurements (all the data were pooled for the rate used in the table). Using these independent rate measurements, the mean rate is 98 × 10−8 and the SD is 18 × 10−8. Second, in addition to the high reproducibility of rate measurements, representative rates were statistically compared: cβ6/cβ6, MMR+ versus cβ6/cβ6, MMR−(χ2 = 6.0, P < 0.05); cβ6/cβ6, MMR+ versus cβ7a/cβ7a, MMR+(χ2 = 10.7, P < 0.01); cβ6/cβ6, MMR− versus cβ7a/cβ7a, MMR−(χ2 = 24, P < 0.01), cβ3b/cβ3b, MMR+ versus cβ3b/cβ3b, MMR−(χ2 = 25.6, P < 0.01); cβ3/cβ3T1531C, MMR+ versus cβ3/cβ3T1531C, MMR−(χ2 = 22, P < 0.01); cβ3b/cβ3b, MMR+ versus cβ3/cβ3T1531C, MMR+ (χ2 = 22, P < 0.01); and cβ3b/cβ3b, MMR− versus cβ3/cβ3T1531C, MMR−(χ2 = 0.18, P > 0.50).
Five substrates containing a single base substitution were constructed to vary the type, sequence context, and position of the potential mismatch. Four were derived from cβ2a and one was derived from cβ3b. For the cβ2a substrates (nucleotides 690-1038 of cβ2; see legend to Table 1), an A to G transition was introduced at position 884, a G to C transversion at position 883, and either a G to A transition or a G to C transversion at position 827. For the cβ3b substrates (nucleotides 1391–1741 of cβ3) a T to C transition was introduced at position 1531. The transitions result in either an A-C or G-T mismatch in heteroduplex recombination intermediates; the transversions yield either a G-G or C-C mismatch. All recombination-generated mismatches with the exception of the C-C mismatch should be recognized efficiently by the yeast MMR machinery (40). For each of the 5 one-mismatch substrates examined, there was a 10- to 20-fold elevation in the recombination rate in the MMR− strain relative to the MMR+ strain. If one compares these rates to those obtained with the 100% control substrates, a single mismatch uniformly lowered the rate of recombination 3- to 4-fold in the MMR+ background but had no effect on the recombination rate in the MMR− strains. We conclude that a single mismatch can trigger the antirecombination function of the MMR machinery in yeast and thereby impede homologous recombination. The MMR system did not appear to discriminate between the potentially different types of mismatches used here.
To examine the antirecombination effect of multiple mismatches, substrates containing either three or four mismatches (cβ2a-3mm and cβ3a-4mm, respectively) were introduced into the MMR+ and MMR− strains. For each of these substrates, there was a 30-fold elevation in the recombination rate in the MMR− strain relative to the MMR+ strain. As with the one-mismatch substrates, a comparison to the 100% control substrates indicates that three or four mismatches impact recombination only in MMR-competent cells.
We reported previously recombination rates between 91%-identical and 77%-identical substrates in wild-type and MMR-defective strains (17). To examine more systematically recombination rates between sequences in the 95% to 75% identity range, additional substrates with identity levels of 94%, 85%, 82%, and 74% were constructed. The 91% substrates (30 mismatches) used in the previous study were derived by combining cβ2a and cβ7b sequences; the 94% substrates (21 mismatches) were obtained by low fidelity PCR using cβ2a as a template. Although the 94% and 91% substrates recombined at similar rates in MMR-competent cells, elimination of MMR elevated recombination between the 94% substrates 140-fold but elevated recombination between the 91% substrates only a 45-fold. This difference in behavior of the 94% versus 91% substrates may be related to the very different distributions of mismatches in these two substrate pairs (see Fig. 2). When compared with the 100% control substrates, 20–30 mismatches reduced recombination rates about 50-fold in MMR-competent cells. There was a relatively modest reduction in recombination rates (2- to 3-fold) associated with this level of divergence in MMR-defective cells.
With the more highly diverged 85%- and 82%-identical substrates (52 and 64 total mismatches, respectively), there was an approximately 80-fold increase in recombination rates in the MMR− strains relative to the MMR+ strains. If one compares these rates to those obtained with the 100% control substrates, however, the 200-fold decrease in recombination rates observed in the MMR+ strains was not eliminated in the MMR− strains. Even in the MMR− strains this level of divergence reduced recombination rates approximately 10-fold. The trend observed with the 85%- and 82%-identical substrates was even more striking with the 74% substrates. When compared with the 100% control substrates, the recombination rate between the 74% substrates was reduced 4000-fold in MMR-competent cells. Although elimination of the MMR system improved recombination between the 74% substrates, the rate was still approximately 200-fold lower than that observed with the 100% control substrates.
DISCUSSION
An intron-based, inverted repeat assay was used to examine recombination between identical substrates and between substrates containing variable numbers of mismatches (Fig. 2). All substrates were derived from cβ cDNAs and were approximately 350 bp long. The substrate length was held constant to insure that recombination rates were affected only by the mismatches present. Because detection of recombination requires inversion of the region between the repeats (Fig. 1), this system only identifies reciprocal exchange events; simple gene conversions cannot be detected. To assess the role of the mismatch repair machinery in regulating recombination between nonidentical sequences, recombination rates were determined in both wild-type and MMR-defective (msh2Δmsh3Δ) strains. Table 1 presents the recombination rates for each substrate pair and these data are summarized in Table 2.
Table 2.
Summary of recombination rates
| Identity | MMR+ rate, ×10−8 | MMR− rate, ×10−8 | MMR−/MMR+ | MMR−/MMR+ normalized to 100% |
|---|---|---|---|---|
| 100% | 110 (1×) | 270 (1×) | 2.7 | 1.0 |
| 99.7% (1 mm) | 27 (4.1× ↓) | 340 (1.3× ↑) | 13 | 4.8 |
| 99% (3-4 mm) | 12 (9.2× ↓) | 310 (1.1× ↑) | 26 | 9.6 |
| 91–94% | 2.2 (50× ↓) | 180 (1.5× ↓) | 82 | 34 |
| 82–85% | 0.33 (330× ↓) | 27 (10× ↓) | 82 | 30 |
| 74% | 0.024 (4600× ↓) | 1.3 (210× ↓) | 54 | 20 |
For each homology level, the rates shown were obtained by averaging the rates observed with each substrate at that level (see Table 1). The number in parentheses following each rate represents the fold increase (↑) or decrease (↓) relative to the rate obtained with the 100% substrates in the same MMR+ or MMR− background.
Recombination rates between five different pairs of identical (100%) substrates were examined and these rates served as controls for subsequent experiments involving mismatched substrates. In MMR-competent strains, the rates varied from 0.7 × 10−6 for the cβ6/cβ6 substrates to 1.7 × 10−6 for the cβ7a/cβ7a substrates. The slight but statistically significant (see legend to Table 1) variation in rate with substrates of the same size indicates that the actual sequence of bases may have subtle effects on recombination. Such subtleties could have their origins in either preferred sites for recombination-initiating lesions or preferred sites for resolving recombination intermediates. Sequence preference for resolution has indeed been observed in the cleavage of Holliday junctions in vitro by the bacterial RuvC protein (41). With each of the five 100% substrates analyzed, the recombination rate in the MMR-defective strain was elevated relative to that in the MMR+ strain; the average increase was 2.7-fold. Although the MMR machinery would not be expected to inhibit recombination between identical sequences, we suggest that the MMR system may be detecting either intrastrand secondary structure or unpaired regions resulting from branch migration of Holliday junctions into the nonhomologous regions that flank the recombination substrates. Regardless of the molecular basis of the increased recombination in MMR− strains seen here, it is important to account for this increase when assessing the effects of mismatches on recombination rates. It should be noted that the inhibitory effect of MMR proteins on recombination seen in this study is in contrast to the small stimulatory role reported elsewhere (42); the conflicting results may reflect differences in the assay systems used.
The effect of a single potential mismatch on recombination rate was determined using five different substrate pairs, each of which contained a defined base substitution. Although both the type and position of the base substitution were varied, all 5 one-mismatch substrates recombined similarly. In the MMR+ strains, the average rate of recombination between the mismatch-containing substrates was approximately 4-fold lower than the average recombination rate between the 100% substrates. In the MMR− strains, however, the average rate of recombination between the mismatch-containing substrates was the same as that between the 100% substrates. These data demonstrate that a single mismatch is sufficient to impair the recombination process in yeast and that this impairment derives solely from action of the MMR machinery. Although a single mismatch within 31 bp substrates also was observed to inhibit recombination in E. coli, the inhibition was not due to the action of the mismatch repair machinery (15).
To more precisely define the impact of sequence divergence on mitotic recombination, additional substrate pairs with identities ranging from 99% (3 mismatches) to 74% (90 mismatches) were examined. In MMR-competent cells there was a cumulative negative effect of mismatches on recombination rates (Table 2). Whereas a single mismatch reduced recombination rates approximately 4-fold, the presence of two to three additional mismatches decreased recombination 9-fold relative to the 100% substrates. This trend continued in the MMR+ strains, with the 91–94% identical substrates recombining 50-fold less efficiently, the 82–84% substrates recombining 300-fold less efficiently and the 74% substrates recombining 4600-fold less efficiently than the 100% control substrates. In striking contrast to MMR-competent strains, there was little effect of sequence divergence on recombination rates in MMR-defective strains until the identity level dropped below the 91–94% range. This indicates that, with sequences of greater than 90% identity, all of the inhibitory effect of sequence divergence on recombination can be attributed to action of the MMR machinery. Once sequences pass a divergence threshold of 5–10%, however, a factor other than the MMR system strongly impairs the recombination process. As suggested previously (17), we believe that this additional impairment likely reflects a general limitation of the yeast recombination machinery, which could correspond to an inability to efficiently initiate strand transfer or resolve intermediates as crossovers. Although the yeast MMR system imposes an antirecombination effect at all levels of sequence divergence examined, the maximum effect exerted by the MMR machinery appears to be about 30-fold (last column of Table 2). This level of inhibition is attained when the sequence divergence is in the 10–15% range.
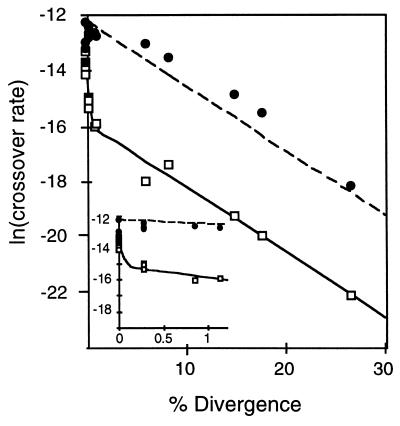
The relationship between sequence identity and recombination rate in both MMR+ and MMR− cells is graphically presented in Fig. 3. For each set of data, a roughly linear relationship is obtained if one plots the ln (recombination rate) versus % sequence divergence. A similar log-linear relationship was observed in studies examining the effects of sequence divergence on the efficiency of bacterial conjugation and transformation (23, 39). A log-linear relationship between recombination rate and sequence divergence can be explained most simply by the concept of a MEPS, which is defined as the minimal length of perfect homology needed for efficient recombination. Shen and Huang (3) originally proposed MEPS to explain the linear relationship observed between recombination rate and substrate size in a bacterial recombination assay. If one considers any piece of DNA as a linear series of overlapping MEPS, each of which recombines at the same efficiency, the MEPS concept postulates that the rate of recombination should be directly proportional to the number of MEPS in a particular sequence. Because the number of MEPS increases linearly with substrate length, the MEPS concept accounts for the observed linearity between substrate length and recombination rate. If the lengths of the substrates are held constant and random mismatches are introduced, the number of MEPS decreases exponentially as the sequence divergence increases.
Figure 3.
Recombination rate versus % sequence divergence. All the data in Table 1 are graphed. (Inset) Data for the 0–4 mismatch substrates only. Open and solid circles correspond to data from MMR+ and MMR− strains, respectively. The curves were derived using the model described in the text. Nonlinear curves were fit using the simplex method of systat 5.0 (Macintosh). Different starting conditions for the nonlinear curve fitting program produced nearly 5-fold variations in the fitted values of R0 and P0, which were statistically indistinguishable from each other; fitted values of the other parameters remained approximately constant with a range <1%.
For substrates with a fixed length and varying numbers of mismatches, as in the data reported here, the length of the MEPS can be estimated according to the equation described in the Materials and Methods. This technique gives a MEPS of 18 bp [14–21 bp 95% confidence interval (C.I.)] for the MMR− strains, and 28 bp (23–33 bp 95% C.I.) for the MMR+ strains. As alluded to above, however, visual examination of the MMR+ data indicates a very rapid dropoff in crossover rates at low sequence divergence, followed by a leveling off to a slope similar to that observed with the MMR− data. This behavior suggests a model in which heteroduplex forms with a probability that declines exponentially with sequence divergence; heteroduplex avoids triggering mismatch repair with a probability that also falls off exponentially with sequence divergence (but at a different rate); and mismatch repair, if triggered, blocks recombination with a certain, fixed probability. In such a model, heteroduplex formation has probability P0e−αx; mismatch repair is triggered with probability 1-R0e−βx and is effective (if triggered) with probability f. In these expressions, x is the divergence (measured on a scale of 0–1) between the recombining sequences; P0 is the probability that identical substrates will form a heteroduplex; and R0 is the probability that a heteroduplex between identical substrates, once formed, will avoid triggering mismatch repair. Within the framework of the MEPS concept, the values α and β correspond to the mismatch-free sequence length required to initiate heteroduplex formation and the mismatch-free sequence length necessary to escape MMR activity, respectively. With these assumptions, the probability of a crossover event is P0e−αx[1 − f(1 − R0e−βx)].
The log-transformed version of above model was fit to the data for the MMR+ strains, and the fitted model is shown as the solid curve in Fig. 3. The fitted values are α = 23, β = 610, f = 0.97, P0 = 5.1 × 10−6, and R0 = 0.18. It should be noted that P0 and R0 are dependent on the substrate length, 350 bp in this case. Although the model is highly consistent with the data, this agreement should be viewed with the knowledge that it has four degrees of freedom and was devised after viewing the data. If the model is correct, it should predict the outcome of the MMR− experiments using the same parameters estimated from the MMR+ data, but setting f = 0 (mismatch repair is never effective). This prediction, shown as the dashed line in Fig. 3, agrees well with the MMR− data. Taken together, these data suggest that ≈20 bp of perfect homology are needed to initiate heteroduplex formation and that ≈610 bp of perfect homology are needed to avoid the antirecombination activity of the MMR machinery. Because the latter estimate is longer than the length of the recombination substrates used here, it can account for the increases in the rates of recombination between 100%-identical substrates that accompanied elimination of MMR.
The model above is based on the number of mismatches rather than on the locations of the mismatches within the recombination substrates. With knowledge of the precise positions of the mismatches in the substrate pairs (see Fig. 2), the number of mismatch-free stretches of any given length can be determined. If the MEPS concept is applicable in its simplest form to yeast recombination, there should be a MEPS length that yields a linear relationship between the number of MEPS and crossover rates, at least in the MMR− strains in which only a single process (heteroduplex formation) may be limiting. When this is attempted with the MMR− data, even the best fit (MEPS = 11 bp) is poor (r2 = 0.62), suggesting that factors other than the length of mismatch-free sequence (e.g., base composition or preferred initiation/resolution sites) affect recombination rate. The observation that different pairs of 100%-identical substrates recombined at slightly different rates supports the notion that recombination is influenced by factors other than substrate length. It is interesting that despite the existence of these other factors, the fit of the model used above to the data is quite good (Fig. 3).
The assay used in this study depends on the resolution of recombination intermediates as crossover events, and so does not provide any information concerning when or how MMR proteins exert their antirecombination role. The regulation could involve some type of steric hindrance resulting from mismatch binding or it could involve the actual destruction of mismatched heteroduplex. Although the destruction of intermediates is easy to imagine in those organisms where a nicking activity has been associated with the MMR machinery (e.g., MutH in E. coli), no comparable nicking activity has been identified in yeast. In relation to the issue of when the antirecombination role of the MMR machinery may be exerted, it should be noted that some assay systems have failed to detect any impact of the MMR machinery on recombination between diverged sequences (21, 43). In contrast to the intrachromatid, inverted repeat crossing-over assay used here, one system examined plasmid-chromosome gene conversion (43) and the other system examined intrachromatid interactions between direct repeats (21). We do not think that our observations are an artifact of using an intrachromatid inverted repeat assay because essentially the same effects have been observed with nonidentical substrates positioned on nonhomologous chromosomes (W. Chen and S.J.-R., unpublished data). Although it is not obvious why the MMR system impacts various recombination systems so differently, it is nevertheless clear the MMR system can exhibit potent antirecombination activity. It is particularly striking that a single mismatch can trigger the antirecombination activity of the yeast mismatch repair machinery. If one extrapolates the results reported here to higher eukaryotes, the accumulation of chromosomal rearrangements via recombination between diverged sequence elements may contribute to the genetic instability of MMR-defective cells and may be important for tumor progression.
Acknowledgments
A.D. and M.H. contributed equally to this work. We thank M. Vulic, F. Taddei, F. Dionisio, and M. Radman for helpful discussions and for communicating results before publication. We also thank J. Majewski and F. Cohan for sharing their data before publication. This work was supported by National Institutes of Health Grant GM38464 to S.J.-R. A.D. was partially supported by the Graduate Division of Biological and Biomedical Sciences, Emory University; M.L. was supported by National Institutes of Health Grant GM33782 to Bruce R. Levin.
ABBREVIATIONS
- MMR
mismatch repair
- cβ
chicken β-tubulin
- MEPS
minimal efficient processing segment
References
- 1.Radman M. Biochimie. 1991;73:357–361. doi: 10.1016/0300-9084(91)90101-6. [DOI] [PubMed] [Google Scholar]
- 2.Singer B S, Gold L, Gauss P, Doherty D H. Cell. 1982;31:25–33. doi: 10.1016/0092-8674(82)90401-9. [DOI] [PubMed] [Google Scholar]
- 3.Shen P, Huang H V. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn B-Y, Dornfeld K J, Fagrelius T J, Livingston D M. Mol Cell Biol. 1988;8:2442–2448. doi: 10.1128/mcb.8.6.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liskay R M, Letsou A, Stachelek J L. Genetics. 1987;115:161–167. doi: 10.1093/genetics/115.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubnitz J, Subramani S. Mol Cell Biol. 1984;4:2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan L-W, Keil R L. Genetics. 1990;124:263–273. doi: 10.1093/genetics/124.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinks-Robertson S, Michelitch M, Ramcharan S. Mol Cell Biol. 1993;13:3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humbert O, Prudhomme M, Hakenbeck R, Dowson C G, Claverys J-P. Proc Natl Acad Sci USA. 1995;92:9052–9056. doi: 10.1073/pnas.92.20.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdulkarim F, Hughes D. J Mol Biol. 1996;260:506–522. doi: 10.1006/jmbi.1996.0418. [DOI] [PubMed] [Google Scholar]
- 11.Zahart T C, Mora G C, Maloy S. J Bacteriol. 1994;176:1527–1529. doi: 10.1128/jb.176.5.1527-1529.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waldman A S, Liskay R M. Proc Natl Acad Sci USA. 1987;84:5340–5344. doi: 10.1073/pnas.84.15.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petit M-A, Dimpfl J, Radman M, Echols H. Genetics. 1991;129:327–332. doi: 10.1093/genetics/129.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matic I, Rayssiguier C, Radman M. Cell. 1995;80:507–515. doi: 10.1016/0092-8674(95)90501-4. [DOI] [PubMed] [Google Scholar]
- 15.Shen P, Huang H V. Mol Gen Genet. 1989;218:358–360. doi: 10.1007/BF00331291. [DOI] [PubMed] [Google Scholar]
- 16.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 17.Datta A, Adjiri A, New L, Crouse G F, Jinks-Robertson S. Mol Cell Biol. 1996;16:1085–1093. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selva E M, New L, Crouse G F, Lahue R S. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Negritto M T, Wu X, Kuo T, Chu S, Bailis A M. Mol Cell Biol. 1997;17:278–286. doi: 10.1128/mcb.17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris S, Rudnicki K S, Haber J E. Genetics. 1993;135:5–16. doi: 10.1093/genetics/135.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran H, Degtyareva N, Gordenin D, Resnick M A. Mol Cell Biol. 1997;17:1027–1036. doi: 10.1128/mcb.17.2.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chambers S, Hunter N, Louis E J, Borts R H. Mol Cell Biol. 1996;16:6110–6120. doi: 10.1128/mcb.16.11.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zawadzki P, Roberts M S, Cohan F M. Genetics. 1995;140:917–932. doi: 10.1093/genetics/140.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radman M. In: Genetic Recombination. Kucherlapati R, Smith G R, editors. Washington, DC: Am. Soc. Microbiol.; 1988. pp. 169–192. [Google Scholar]
- 25.Worth L, Jr, Clark S, Radman M, Modrich P. Proc Natl Acad Sci USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 27.Umar A, Kunkel T A. Eur J Biochem. 1996;238:297–307. doi: 10.1111/j.1432-1033.1996.0297z.x. [DOI] [PubMed] [Google Scholar]
- 28.Kolodner R. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 29.Crouse G F. In: DNA Damage and Repair: DNA Repair in Prokaryotes and Lower Eukaryotes. Nickoloff J A, Hoekstra M F, editors. Vol. 1. Totowa, NJ: Humana; 1997. , in press. [Google Scholar]
- 30.Sherman F. Methods Enzymol. 1991;194:3–20. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 31.Monteiro M J, Cleveland D W. J Mol Biol. 1988;199:439–446. doi: 10.1016/0022-2836(88)90616-x. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan K F, Lau J T Y, Cleveland D W. Mol Cell Biol. 1985;5:2454–2465. doi: 10.1128/mcb.5.9.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy D B, Wallis K T, Machlin P S, Ratrie H, III, Cleveland D W. J Biol Chem. 1987;262:14305–14312. [PubMed] [Google Scholar]
- 34.Sullivan K F, Machlin P S, Ratrie H, III, Cleveland D W. J Biol Chem. 1986;28:13317–13322. [PubMed] [Google Scholar]
- 35.Leung D W, Chen E, Goeddel D V. Technique. 1989;1:11–15. [Google Scholar]
- 36.Geitz D, St. Jean A, Woods R A, Schiestl R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lea D E, Coulson C A. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 38.Wierdl M, Greene C N, Datta A, Jinks-Robertson S, Petes T D. Genetics. 1996;143:713–721. doi: 10.1093/genetics/143.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vulic, M., Dionisio, F., Taddei, F. & Radman, M. (1997) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 40.Detloff P, Sieber J, Petes T D. Mol Cell Biol. 1991;11:737–745. doi: 10.1128/mcb.11.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah R, Bennett R J, West S C. Cell. 1994;79:853–864. doi: 10.1016/0092-8674(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 42.Saparbaev M, Prakash L, Prakash S. Genetics. 1996;142:727–736. doi: 10.1093/genetics/142.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porter G, Westmoreland J, Priebe S, Resnick M A. Genetics. 1996;143:755–767. doi: 10.1093/genetics/143.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]