Abstract
A better knowledge of peptide structures interacting with major histocompatibility complex (MHC) molecules is of great interest for better understanding of the molecular basis of immune recognition. We have isolated naturally processed peptides from a continuously growing antigen-presenting Epstein–Barr virus-transformed human B-cell line. HLA-DR complexes were purified by specific affinity chromatography and complexed peptides were released by acid treatment. The isolated peptides were separated by reversed phase chromatography and fractions were analysed by Edman degradation at picomolar ranges. From 30 fractions that were examined seven peptides bound to the HLA-DRB1*0405 and two peptides from the human leucocyte antigen (HLA) class II associated invariant chain bound to HLA-DRB1*1302. In addition, a N-terminal β-chain peptide of the 0405 allele was identified. Evaluation of amino acid sequences revealed a refined FXXL motif for the 0405 allele, in which F (phenylalanine) stands for any aromatic amino acid and L (leucine) can be exchanged by either I (isoleucine) or V (valine). In total, three fractions contained a peptide derived from the human migration inhibition factor (MIF), a pro-inflammatory cytokine that is normally produced by activated T lymphocytes and monocytes/macrophages. Indeed, cytokine analysis revealed high amounts of MIF secreted by the B-cell line, confirming that MHC class II expressing cells can present any intrinsic peptide that contains the distinct motif for HLA-binding. For MIF, the amino acid sequence Y36IAV39 represents the required binding motif for HLA-DRB1*0405. Nevertheless, it is the first time that cytokine fragments were found to bind to HLA molecules on human B cells.
INTRODUCTION
Major histocompatibility complex (MHC) molecules possess the intrinsic feature to bind peptide ligands from antigens processed in cellular compartments. These complexes are transported to the cell surface where they present the antigenic fragment to the specific T cells.1 MHC class II molecules bind fragments from proteins occurring in the endosomal/lysosomal compartment, including endogenous as well as phagocytosed antigens.2 The class II molecules are synthesized in the endoplasmic reticulum and nonameric complexes are formed, consisting of three dimers associated with trimerized invariant chain (Ii).2,3 The α/β dimers consist of two noncovalently linked 34 000 MW and 27 000 MW transmembrane chains. Ii directs this complex into the endosomal system and prevents class II molecules from interacting with peptides or segments of proteins during transport.4 Proteolytic cleavage of the Ii in a post-Golgi compartment releases α/β dimers, making the class II binding site susceptible for peptide binding. These dimers are derived from internalized antigens and delivered to endosomes after being processed in lysosomes.5,6 The class II/peptide complex is then released to the plasma membrane for recognition by specific CD4+ T cells.
The human MHC class II molecules are classified into human leucocyte antigen (HLA)-DR, DP, and DQ, depending on the allelic form of the β-chain. Each allelic form of the highly polymorphic MHC molecules can bind a large array of different peptides to ensure T-cell mediated immunity to the antigenic universe. The identification of a large number of peptides that bind to a distinct MHC molecule makes it possible to reveal the molecular prerequisites for MHC/peptide interaction. Knowledge of the structural features of MHC-presented T-cell epitopes provides attractive applications in the field of specific immunotherapy.7,8
Anchor residues are amino acids with similar side chains frequently occurring at certain positions in a ligand. Whereas type 1 anchors mainly contribute to the binding energy of the peptide, type 2 anchors mostly reflect steric constraints.9 Typically, type 1 anchors are amino acid residues with distinct physicochemical characteristics, reflecting the chemical environment of a given pocket. On the other hand, type 2 anchors are mostly small amino acid residues that are relatively enriched, if the pocket size limits the interaction with bulky residues.10 Thus, steric constraints rather than major contributions to the binding energy of the peptide represent the mechanism underlying their enrichment.
Sequence analysis of pooled endogenous peptides eluted from MHC class I molecules was a successful approach to define allele-specific MHC class I binding motifs.11 Single peptide analysis of human and murine MHC class II eluted fragments revealed larger peptides of variable size.12,13 Putative binding motifs were determined for various HLA-DR specificities by alignment of DR-binding peptides in combination with functional analyses,14 by alignment of sequenced endogenous peptides,13 and the usage of peptide libraries displayed on phage surface.10 Although ≈85% of class II molecules are of HLA-DR type, not all DR subtypes could be associated to a binding motif. Using the human Epstein–Barr virus (EBV)-B-cell line BuB1 and pool sequencing at picomolar levels, we isolated and identified naturally processed peptides bound to HLA-DRB1*0405 (DR4) and -DRB1*1302 (DR13(6)) molecules. We could define a FXXL motif for the 0405 allele, and identified a DRB1-binding peptide derived from migration inhibition factor (MIF), a cytokine, which simultaneously was secreted by the antigen-presenting B cell itself.
MATERIALS AND METHODS
Reagents
The anti-HLA-DR monoclonal antibody (mAb) L243 (ATCC, Rockville, MD) was purified from L243 hybridoma supernatant using a MAbTrapTMGII (Pharmacia LKB Biotechnology, Uppsala, Sweden). Zwittergent 3-12 was purchased from Calbiochem (La Jolla, CA). CNBr-activated sepharose was from Pharmacia. High pressure liquid chromatography (HPLC) solvents (LiChrosolv gradient grade) were from Merck (Darmstadt, Germany). All other chemicals were purchased from Fluka Chemie AG, Buchs, Switzerland at the highest purity available.
Cells and cell cultures
The human B-cell lines BuB1 and BuB were derived from EBV-infected peripheral blood mononuclear cells (PBMC) from a honeybee venom sensitized individual (Bu) by repeated selection and subcloning.15 BuB cells secrete Abs of unknown specificity and BuB1 secrete immunoglobulin G (IgG)4/λ Abs to bee venom phospholipase A2 (PLA). BuB1 and BuB represent B-cell lines, which can effectively act as antigen-presenting cells (APC) for the bee venom antigen phospholipase A2in vitro.15 Both lines were maintained for >4 years in culture without changes in properties. The cells express DRB1*0405, DRB1*1302, DQB1*0302, DQB1*0604, DPB1*0101, and DPB1*02012, as determined by oligonucleotide hybridization after polymerase chain reaction (PCR)amplification. HLA class II peptides were isolated from the BuB1 line. Cells were cultured in 750 ml-spinner bottles (Technomara AG, Wallisellen, Switzerland) starting with 4×105 cells/ml in 200 ml RPMI-1640 medium (Gibco BRL, Life Technologies, Basel, Switzerland) supplemented as described.8 The cell suspension was continuously stirred at 40 r.p.m. using a rotation angle of 540°. The cell density was monitored daily and the suspension was diluted 1:2 with fresh medium when a density of 1·5×106 cells/ml was reached. After 12–15 days 1·5×109 cells/l were obtained with a viability >90%. All the cells were regularly checked for Mycoplasma infection by using RIDASCREEN® Mycoplasma IFA (Vitromex, Vilshofen, Germany). There was no contamination detectable throughout maintenance of the cell lines.
Cell surface expression of MHC class II molecules
BuB1 and BuB cells, both from the same donor, were incubated for 30 hr at different cell concentrations. HLA class II expression was measured by using the fluoroscein isothiocyanate (FITC)-labelled anti-HLA class II mAb CR3/43 (Dako, Denmark). Mouse IgG1–FITC was used as control. Cells were incubated for 30 min at room temperature and fluorescence analysis was performed on an Epics Profile flow cytometer (Coulter Corp., Hialeah, FL).
Isolation of the HLA-DR/peptide complexes
Isolation of the HLA-DR/peptide complex was performed as described by Halder et al.16 In brief, 3×109 cells were incubated for 7 hr in 40 ml fetal calf serum (FCS)-free RPMI-1640 medium at 37° and extensively washed with Tris-buffered saline (TBS: 50 mm Tris–HCl, 0·9% NaCl, pH 7·6). The cell pellet was stored at 20°. Cells were lysed by three freeze–thaw cycles and extensive stirring on ice in a fivefold volume of lysis buffer (10 mm Tris, 140 mm NaCl, 0·2 mg/ml NaN3, 0·2 mm phenylmethanesulphonyl fluoride (PMSF), 5 μm leupeptin, 10 μm pepstatin, 10 μm chymostatin, pH 8·0) supplemented with 2% Triton-X-100. Fragments were separated after 3 hr of lysis by centrifugation at 2000 g and the procedure was repeated. The supernatants were centrifuged at 48 000 g and filtered through a 0·45-μm filter (Millipore Corp., Bedford, MA). HLA-DR was isolated on a 1×2·5 cm anti-HLA-DR L243-coupled sepharose column at 4°. The use of a bovine serum albumin (BSA)-coupled sepharose precolumn minimized non-specific matrix binding of proteins. The following steps were performed at 4° at a flow rate of 72 ml/min. After loading, the L243-sepharose column was washed with washing buffer 1 (50 mm Na2HPO4, 150 mm NaCl, 0·1% Zwittergent 3-12, pH 8·0) and washing buffer 2 (100 mm Na2HPO4, 0·1% Zwittergent 3-12, pH 8·0) until UV absorption at 278 nm was constant. The HLA-DR/peptide complex was eluted with elution buffer (100 mm Na2HPO4, 0·1% Zwittergent 3-12, pH 11·0). The pH was adjusted to 7·0 with 6 n HCl and the solution was stored at 4°.
Release of HLA-bound peptides
The eluate from the affinity column was concentrated with Centriprep-30 (Grace Co, Amicon Division, MA), washed 10 times with water (HPLC grade) to remove detergent, and adjusted to a final protein concentration of 0·2–0·5 mg/ml (Bradford assay, BioRad, Hercules, CA). Normally, 3×106 cells yielded 1·9–2·5 mg of HLA-DR/peptide complex. The pH was adjusted to 2·0–2·2 with trifluoracetic acid (TFA), and after 2 hr of incubation at 37° the peptides were separated from the DR molecule by filtering through a 30 000 MW cut-off membrane Centricon-30 (Grace Inc., Beverley, CA). The filter was washed three times with water/TFA pH 2·0 and the volume of the filtrate was reduced to 100 μl in a SpeedVac (Savant Instruments Inc., Farmingdale, NY).
Separation of naturally processed peptides
The peptides (≤100 μg) were separated at room temperature by HPLC (pump: 140 B, Applied Biosystems, CA; detector: SPD 10 A, Shimazu Corp., Kyoto, Japan) using a flowsplitter that allowed a flow rate of 4–5 μl/min, a C18 column (LC Packings, 300 μm×150 mm), and a linear TFA/acetonitrile gradient.16
Detection of MIF
BuB1 cells (5×105/ml) were grown to a density of 1·6×106 cells/ml in a tissue culture flask (Falcon, Becton Dickinson, NJ). MIF concentration was determined in cell supernatants by solid phase sandwich enzyme-linked immunosorbent assay (ELISA). In brief, 96-well microtitre plates (Maxisorp, Nunc, Roskilde, Denmark) were coated over night at 4° with 5 μg/ml of IgG1 antihuman MIF mAb (Clone: 12302.2; R & D Systems, Abingdon, UK) in phosphate-buffered saline (PBS). The plate was blocked for 1 hr with 1% BSA and 5% sucrose in PBS. Recombinant human MIF (R & D Systems) was used as standard. The plate was incubated for 2 hr at room temperature with a biotinylated goat antihuman MIF-specific IgG antibody (R & D Systems). After incubation with streptavidin–peroxidase (Sigma) for 1 hr, the plate was developed with 1·5 mg/ml of o-phenylendiamine (OPD) in 0·05 m phosphate–citrate buffer (pH 5·0) supplemented with 1/1000 (v/v) 30% H2O2. The reaction was stopped after 30 min with 50 μl 0·5 m H2SO4. Optical density at 490 nm was measured with a microplate reader (V max, Molecular Devices, Menlo Park, CA).
RESULTS
HLA expression on B cells
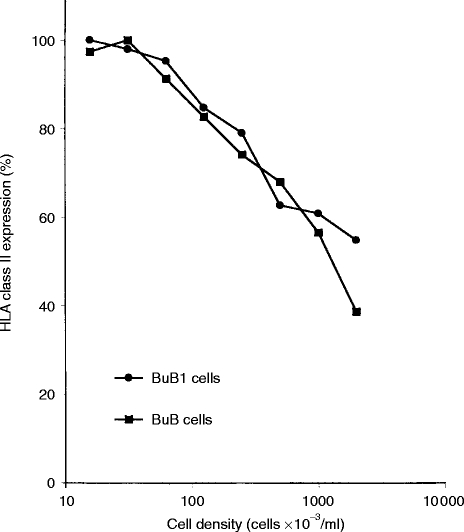
The amount of naturally processed peptides that yields from the cells depends mainly on the degree of HLA expression on the cell surface. Therefore, we first measured the HLA expression on BuB1 and BuB cells at different cell densities by fluorescence-activated cell sorting (FACS) analysis (Fig. 1). We found that the HLA class II expression decreased on both BuB1 and BuB cells with increasing cell density. At the highest cell concentration (1·5×106 cells/ml) the class II expression was ≈50% of the highest HLA expression measured at optimal conditions. On the other hand, this higher cell density results in 150 times more cells from the same culture volume, giving finally a 75 times higher HLA yield.
Figure 1.
Expression of HLA class II in BuB1 and BuB cells at different cell densities. Cells were incubated for 30 hr at different cell concentrations. The final cell density was determined after the incubation time. The viability was >90% in all experiments. HLA class II was measured with FITC-labeled anti-HLA class II mAb. The highest expression obtained was set as 100%
Analysis of naturally processed peptides
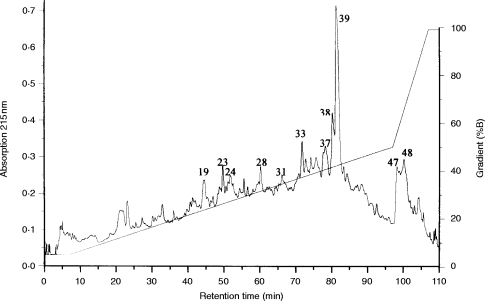
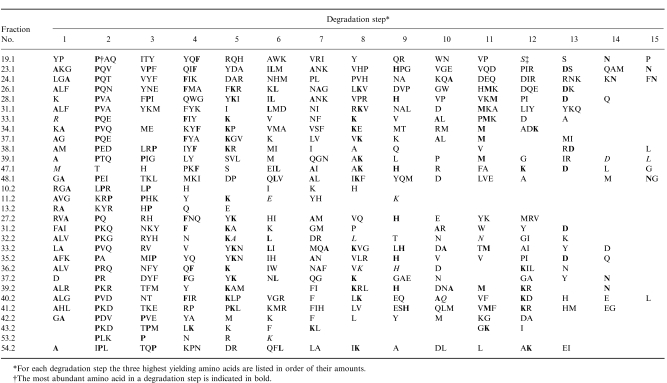
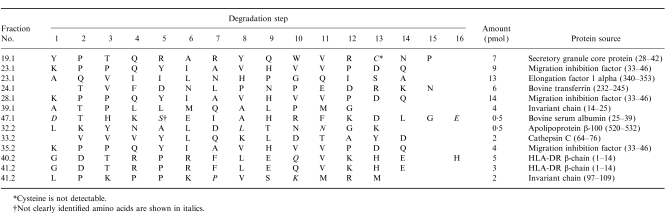
The peptide mixtures isolated from HLA-DR were separated by HPLC. Figure 2 shows a chromatogram representative for different batches. Only a few distinct peaks could be detected due to the complexity of the mixture. Most of the peaks show shoulders or asymmetric shapes, indicating a large heterogeneity of the peptides. Fractions collected after HPLC separation were analysed using the protein sequencer ABI 476 A (Applied Biosystems) in order of their relative absorbency. In Table 1 the results of 30 fractions analysed are summarized. Mainly multiple sequences were found, with the common attribute that proline appears significantly often in the second degradation step, indicating cleavage properties of an aminopeptidase N-like enzyme. From the established peptide sequences, the respective proteins were determined by the databases Swissprot, PIR, and GenEMBL with programs from the Genetics Computer Group (GCG, Wisconsin Package, Madison, WI). In 11 out of 30 fractions, a sequence filed in an available database could be identified. The reliability of the found sequences was controlled by comparing the amounts of corresponding amino acids in each degradation step. Table 2 summarizes the peptide sequences and the appropriate proteins. Two exogenous peptides derived from bovine transferrin and bovine serum albumin could be identified in the analysed fractions. Both proteins are constituents of FCS contained in the medium. Furthermore, seven endogenous peptides were found, of which two were derived from the HLA class II invariant chain. In three fractions, a peptide derived from the migration inhibition factor was identified.
Figure 2.
HPLC separation of peptides released from HLA-DRB1*0405 and -DRB1*1302. The absorption at 215 nm exhibited a complex mixture of peptides in little amounts. For separation a gradient of 5% B to 50% B in 90 min and 50% to 100% B in 10 min (A: 0·1% TFA; B: 84% acetonitrile, 0,08% TFA) was used. Single peaks in 56 fractions of at least 5 μl were collected. Numbers correspond to the fraction numbers of batch 1.
Table 1.
Sequence analysis of peptides isolated from HLA-DRB1*0405 and DRB1*1302
Table 2.
Amino acid sequences related to database-filed proteins
BuB1 cells secrete MIF and present MIF-derived peptides
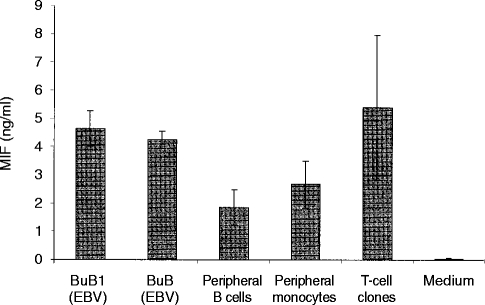
MIF is a cytokine normally secreted by activated T lymphocytes and monocytes/macrophages however, MIF-secreting unstimulated B cells had not been reported so far. Because BuB1 cells presented a peptide from MIF protein, the cell cultures were tested for MIF production. After incubation of 5×105 cells for 72 hr, a final density of about 1·5×106 cells/ml was reached, and MIF was detected in the supernatants by solid phase sandwich ELISA. As shown in Fig. 3, BuB1 cells secreted MIF in amounts of 3·9–5·2 ng/ml. Similar amounts of MIF were also secreted by the non-specific EBV-B cell line BuB originating from the same donor. In addition, the secretion of MIF by peripheral B cells, peripheral monocytes and three T-cell clones was determined. Peripheral B cells, freshly isolated from monocyte-depleted PBMCs by magnet activated cell sorter (MACS) and anti-CD19 (Miltenyi Biotec, Bergisch Gladbach, Germany), secreted about 50% less MIF than the EBV-transformed B cells under similar culture conditions. Monocytes, freshly isolated by the MACS procedure, were cultured for 72 hr at a cell concentration of 1·5×106 cells/ml. They secreted MIF in amounts of 1·8–3·5 ng/ml. Three PLA-specific human T-cell clones from three donors were washed with medium at day 6 after restimulation with antigen and further incubated for 72 hr at 1·5×106 cells/ml in interleukin-2 (IL-2) (25 U/ml) containing medium. Supernatants of these cultures contained MIF in the range of 2·8–7·9 ng/ml.
Figure 3.
Secretion of MIF by the EBV-B cell lines BuB1 and BuB, by freshly isolated peripheral blood B cells and monocytes, and by three antigen-specific human T-cell clones from three different donors. The data represent the average of three experiments performed in duplicates (±standard deviation). EBV-B cells (5×105 cells/ml) were incubated for 72 hr to a cell density of 1·6×106 cells/ml. The other cell types were maintained for 72 hr at 1·5×106 cells/ml. T cells were washed at day 6 after last restimulation with the antigen and cultured in IL-2 (25 U/ml) containing medium. MIF was measured in the supernatants by solid phase sandwich ELISA.
Identification of anchor residues
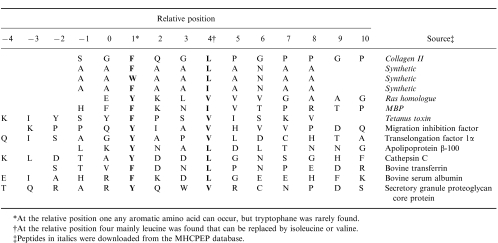
Pool sequencing17 and competition assays with polyalanine substituted analogues18 allowed the definition of several motifs for HLA-DRB1*0405 and HLA-DRB1*1302.19 These motifs describe up to nine possible amino acids for each anchor position. Therefore, we attempted to elucidate a more restricted motif for these MHC types. In contrast to peptides released from MHC class I molecules, class II derived peptides vary much more in size, with a length ranging from 13 to 25 amino acids.13 The detection limit of the sequencer used enabled the determination of 13–18 amino acid residues for each peptide analysed. Moreover, only N-terminal sequences of isolated peptides that are not necessarily part of the binding cleft may have been found. Therefore, we extended in a first step the sequences to a length of 30 amino acids according to the database entries. Peptides that are known to bind with high affinity to DRB1*0405 were aligned. These peptides were downloaded from the MHCPEP database (http://wehih.wehi.edu.au/mhcpep).20 The motif [FYTW]XX[LIV] was determined, in which any aromatic amino acid can occur at the first position. Phenylalanine was mainly observed at the first position, whereas tryptophan occurred rarely. At position four, leucine appears to be preferred, but other hydrophobic amino acids with a medium sized sidechain, such as isoleucine or valine, can also be observed. Table 3 shows the sequences from the MHCPEP database together with our results, aligned according to the proposed motif for DRB1*0405. The FXXL motif was compared with sequences from the MHCPEP database expressing moderate or low affinity to DRB1*0405. Accordingly, at the relative position one, amino acids with less bulky side chains show low affinity to DRB1*0405. The affinity is also lower if bulky side chains are contained at the relative position four of the motif.
Table 3.
Known HLA-DRB1*0405 binding peptides, aligned for a possible motif
Similar motifs were found for DRB1*0101 ([YVLF]XX[LAIVM] and DRB1*0401 ([WY]XX[VLMA]).21,22 A comparison of the pocket-forming amino acids in the binding cleft (published at http://histo.cryst.bbk.ac.uk) demonstrated similar pockets for DRB1*0405 and DRB1*0401, but greater differences to DRB1*0101. Pocket 1 shows no differences, in pocket 4 the residue H13 in DRB1*0405 changed to F in DRB1*0101, in pocket 6 V11 to L, in pocket 7 D28 and Y30 to E and C, and in pocket 9 E9, Y37, and S57 to W, S and D, respectively. In DRB1*0401 only S57 and R71 are exchanged by D and K. This further suggests that the anchor residue one is of type 1, because its sidechain is bulky and does not fit exactly in a pocket formed by the β-chain of the HLA molecule. On the other hand, the anchor residue at position four is likely to be of type 2 and may not substantially contribute to the binding energy of the peptide.
Out of the 10 different peptides identified, three did not display the FXXL motif. One of them corresponded to the N-terminus of the HLA-DRB1*0405 β-chain. The other two were derived from Ii and probably bound to DRB1*1302.
DISCUSSION
We have isolated naturally processed peptides from the EBV-transformed human B-cell line BuB1 and determined the amino acid sequence of 11 different peptides from nine proteins. Isolation of naturally processed antigens normally yields a complex mixture of various peptides. At least 654 different peptides were detected from the murine class II molecule IAd,23 which only account for about 10% of the class II molecules. This relatively low recovery can be explained either by a loss of peptides during isolation, the presence of undetectable peptides, or ‘empty’ MHC molecules. In any case, high sensitive analytic tools like capillary HPLC combined with either high sensitive Edman degradation or nano-spray mass spectrometry are needed to identify naturally processed peptides. Nevertheless, the most important peptides are probably selected by this approach.
Of class II ligands, both the N- and C-termini are ragged and hang out of the binding groove, and are thought to be not required for MHC-binding.12,13,24 On the other hand, it was reported25 that the length of the peptide determines the lifespan of the MHC/peptide complex, whereas trimming of bound peptides by aminopeptidase N has consequences for T-cell antigen recognition.26 From our sequence data, it is obvious that many of the peptides found contain a proline at the N-terminal position two. This was also reported by Falk et al.,22 who suggested that class II derived peptides are processed by an aminopeptidase N-like enzyme that specifically cuts one residue before proline. Such an enzyme, known as CD13, is expressed on the surface of antigen presenting cells. Breloer et al.27 demonstrated that proline at position two enhances the recognition capacity of MHC class II-bound peptides by T-cells. Therefore, antigen processing is not finished after the peptide has bound to the MHC molecule and may continue on the cell surface.
It has been suggested that T-cell recognition of presented peptides is independent of peptide affinity to the MHC molecule.28 Furthermore, it was shown29 that peptides are still able to bind to HLA molecules even after substitution of anchor residues with amino acids having contrary physicochemical properties. A higher affinity could be obtained in optimized interaction between non-anchor residues and binding cleft-forming amino acids from the β-chain. Therefore, studies using synthetic peptides do not necessarily lead to a motif used in nature as well.
In our analysis, we repeatedly identified a MIF derived peptide presented by the BuB1 cells. MIF was secreted in high amounts by both BuB1 and BuB cells. This cytokine is known to be produced mainly by activated T lymphocytes and monocytes/macrophages and has both macrophage- and T-cell-activating properties.30 MIF is also a major constituent of corticotropic cells within the anterior pituitary gland, is secreted into the circulation in a hormone-like fashion, and is thus believed to integrate central and peripheral inflammatory responses.31*sref31* Other studies describe that MIF can be released by B cells after stimulation with polyclonal activators.32,33 BuB1 cells are effective APC in specific T-cell responses,34 but so far it was not shown that they simultaneously present peptide fragments from a cytokine secreted by the B cells themselves.
In conclusion, we showed that MIF derived peptides bind to HLA-DRB1*0405 molecules of MIF-secreting human B cells. Furthermore, we identified seven different peptides possessing the structural requirements for HLA-DRB1*0405 binding, which contain a FXXL motif for this allele. Two peptides derived from the Ii were probably bound to HLA-DRB1*1302.
Acknowledgments
The authors thank Professor Dr E. D. Albert, Kinderpoliklinik LMU, München, Germany for determining the HLA subtypes of BuB1 cells.
This work was supported by the Swiss National Foundation, Grant no. 3100·0529·86·97/1, the Theodor and Ida Herzog-Egli Foundation, and the Deutsche Forschungsgesellschaft ME 765/5–1.
REFERENCES
- 1.Ziegler HK, Unanue ER. Identification of a macrophage antigen processing event required for I-region restricted antigen presentation to T lymphocytes. J Immunol. 1991;127:1869. [PubMed] [Google Scholar]
- 2.Germain RN. MHC-dependent antigen processing and peptide presentation: providing ligands for T lymphocyte activation. Cell. 1994;76:287. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- 3.Cresswell P. Chemistry and functional role of the invariant chain. Curr Opin Immunol. 1992;4:87. doi: 10.1016/0952-7915(92)90131-w. [DOI] [PubMed] [Google Scholar]
- 4.Lotteau V, Teyton L, Peleraux A, et al. Intracellular transport of class II MHC molecules directed by invariant chain. Nature. 1990;348:600. doi: 10.1038/348600a0. [DOI] [PubMed] [Google Scholar]
- 5.Roche PA, Cresswell P. Proteolysis of the class II-associated invariant chain generates a peptide binding site in intracellular HLA-DR molecules. Proc Natl Acad Sci USA. 1993;88:3150. doi: 10.1073/pnas.88.8.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blum JS, Cresswell P. Role of intracellular proteases in the processing and transport of class II HLA antigens. Proc Natl Acad Sci USA. 1988;85:3975. doi: 10.1073/pnas.85.11.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruppert J, Franco A, Alexander J, et al. MHC blocking peptides and T-cell receptor antagonists: novel paths to selective immunosuppression? Chem Immunol. 1995;60:61. [PubMed] [Google Scholar]
- 8.Akdis AC, Akdis M, Blesken T, Wymann D, Alkan SS, Blaser K. Epitope specific T cell tolerance to phospholipase A2 in bee venom immunotherapy and recovery by IL-2 and IL-15. In Vitro J Clin Invest. 1996;98:1676. doi: 10.1172/JCI118963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammer J, Belunis C, Bolin D, et al. High-affinity binding of short peptides to major histocompatibility complex class II molecules by anchor combinations. Proc Natl Acad Sci USA. 1994;91:4456. doi: 10.1073/pnas.91.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammer J, Takacs B, Sinigaglia F. Identification of a motif for HLA-DR1 binding peptides using M13 display libraries. J Exp Med. 1992;176:1007. doi: 10.1084/jem.176.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falk K, Rötzschke O, Stevanovic S, Jung G, Rammensee H-G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 12.Hunt DF, Michel H, Dickinson TA, et al. Peptides presented to the immune system by the murine class II major histocompatibility complex molecule I-Ad. Science. 1992;256:1817. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- 13.Chicz RM, Urban RG, Lane WS, et al. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. J Immunol. 1992;358:764. doi: 10.1038/358764a0. [DOI] [PubMed] [Google Scholar]
- 14.O'Sullivan D, Arrhenius T, Sidney J, et al. On the interaction of promiscuous antigenic peptides with different DR alleles. Identification of common structural motifs. J Immunol. 1991;147:2663. [PubMed] [Google Scholar]
- 15.Carballido JM, Faith A, Carballido-Perrig N, Blaser K. The intensity of T cell receptor engagement determines the cytokine pattern of human allergen-specific T helper cells. Eur J Immunol. 1997;27:515. doi: 10.1002/eji.1830270224. [DOI] [PubMed] [Google Scholar]
- 16.Halder T, Pawelec G, Kirkin AF, et al. Isolation of novel HLA-DR restricted potential tumor-associated antigens from the melanoma cell line FM3. Cancer Res. 1997;57:3238. [PubMed] [Google Scholar]
- 17.Friede T, Gnau V, Jung G, Keilholz W, Stevanovic S, Rammensee H-G. Natural ligand motifs of closely related HLA-DR4 molecules predict features of rheumatoid arthritis associated peptides. Biochim Biophys Acta 1316. 1996:85. doi: 10.1016/0925-4439(96)00010-5. [DOI] [PubMed] [Google Scholar]
- 18.Matsushita S, Takahashi K, Motoki M, Komoriya K, Ikagawa S, Nishimura Y. Allele specificity of structural requirement for peptides bound to HLA-DRB1*0405 and -DRB1*0406 complexes: Implication for the HLA-associated susceptibility to methimazole-induced insulin autoimmune syndrome. J Exp Med. 1994;180:873. doi: 10.1084/jem.180.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boitel B, Blank U, Mège D, et al. Strong similarities in antigen fine specificity among DRB1*1302-restricted tetanus toxin tt830-specific TCRs in spite of highly heterogeneous CDR3. J Immunol. 1995;154:3245. [PubMed] [Google Scholar]
- 20.Brusic V, Rudy G, Kyne AP, Harrison L. MHCPEP – a database of MHC-binding peptides: update 1995. Nucl Acids Res. 1996;24:242. doi: 10.1093/nar/24.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammer J, Valsasnini P, Tolba K, et al. Promiscuous and allele-specific anchors in HLA-DR-binding peptides. Cell. 1993;74:197. doi: 10.1016/0092-8674(93)90306-b. [DOI] [PubMed] [Google Scholar]
- 22.Falk K, Rötzschke O, Stevanovic S, Jung G, Rammensee H-G. Pool sequencing of natural HLA-DR, DQ, and DP ligands reveals detailed peptide motifs, constraints of processing, and general rules. Immunogenetics. 1994;39:230. doi: 10.1007/BF00188785. [DOI] [PubMed] [Google Scholar]
- 23.Sette A, Demars R, Grey HM, et al. Isolation and characterization of naturally processed peptides bound by class II molecules and peptides presented by normal and mutant antigen-presenting cells. Chem Immunol. 1993;57:152. [PubMed] [Google Scholar]
- 24.Rudensky AY, Preston-Hurlburt P, Hong SC, Barlow A, Janeway CA. Sequence analysis of peptides bound to MHC class II molecules. Nature. 1991;353:622. [Google Scholar]
- 25.Nelson CA, Petzold SJ, Unanue ER. Peptides determine the lifespan of MHC class II molecules in the antigen-presenting cell. Nature. 1994;371:250. doi: 10.1038/371250a0. [DOI] [PubMed] [Google Scholar]
- 26.Larsen SL, Pedersen LO, Buus S, Stryhn A. T cell response affected by aminopeptidase N (CD13) -mediated trimming of major histocompatibility complex class II-bound peptides. J Exp Med. 1996;184:183. doi: 10.1084/jem.184.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breloer M, Ehrlich S, Fleischer B, Von Bonin A. A biological function for the XP motif within the N terminus of major histocompatibility complex class II-associated peptides. Eur J Immunol. 1996;26:1825. doi: 10.1002/eji.1830260824. [DOI] [PubMed] [Google Scholar]
- 28.Wu S, Gorski J, Eckels DD, Newton-Nash DK. T cell recognition of MHC class II-associated peptides is independent of peptide affinity for MHC and sodium dodecyl sulfate stability of the peptide/MHC complex. J Immunol. 1996;156:3815. [PubMed] [Google Scholar]
- 29.Stryhn A, Pedersen LO, Romme T, Holm CB, Holm A, Buus S. Peptide binding specificity of major histocompatibility complex class I resolved into an array of apparently independent subspecificities: quantitation by peptide libraries and improved prediction of binding. Eur JImmunol. 1996;26:1911. doi: 10.1002/eji.1830260836. [DOI] [PubMed] [Google Scholar]
- 30.Bacher M, Metz C, Calandra T, et al. Macrophage migration inhibitory factor is a neuroendocrine mediator of endotoxaemia. Trends Microbiol. 1994;2:198. doi: 10.1016/0966-842x(94)90111-h. [DOI] [PubMed] [Google Scholar]
- 31.Bernhagen J, Calandra T, Cerami A, Bucala R. Macrophage migration inhibitory factor is a neuroendocrine mediator of endotoxaemia. Trends Microbiol. 1994;2:198. doi: 10.1016/0966-842x(94)90111-h. [DOI] [PubMed] [Google Scholar]
- 32.Rocklin RE, Chess L, MacDermott RP, Schlossman SF, David JR. Studies on the production of MIF and mitogenic factor using highly purified human T and B lymphocytes. Rheumatology. 1975;6:98. [PubMed] [Google Scholar]
- 33.Sugane K, Dasahara T, Shioiri-Nakano K. Release of migration inhibitory factor from mouse T and B cells activated by insoluble phytomitogens. Jpn J Exp Med. 1975;45:19. [PubMed] [Google Scholar]
- 34.Santamaria LF, Bheekha R, Van Reijsen FC, et al. Antigen focusing by specific monomeric immunoglobulin E bound to CD23 on Epstein-Barr virus-transformed B cells. Hum Immunol. 1993;37:23. doi: 10.1016/0198-8859(93)90139-r. [DOI] [PubMed] [Google Scholar]