Abstract
The het-s locus of Podospora anserina is a heterokaryon incompatibility locus. The coexpression of the antagonistic het-s and het-S alleles triggers a lethal reaction that prevents the formation of viable heterokaryons. Strains that contain the het-s allele can display two different phenotypes, [Het-s] or [Het-s*], according to their reactivity in incompatibility. The detection in these phenotypically distinct strains of a protein expressed from the het-s gene indicates that the difference in reactivity depends on a posttranslational difference between two forms of the polypeptide encoded by the het-s gene. This posttranslational modification does not affect the electrophoretic mobility of the protein in SDS/PAGE. Several results suggest a similarity of behavior between the protein encoded by the het-s gene and prions. The [Het-s] character can propagate in [Het-s*] strains as an infectious agent, producing a [Het-s*] → [Het-s] transition, independently of protein synthesis. Expression of the [Het-s] character requires a functional het-s gene. The protein present in [Het-s] strains is more resistant to proteinase K than that present in [Het-s*] mycelium. Furthermore, overexpression of the het-s gene increases the frequency of the transition from [Het-s*] to [Het-s]. We propose that this transition is the consequence of a self-propagating conformational modification of the protein mediated by the formation of complexes between the two different forms of the polypeptide.
Keywords: filamentous ascomycete, vegetative incompatibility, structural heredity
The het-s locus of the filamentous fungus Podospora anserina is one of the nine known loci controlling heterokaryon incompatibility in that species (for review, see ref. 1). Coexpression of the antagonistic het-s and het-S alleles in the same cytoplasm triggers an adverse reaction that prevents the formation of viable heterokaryotic cells between strains that contain the incompatible alleles (2). This locus encodes a 289-aa protein that is not essential for cell viability or completion of the life cycle of the fungus (3, 4). The proteins encoded by the incompatible alleles differ by 14 amino acid substitutions, but one difference is sufficient for expression of the antagonistic [Het-s] and [Het-S] specificities (5).
It has been reported by Rizet (ref. 2; for review, see ref. 6) that haploid strains of the het-s genotype can exhibit two different phenotypes: they either are incompatible with het-S strains (this phenotype will herein be designated [Het-s]), or they are neutral in incompatibility and display the [Het-s*] phenotype. This latter phenotype is stably maintained during vegetative growth but a transition from [Het-s*] to [Het-s] can occur spontaneously at a very low frequency, estimated under 10−7 per nucleus (7). This phenotypic conversion is, however, invariably induced after anastomosis and cytoplasmic mixing with a [Het-s] strain. This transition cannot be induced by strains that contain het-S or the null alleles het-sx and het-s°. The [Het-s] character is dominant and can propagate in [Het-s*] strains in the absence of nuclear transmission. When it has been induced, the [Het-s*] → [Het-s] transition spreads very rapidly as an infectious process from the region of anastomosis throughout the mycelium. Both the [Het-s] and [Het-s*] characters are transmitted as nonmendelian elements through meiosis. The phenotype of the offsprings of the het-s genotype depends on the phenotype of the female parent, suggesting that these phenotypes are controlled by cytoplasmic elements. It has been proposed from these different results that the expression of the het-s gene might be positively controlled by its protein product (7).
We report here that a polypeptide encoded by het-s is present in similar amounts in [Het-s*] and [Het-s] strains. So the absence of reactivity of [Het-s*] strains in incompatibility does not result from the lack of expression of the het-s-encoded protein but to a posttranslational modification of the protein. We also show that several properties of the protein encoded by the het-s gene suggest that it may have a behavior similar to that of a prion protein: (i) overexpression of the protein increases the rate of the spontaneous [Het-s*] → [Het-s] transition; (ii) the protein present in [Het-s] and [Het-s*] strains displays different sensitivity to a protease; (iii) a physical interaction between monomers can be detected using the two-hybrid yeast system. Similar properties have already been described for yeast prions [URE3] and [PSI] (8–12). We propose that the protein encoded by the het-s gene can exist under two different conformations, pHET-s and pHET-s*, and that the stable form present in the [Het-s] strain is able to propagate by transmitting its conformation to the form present in [Het-s*] strains.
MATERIALS AND METHODS
P. anserina Strains.
P. anserina is a filamentous ascomycete. The mycelium has a coenocytic structure. It is formed by multinucleate cells isolated from each other by incomplete cell walls. Life cycle and methods for genetic analysis have been described (13). Compatibility between strains can be determined by confrontation on cornmeal agar medium. Incompatibility results in the formation of a barrage, a dense and unpigmented line in the region where the strains meet (2). Three different alleles of the het-s locus have been found in wild-type isolates: het-s and het-S are reactive, and incompatible alleles, het-sx is neutral in incompatibility; het-sx is compatible with both het-s and het-S. The DNA sequence of these wild-type alleles has been reported (3, 5). The het-s° allele was derived from het-s, the promoter and the 5′ end of the coding sequence having been deleted by gene replacement (4). Strains that display the [Het-s*] phenotype were obtained in the offspring of crosses between het-s and het-S strains. These strains are compatible with both parents. All strains used in this study are isogenic except for the allele present at the het-s and mating type loci.
Transformation of P. anserina.
Protoplasts were prepared and transformed as described (14). The pMOcosX, containing the bacterial hph gene coding for hygromycin resistance, was used in cotransformations (15). Transformants were screened for hygromycin B resistance at 100 μg/ml.
DNA Analysis.
General methods for nucleic acid analysis and vector construction were as described (16). PCR amplification of DNA (17) was achieved in a 50-ml reaction mixture: 10 mM Tris⋅HCl, pH 8.4/5 mM KCl/1.5 mM MgCl2/0.2 mM of each dNTP/100 ng of each primer/2 ng of plasmids pSKPS, which contains the het-s allele, or pSKGS, which contains the het-S allele. After 10 min at 95°C, 1 unit of Taq polymerase (AmpliTaq, Cetus) was added, and DNA was amplified for 35 cycles in a Perkin–Elmer/Cetus Thermocycler. The cycling parameters were: denaturation at 95°C for 30 sec, annealing at 58°C for 2 min, and extension at 72°C for 2 min.
Site-specific mutagenesis was carried out using the Transformer Site-Directed Mutagenesis Kit (CLONTECH) according to the manufacturer’s protocols. Two primers were used in the mutagenesis. For het-s modification the oligonucleotide (5′-ACGGTTCTGCCATGGCAGTTTG-3′) produced a TCA → GCA modification, which creates a NcoI restriction site overlapping the initiator ATG codon. The second primer produced a mutation in the amp gene, which restores the resistance to ampicillin. This modification was used for the selection of mutant plasmids.
Protein Extraction and Analysis.
For preparation of cell-free extracts, the mycelia were grown at 26°C in 100 ml of liquid medium in 1,000-ml Roux bottles. Mycelia from 2-day-old cultures were harvested, rinsed twice with cold distilled water, and frozen at −80°C for 1 hr. The mycelium then was lyophilized and ground. The powder was resuspended in the extraction buffer (50 mM NaH2PO4, pH 8/10 mM Tris⋅HCl, pH 8/100 mM NaCl/0.25% Triton/4.3 mM phenylmethylsulfonyl fluoride/1 mM l-1-tosyl amide-2-phenylethyl chloromethyl ketone/1 mM Nα-p-tosyl-l-lysine chloromethyl ketone/5 μg/ml leupeptin/5 μg/ml pepstatin A). The mixture was sonicated for 2 min at 4°C by applying 120-watt short pulses of 20 sec each. The homogenate then was incubated at 4°C for 1 hr with gentle agitation. The homogenate was centrifuged for 15 min at 10,000 × g. The supernatant constituted the crude extract. Conditions for SDS/PAGE and immunoblotting were as described (5). Sample preparation for proteinase K digestion was as described above except that the extraction buffer was devoid of protease inhibitors. Ten milliliters of crude extract was incubated with 50 μg/ml of proteinase K at 37°C for 25 min. The digestion was stopped by addition of final concentrations of 50 mM EDTA and 4.3 mM phenylmethylsulfonyl fluoride after 0- to 25-min incubation. Proteins were precipitated by adding ammonium sulfate to 662 mg/ml to the samples. Proteins were collected by centrifugation for 15 min at 10,000 × g, and the pellet was resuspended and dialyzed overnight in 0.1 M phosphate buffer at pH 8.
Yeast Strains and Methods.
Vectors for construction of GAL4 fusions were pDBT and pTAL (P. Navarro, personal communication) constructed by exchanging the ApaI–BamHI fragment between pPC62 and pPC86 (18). A PCR amplification on pSKPS or pSKGS plasmids with DH1 oligonucleotide (5′-ACTGCCCCGGGGAACCGTTC-3′), which creates the SmaI restriction site overlapping the initiator ATG codon, and DH2 oligonucleotide (5′-ACATTCTAGCGGCCGCCCGTTAAT-3′), which creates the NotI restriction site downstream to the termination codon, was performed as described above. The 950-bp PCR fragments were ligated in pDBT and pTAL after digestion of fragments and vectors by SmaI and NotI. The fusion vectors carrying the GAL4 DNA-binding domain in-frame with the ORFs of het-s and het-S were designated pDBT-s and pDBT-S, respectively. The fusion vectors carrying the GAL4 transcription activator domain in-frame with the het-s and the het-S ORFs were named pTAL-s and pTAL-S, respectively. These fusion vectors were verified by restriction mapping and sequencing across recombinant joints and PCR products. Yeast strain Y190 (MATa gal4 gal80 his3 trp1–901 ade2–101 ura3–52 leu2–3,-112+URA3::GAL → lacZ, LYS2::GAL → HIS3 cyhr) was used in the two-hybrid assay. Yeast strains were grown in yeast extract/peptone/dextrose liquid medium for the transformation step and on supplemented synthetic dextrose medium lacking tryptophan and leucine and supplemented or not with histidine. These media were noted SD-leu-trp and SD-leu-trp+his, respectively. A combination of two fusion vectors was used for each transformation. Transformants were first tested for histidine prototrophy by plating cells on SD-leu-trp containing 0–100 mM 3-amino-1,2,4-triazole. The presence of 3-amino-1,2,4-triazole limits the growth of the recipient strain on minimal medium due to leakage of the HIS3 promoter, so that the interaction between proteins could be estimated by the level of resistance to 3-amino-1,2,4-triazole. The transformants also were assayed for β-galactosidase activity as described (19). Transformation with pPC76 and pPC79 (18), which contain, respectively, the GAL4-DB in frame with Fos oncoprotein and the GAL4-TA in frame with Jun oncoprotein, were used as positive controls. Combinations of each fusion vector with pDBT or pTAL were used as negative controls.
RESULTS
Analysis of the Protein Encoded by the het-s Gene in [Het-s] and [Het-s*] Strains.
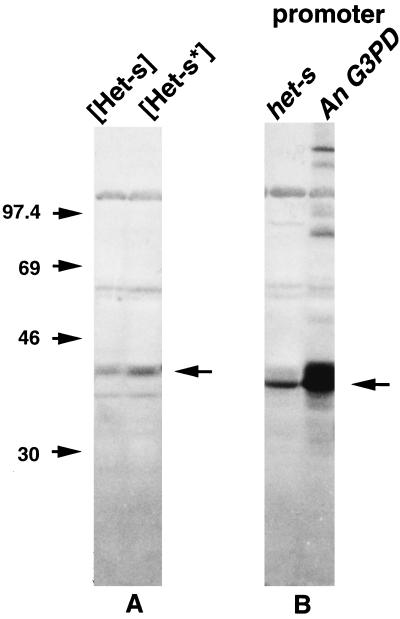
[Het-s*] strains contain the het-s gene but display a neutral incompatibility phenotype (2). This absence of reactivity against antagonistic het-S strains could result from the lack of expression of the het-s gene. Alternatively, the protein encoded by the het-s gene might be expressed as an inactive form in [Het-s*] strains. The expression of the het-s gene has been examined by Northern and Western blotting in [Het-s] and [Het-s]* strains. The results show that the gene is expressed at the same level in both strains. No significant differences were observed either in the amount or apparent size of the RNA (not shown) or of the protein. A protein with the same electrophoretic mobility was detected in extracts from [Het-s] and [Het-s*] strains (Fig. 1A). Therefore, the difference of reactivity of the two strains in incompatibility against a het-S strain is not due to a differential expression of the het-s gene but to a posttranslational difference. Furthermore, as the two polypeptides display the same mobility in SDS/PAGE, they differ very little or not at all in molecular mass. It can be estimated that this difference in molecular mass, if it exists, cannot be greater than 500 to 1,000 Da. The difference in reactivity cannot be attributed to a maturation process such as an intensive glycosylation or the cleavage of a long peptide. The two polypeptides will be tentatively designated pHET-s and pHET-s*. The presence of the protein in [Het-s*] strains suggests that the phenotypic transition [Het-s*] → [Het-s] is related to a modification of pHET-s* that can be converted to the pHET-s form.
Figure 1.
Immunoblot analysis of proteins isolated from [Het-s] and [Het-s*] strains (A) or from strains that contain the het-s ORF under the control of its promoter or under the control of the promoter of the glyceraldehyde 3-phosphate dehydrogenase (G3PD) gene of A. nidulans (B). Markers used for gel calibration were muscle rabbit phosphorylase B (97.4 kDa), BSA (66 kDa), egg albumin (45 kDa), and carbonic anhydrase from bovine erythrocytes (29 kDa). Arrowheads indicate position of protein encoded by the het-s gene.
Analysis of the Propagation of the [Het-s] Character.
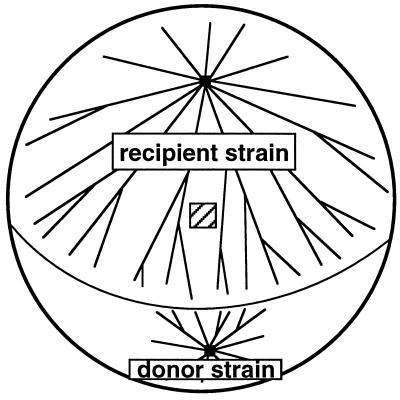
Some aspects of the transmission of the [Het-s] character to [Het-s*] strains have been analyzed using the experimental procedure described by Beisson-Schecroun (7) and shown in Fig. 2. Recipient and donor strains were grown on cellophane pads laid on corn meal agar. Formation of anastomosis between the mycelia was followed under a binocular microscope. Immediately after the fusion of filaments, the cellophane pads were transferred to fresh medium supplemented or not with cycloheximide at a concentration of 25 μg/ml, which is known to inhibit more than 90% of growth of the mycelium (20). Propagation of the [Het-s] character in the recipient strains was measured 15 hr later by sampling small pieces of mycelium on the recipient strain at 3 cm behind the line of contact between the strains. The phenotype of the mycelia regenerated from these samples was determined in a barrage test against a het-S strain. No significant difference was observed in the rate of propagation of the [Het-s*] → [Het-s] transition whether the medium contained cycloheximide or not (Table 1). As previously described (7), we have verified that the transition is not due to the migration of nuclei from the donor to the recipient. This possibility was eliminated using donor and recipient strains that differ in their mating type. In no case had samples of mycelium taken from the recipient become self-fertile, thus showing that no nuclei had migrated from the donor into the recipient mycelium. We never observed the propagation of the [Het-s] character if the recipient strain contained the null allele het-s°. Furthermore, we observed that het-s° strains that have been previously fused to a [Het-s] donor strain cannot induce the [Het-s*] → [Het-s] transition when they are used as donor strains in confrontation with a [Het-s*] recipient (Table 1). These results suggest that the molecular determinant responsible for the [Het-s] character and for the [Het-s*] → [Het-s] transition cannot be maintained in the het-s° genetic background and that the presence of the het-s gene is necessary for the propagation of the [Het-s] character. As the propagation of the [Het-s] character in the [Het-s*] strain is independent of protein synthesis, it can be interpreted as the consequence of the conversion of the pre-existing inactive pHET-s* form of the protein into the reactive pHET-s form.
Figure 2.
Schematic drawing of the experimental device used to analyze the [Het-s*] → [Het-s] transition. The striped square shows the position where a piece of mycelium was picked up 15 hr after the fusion of donor and recipient mycelia to determine the phenotype.
Table 1.
Analysis of the [Het-s*] → [Het-s] transition
| Number of strains that have gained the [Het-s]
phenotype
|
|||
|---|---|---|---|
| Donor strain = [Het-s] Recipient strain = [Het-s*] | Donor strain = [Het-s] Recipient strain = het-s° | Donor strain = het-s° Recipient strain = [Het-s*] | |
| Absence of cycloheximide | 17/17 | 0/12 | 0/10 0/7† |
| Presence of cycloheximide | 13/13 | nd | nd |
Conditions were as described in Materials and Methods, Fig. 2, and in the text. The values are given as the ratio between the number of strains that have gained the [Het-s] phenotype and the number of tested recipient strains. nd, not done.
In this experiment, the het-s° strain was first fused to a [Het-s] strain then pieces of mycelium of the het-s° strain were used to regenerate mycelia that were used as donors.
Differential Sensitivity of pHET-s and pHET-s* to Proteinase K.
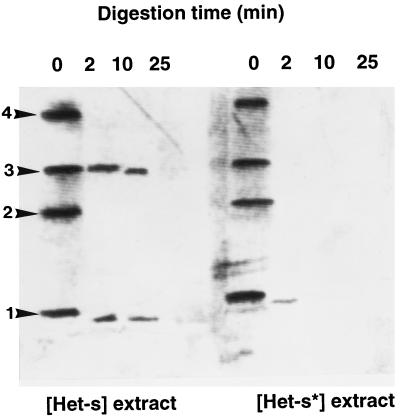
Biochemical evidence of a structural difference between pHET-s and pHET-s* has been obtained by comparing their sensitivity to a proteolytic enzyme. Crude extracts were prepared from [Het-s] and [Het-s*] strains, and equivalent amounts of total protein were incubated with proteinase K for different times. Control experiments have shown that the kinetics of degradation of total protein is similar for both extracts (not shown). Western blot analysis revealed that the protein present in the [Het-s] strain is more resistant to the digestion (Fig. 3). The protein is still clearly detectable in the extract from the [Het-s] strain after 10-min incubation. In contrast, the protein is almost completely degraded after 2 min in [Het-s*] extracts. Similar results have been observed with prions such as the Ure2p and Sup35p proteins in yeast (10, 12). As for these yeast proteins and for mammal prion protein (PrP), the more resistant protein is the form that is associated with the transmissible and stable phenotype. As for yeast proteins, the difference in resistance between the two polypeptides was not as high as between PrPSc and PrPC mammalian proteins, and no protease-resistant core was observed for pHET-s. Comparison with the results reported in Fig. 1 shows that the formation of multimeric aggregates, resistant to SDS/PAGE conditions, is induced by the conditions used in this experiment (incubation and/or concentration by ammonium sulfate precipitation). However, some high molecular weight aggregates are also present in crude extracts, especially when the protein is overexpressed in vivo (Fig. 1B). It has been proposed that the difference in protease resistance of the two forms of Sup35p could be explained by a poor accessibility of the Sup35 ppsi+ due to its aggregation. This does not seem to be the case for the proteins encoded by the het-s gene as the two forms display similar aggregation patterns after incubation of extracts in vitro. The fact that monomeric and trimeric forms of pHET-s are preferentially present after proteinase K treatment may result from the proteolysis of pHET-s leading to the production of a shorter polypeptide, which is visible on Western blots, that would preferentially produce trimeric aggregates rather than dimeric or tetrameric complexes as does the full-length polypeptide.
Figure 3.
Immunoblot of protein extracts from [Het-s] and [Het-s*] strains after digestion for various times with proteinase K. Numbers on the left show the position of monomers, 1; dimers, 2; trimers, 3; and tetramers, 4.
Effect of the Overexpression of the het-s Gene.
Inactivation of the het-s gene abolishes the incompatibility reaction but it does not impair the growth or the life cycle of the fungus. This was deduced from gene replacement by a disrupted allele (4) and from analysis of the neutral het-sx allele, which was shown to contain a small duplication within the coding sequence leading to a frame-shift and the expression of a putative truncated polypeptide (5). The effect of overexpression was then examined. First, a NcoI site overlapping the initiator ATG codon of the het-s gene was introduced by site-specific mutagenesis. This mutation replaces the second codon for serine by a codon for alanine. We determined that this mutation does not alter the expression of the het-s gene. Expression of this gene in het-s° protoplasts produces the same proportion of [Het-s*] and [Het-s] transformants as does the expression of the wild-type gene. A chimeric gene was then constructed, using this NcoI site, by fusing the het-s ORF to the noncoding 5′ end and promoter of the gene encoding the glyceraldehyde 3-phosphate dehydrogenase gene from Aspergillus nidulans. This gene is constitutively and strongly expressed in Aspergillus, and it has been used for heterologous expression in other fungi (21).
Protoplasts of a strain containing the inactive het-s° allele were cotransformed with the plasmid containing the chimeric gene and the pMOcosX vector carrying the hph gene as selectable marker. As a control, protoplasts were cotransformed with a plasmid containing the wild-type het-s gene instead of the chimeric one. The hygromycin-resistant transformants were tested against the het-S strain either directly after the selection or after a contact with a [Het-s] strain, a procedure that induces the [Het-s*] → [Het-s] conversion. Three classes of transformants were obtained (Table 2). Some displayed a stable neutral incompatibility phenotype; they account for about 20% of transformants and are likely to correspond to transformants that have not been cotransformed. The remaining strains exhibited the [Het-s] character either spontaneously or only after contact with the [Het-s] strain. The latter strains correspond to transformants that expressed the [Het-s*] phenotype upon transformation.
Table 2.
Effect of overexpression of the het-s gene on the expression of the [Het-s*] phenotype in transformants
| het-s gene present on the transforming vector | Number of hygromycin resistant transformants tested | Number of het-s transformants
|
|
|---|---|---|---|
| With [Het-s] phenotype | With [Het-s*] phenotype | ||
| Wild type | 170 | 2 (1.5%) | 136 |
| Chimeric, A. nidulans G3DP promoter | 250 | 59 (29.8%) | 139 |
The values in parentheses give the percentage of strains with the [Het-s] phenotype among transformants that contain the het-s gene. G3DP, glyceraldehyde 3-phosphate dehydrogenase.
The proportion of transformants that display spontaneously the [Het-s] character is about 20 times higher than in the control when the het-s coding sequence is expressed from the glyceraldehyde 3-phosphate dehydrogenase promoter. We also observed that transformants that exhibit the [Het-s*] phenotype are much more stable when they contain the wild-type het-s gene as opposed to the chimeric gene. One hundred percent of the latter strains had spontaneously switched to the [Het-s] phenotype after 3 days growth when the former could be stably propagated vegetatively, and these rarely switched spontaneously to the [Het-s] phenotype, as described previously. Western blotting was used to compare expression levels of the protein from the native and chimeric genes. From the results shown in Fig. 1B the amount of the protein is about 10 times higher when the ORF is under the control of the A. nidulans promoter. Except for the high rate of spontaneous [Het-s*] → [Het-s] transition, no change of the phenotype could be observed for transformants in which the protein was overexpressed.
Detection of Interactions Between the Proteins Encoded by the het-s Locus.
It has been suggested that the proteins encoded by the het loci are active as multimeric complexes and that the incompatibility reaction is triggered by the presence of poisonous complexes formed by the interaction of the proteins encoded by incompatible genes (1). We have tested the formation of complexes between proteins encoded by the het-s locus using the two-hybrid system in yeast. The ORFs of both het-s and het-S genes were fused in-frame with the DNA-binding domain and to the transactivator region of GAL4 on plasmids (18). The fusion plasmids were transferred in yeast strain Y190 in various combinations, and transformants were scored for β-galactosidase activity and resistance to 3-amino-1,2,4-triazole (Table 3). Activation of reporter genes was undetectable when any plasmid was tested alone. Positive results observed with the different combinations of fusion plasmids lead to the conclusion that proteins encoded by het-s and het-S genes can interact to form heterodimers and that each protein also can produce homodimers.
Table 3.
β-galactosidase activity in yeast cells cotransformed with various combinations of fusion vectors to detect interactions between products of the het-s locus
| ORF fused the GAL4 transactivator domain | ORF fused to the
GAL4 DNA-binding domain
|
|||
|---|---|---|---|---|
| None | het-s | het-S | c-jun | |
| None | 4 (−) | 3 (−) | 3 (−) | nd |
| het-s | 4 (−) | 157 (+) | 149 (+) | nd |
| het-S | 4 (−) | 144 (+) | 155 (+) | nd |
| c-fos | nd | nd | nd | 200 (+) |
Numbers give β-galactosidase activity in units/μg of protein, (+) and (−) indicate that strains can or cannot grow on medium supplemented with 100 μg/ml of 3AT. nd, not done.
DISCUSSION
Prions are transmissible agents responsible for spongiform encephalopathies such as scrapie, bovine spongiform encephalopathy, or Creutzfeldt–Jakob disease. Many lines of evidence suggest that the prion is devoid of nucleic acid and is identical to PrPSc, a modified form of PrPC. PrPC is a normal cell protein found predominantly at the surface of neurons. PrPSc in contrast to PrPC is relatively resistant to proteases and accumulates in cytoplasm. Studies of both forms have shown that PrPSc and PrPC have identical chemical composition but differ in their content of β-sheet and α-helix conformations (for review, see ref. 22). The current hypothesis to explain the propagation of prions is that the PrPSc molecule is able to convert the PrPC isoform into PrPSc. Two models for this conformational conversion of PrPC to PrPSc have been proposed. In the refolding model, the conversion occurs between monomers and aggregation is a secondary process. In the seeding model, the PrPSc conformation is acquired during binding of PrPC molecules to cristal-like seed or aggregate of PrPSc (for review, see ref. 23). The prion phenomenon is apparently not restricted to PrP. In the yeast S. cerevisiae, [URE3] and [PSI] are nonchromosomal genetic elements that display some prion-like modes of inheritance. [URE3] derepresses nitrogen catabolic enzymes and [PSI] increases the efficiency of several nonsense suppressor tRNAs. These phenotypes are identical to those conferred by mutations of the chromosomal genes URE2 and SUP35, which encode homologues of glutathione S-transferase and eukaryotic polypeptidic chain release factor eFR3, respectively. Data from several experiments indicate that [URE3] and [PSI] are prion-like isoforms of the polypeptides encoded by URE2 and SUP35 genes. They are inherited as cytoplasmic determinants, but their maintenance and propagation depend on the chromosomal genes URE2 and SUP35, respectively, and overexpression of the genes increases the generation of the nonmendelian elements (8, 9, 11). Moreover, as described for PrP, the proteins encoded by URE2 and SUP35 are more resistant to proteases in strains with [URE3] and [PSI] phenotypes (10, 12). By analogy to prions, it was proposed that [URE3] and [PSI] are altered forms of the proteins encoded by URE2 and SUP35 that are inactive and can convert the normal form of the proteins to the altered conformation.
A similar situation is suggested by the properties of the het-s gene of the P. anserina. The het-s locus is involved in vegetative incompatibility. Coexpression of the two antagonistic alleles het-s and het-S in heterokaryotic cells triggers a lethal and degenerative reaction. A search for interactions between the proteins encoded by the het-s locus, using the yeast two-hybrid system, revealed that proteins encoded by the two alleles can form homodimers and heterodimers. So, as proposed previously (1), the complex between the protein products of these incompatible genes may be poisonous, inducing a lethal disorder of some cellular function. Two forms of the protein encoded by the het-s gene have been identified. One, pHET-s, is present in strains that are incompatible with those containing the het-S allele. The other form, pHET-s*, is not reactive in incompatibility and is present in strains that display the neutral [Het-s*] incompatibility phenotype. These two forms also can be differentiated from each other by their sensitivity to proteinase K. The pHET-s version is able to be transferred into a strain that displays the [Het-s*] phenotype and to be propagated there, leading to a [Het-s*] → [Het-s] transition. This transition spreads very rapidly from the region of anastomosis to all the mycelium. It may be greatly facilitated by the coenocytic structure of the mycelium. The propagation of the [Het-s] character is independent of protein synthesis but depends on presence of the het-s gene; it was not observed in strains that contain a null allele of het-s, and the propagation and the maintenance of the molecular determinant responsible for the [Het-s] phenotype cannot be achieved in het-s° strains. We propose that, as for the PrP, the two forms of the protein encoded by the het-s gene differ in their conformation and that the pHET-s form is able to confer its conformation to pHET-s*, probably as the result of interaction between the molecules. This conformational change also could occur spontaneously, but at a very low frequency. It could account for the observed spontaneous [Het-s*] → [Het-s] transition. The frequency of this spontaneous event is increased by overexpression of the protein, as expected in a prion-like mechanism. All transformants that overexpress the het-s gene are rapidly and spontaneously converted to the [Het-s] phenotype. Such a high rate of conversion compared with the effect of overexpression of URE2 in yeast may be the consequence of the coenocytic structure of the mycelium. A pHET-s* → pHET-s conversion occurring in any cell will rapidly spread within all the mycelium. Such an infectious propagation cannot occur for an intracellular protein in a unicellular organism.
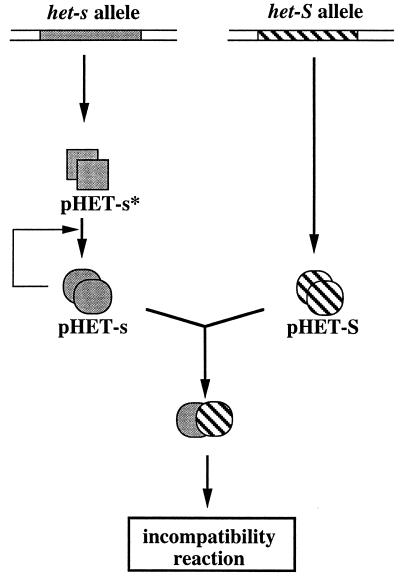
The properties of the protein expressed from the het-s gene are entirely compatible with the prion hypothesis as already proposed for [URE3] and [PSI] characters in yeast. In yeast, curing of [PSI] and [URE3] by guanidine has been obtained. It is reversible as it produces cells from which [URE3] and [PSI] can be isolated again (9, 24). We could not cure [Het-s] from mycelia to produce [Het-s*] strains in the presence of various concentrations of guanidium chloride (data not shown). However, it has been shown that regeneration of protoplasts prepared from [Het-s] mycelium produces about 8% of [Het-s*] strains that can be reversibly converted to the [Het-s] strains either spontaneously or after fusion to the [Het-s] strain (25). Such curing may be induced by the treatment of the mycelium at high osmotic strength (0.8 M sucrose) necessary for protoplast stabilization. Curing in hypertonic media has been observed for [PSI] in yeast (26). A difference between the [Het-s] and the yeast [URE2] and [PSI] traits is that here the transmissible prion isoform displays a biological activity; it is reactive in incompatibility. However, we have no information yet on the function of the het-s gene in the biology of the fungal cell. It is possible that the pHET-s* form of the protein, which is not reactive in incompatibility, has a biological function that might be lost upon conversion into the pHET-s form. This latter protein should be inactive in this putative cellular function but has acquired a conformation that induces the formation of poisonous complexes with the protein encoded by the antagonistic het-S gene (Fig. 4).
Figure 4.
Model of relations between the proteins encoded at the het-s locus. The het-S allele encodes a protein that can form complexes with the pHET-s polypeptide; coexpression of the two proteins triggers the incompatibility reaction. The het-s allele encodes the pHET-s* polypeptide, which is not reactive in incompatibility, and can be converted into a structurally different form, the prion form pHET-s, which is reactive against pHET-S, and can catalyze the conversion of pHET-s* to the pHET-s conformation.
Preliminary data (V. C. and S. S., unpublished results) show that it is possible to isolate mutations that modify or suppress the [Het-s*] → [Het-s] transition, providing a biological model to characterize protein domains that could be involved in structural transition. Various proteins involved in quite different cellular functions and with no significant sequence similarity can display prion-like properties. It is predictable that other proteins that are able to exist under metastable conformations and to self-modify their conformation will be described. This heritable protein conversion model opens new perspectives for understanding the epigenetic transmission of metastable phenotypes.
Acknowledgments
This work was supported by grants from the Centre National de la Recherche Scientifique (Action Concerte Science de la Vie 10 number 9510001). V.C. is funded by Ministere del Education Nationale de l’Enseignement Superieur et de la Recherche.
ABBREVIATION
- PrP
prion protein
References
- 1.Bégueret J, Turcq B, Clavé C. Trends Genet. 1994;10:441–446. doi: 10.1016/0168-9525(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 2.Rizet G. Rev Cytol Biol Veg. 1952;13:51–92. [Google Scholar]
- 3.Turcq B, Denayrolles M, Bégueret J. Curr Genet. 1990;17:297–303. [Google Scholar]
- 4.Turcq B, Deleu C, Denayrolles M, Bégueret J. Mol Gen Genet. 1991;288:265–269. doi: 10.1007/BF00282475. [DOI] [PubMed] [Google Scholar]
- 5.Deleu C, Clavé C, Bégueret J. Genetics. 1993;135:45–52. doi: 10.1093/genetics/135.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ephrussi B. Nucleocytoplasmic Relations in Micro-organisms. Oxford: Clarendon; 1953. pp. 79–108. [Google Scholar]
- 7.Beisson-Schecroun J. Ann Genet. 1962;4:3–50. [PubMed] [Google Scholar]
- 8.Wickner R B, Masison D C, Edskes H K. Yeast. 1995;11:1671–1685. doi: 10.1002/yea.320111609. [DOI] [PubMed] [Google Scholar]
- 9.Wickner R B. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 10.Masison D C, Wickner R B. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 11.Pätino M M, Liu J J, Glover J R, Lindquist S. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 12.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 13.Esser K. In: Handbook of Genetics. King R C, editor. New York: Plenum; 1974. pp. 531–551. [Google Scholar]
- 14.Bergès T, Barreau C. J Gen Microbiol. 1989;135:601–604. doi: 10.1099/00221287-135-3-601. [DOI] [PubMed] [Google Scholar]
- 15.Orbach M J, Sweigard A, Walter A, Farral L, Chumley F G, Valent B. Fungal Genet Newsletter. 1991;38:16. [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higushi R, Horn R, Mullis K B, Erlich H A. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 18.Chevray P M, Nathan D. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. pp. 352–355. [Google Scholar]
- 20.Crouzet M, Perrot M, Nogueira M, Bégueret J. Biochem Genet. 1978;16:271–278. doi: 10.1007/BF00484084. [DOI] [PubMed] [Google Scholar]
- 21.Punt P J, Dingemanse M A, Jacobs-Meijsing B J M, Pouwels P H, Van den Hondel C A M J J. Gene. 1988;69:49–57. doi: 10.1016/0378-1119(88)90377-0. [DOI] [PubMed] [Google Scholar]
- 22.Prusiner S B. Annu Rev Microbiol. 1994;48:455–486. doi: 10.1146/annurev.mi.48.100194.003255. [DOI] [PubMed] [Google Scholar]
- 23.Weissmann C. FEBS Lett. 1996;389:3–11. doi: 10.1016/0014-5793(96)00610-2. [DOI] [PubMed] [Google Scholar]
- 24.Tuite M F, Mundi C R, Cox B S. Genetics. 1981;98:691–701. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belcour L. Neurospora Newsletters. 1975;23:26–27. [Google Scholar]
- 26.Singh A C, Helms C, Sherman F. Proc Natl Acad Sci USA. 1979;76:1952–1956. doi: 10.1073/pnas.76.4.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]