Abstract
The expression of alternatively spliced mRNAs from genes is an ubiquitous phenomenon in metazoa. A screen for trans-acting factors that alter the expression of alternatively spliced mRNAs reveals that the smg genes of Caenorhabditis elegans participate in this process. smg genes have been proposed to function in degradation of nonsense mutant mRNAs. Here we show that smg genes affect normal gene expression by modulating the levels of alternatively spliced SRp20 and SRp30b mRNAs. These SR genes contain alternatively spliced exons that introduce upstream stop codons. The effect of smg genes on SR transcripts is specific, because the gene encoding the catalytic subunit of the cAMP-dependent protein kinase, which also contains an alternatively spliced exon that introduces upstream stop codon, is not effected in a smg background. These results suggest that the levels of alternatively spliced mRNAs may, in part, be regulated by alternative mRNA stability.
Keywords: alternative pre-mRNA splicing, alternative stop codons, RNA stability
In multicellular organisms many genes express developmentally regulated or tissue-specific alternative messenger RNAs. Through the use of different splice sites, alternative pre-mRNA splicing can dramatically alter the coding capacity of a gene (1). Many proteins have been identified as alternative splicing factors, and cis-acting elements are important for splice site selection (2–4). Though modulation of splicing is required for the synthesis of alternatively spliced mRNAs, differential stability of the resulting mRNAs could determine the level of expression of any particular alternative message.
Regulation of mRNA stability is thought to play a critical role in a variety of processes, including development (5), growth and differentiation (6), and cytoskeleton dynamics (7). Untranslated sequences as well as sequences within coding regions are necessary and/or sufficient for proper mRNA turnover (8–12). Trans-acting factors have been identified that positively or negatively regulate mRNA degradation (8, 13). However, genetic description of trans-acting factors involved in metazoan mRNA turnover is limited to the smg genes of Caenorhabditis elegans. The smg mutants (suppressor with morphogenetic effects on genitalia) exhibit allele-specific suppression of certain nonsense mutations (14). Molecular analysis suggested that some unc-54 alleles containing premature stop codons exhibit reduced mRNA expression. This reduced expression could be restored by mutations in any of the six smg genes (15). For this reason it has been proposed that the smg genes participate in the degradation of mutant mRNAs.
We present results of a genetic screen designed to identify factors involved in alternative pre-mRNA processing. Using a method analagous to yeast screens for temperature-sensitive pre-mRNA splicing mutants (16, 17), we screened mRNA from temperature-sensitive embryo and early larval lethal worm mutants by Northern blotting with alternative exon probes. Through this analysis we found that smg mutants affect the expression of alternatively spliced mRNAs. These results indicate that, in addition to their role in degradation of mutant mRNAs, smg genes may play a role in the expression of wild-type genes.
MATERIALS AND METHODS
C. elegans Culture.
Worm handling and husbandry were described previously (18). The strains used in these experiments were derived from N2 strain Bristol (18). The g43 allele of emb-14 was described previously (19). smg mutants were kindly provided by the Caenorhabditis Genetics Center (University of Minnesota, St. Paul) and the laboratory of Phil Anderson (University of Wisconsin, Madison).
RNA Isolation and Analysis.
Mixed stage worms were washed in M9 buffer and floated on 30% sucrose (20). Embryos were collected by hypochlorite treatment of gravid adults (20). RNA isolation (21), poly(A)+ mRNA enrichment (22), Northern blotting (22), and reverse transcription–PCR (RT-PCR) (23) were described previously. Signals were quantitated on a Molecular Dynamics PhosphorImager. Oligonucleotides used for SRp30b RT-PCR are 105-GTATGATCGACCATC and 110-CACGACGTACCGGTG.
Cloning of SRp20 and 30b Genes and Generation of CeCat and Actin Hybridization Probe.
cDNAs encoding C. elegans SR proteins (SRp20 and SRp30b) were identified in expression sequence tag libraries by homology with mammalian SR cDNAs (24). These cDNAs were used to isolate genomic fragments containing these genes from a genomic library (Stratagene). DNA sequences of the relevant DNAs appear in GenBank under accession nos. U23484 and U41007. Chromosome mapping of SR genes was done by Southern hybridization to filters of cloned genomic DNAs provided by Alan Coulson (Sanger Center, Cambridge, U.K.). A hybridization probe for both isoforms of CeCat was made by random-priming a fragment of a CeCat cDNA that contains the alternative exon and the two flanking common exons. The following oligonucleotides were used for this work: CR-63-GACTTGCTGAAGAATTTGCTC and CR64-GTGCCCGAGATACGAAGCGG (25).
RESULTS
To better understand the mechanisms that affect alternative mRNA expression we screened a collection of temperature-sensitive C. elegans strains for mutants with altered expression of alternatively spliced mRNAs. As test genes we chose two genes that encode SR proteins (SRp20 and SRp30b) and the gene that encodes the catalytic subunit of the cAMP-dependent protein kinase (CeCat) (25). Each is subject to alternative pre-mRNA splicing of a single exon bearing an in-frame stop codon (Fig. 1). We were able to obtain probes to C. elegans SR protein genes because the primary sequence of SR proteins is highly conserved in the animal kingdom (24). For example, human and C. elegans SRp20 are more related to each other than C. elegans SRp20 is to C. elegans SRp30b. cDNAs encoding C. elegans SRp20 and SRp30b were identified in expression sequence tag databases (26, 27). The cDNAs were used to isolate genomic clones and to determine that SRp20 is located on chromosome IV and SRp30b is located on chromosome II.
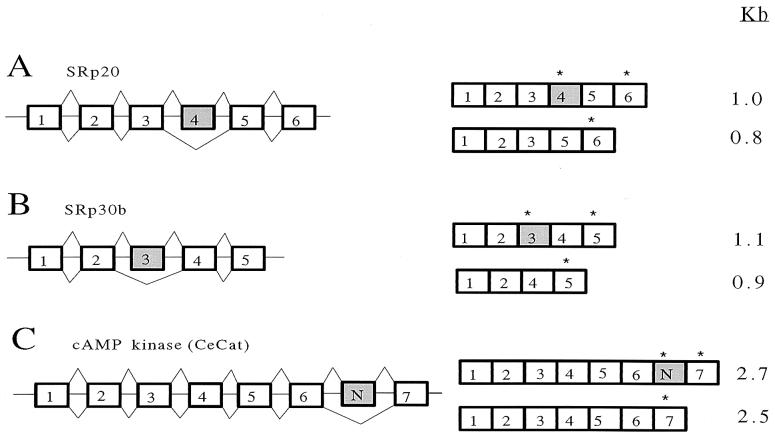
Figure 1.
SR pre-mRNAs are alternatively spliced. Schematic representation of the genomic organization, alternative splicing, and message expression from (A) SRp20, (B) SRp30b, and (C) catalytic subunit of the cAMP-dependent protein kinase (CeCat) genes. (Left) Gene structures. (Right) mRNAs. Exons are boxes and introns are lines. The sizes of the exons and introns are not drawn to scale. ∗ indicate positions of alternative stop codons. The RRM and SR domain of SRp20 are encoded by exons 1–3 and 5–6, respectively. The RRM and SR domain of SRp30b are encoded by exons 1–2 and 3–4, respectively.
SR proteins are essential pre-mRNA splicing factors that contain amino terminal RNA recognition motifs (RRM) and a carboxy terminal domain composed of alternating serine and arginine (SR). In mammals, SRp30a pre-mRNA is alternatively spliced, and alternative stop codons are used to encode proteins containing either the RRM domain alone or the RRM and an SR domain (28). These proteins are expressed in vivo and have distinct functions (J. L. Manley, personal communication; ref. 29). The organization of exons and isolation of cDNAs suggest that pre-mRNAs from C. elegans SRp20 and SRp30b genes are alternatively spliced to encode similar protein isoforms (Fig. 1).
In wild-type animals the exon-skipped mRNAs for SRp20 and SRp30b are more abundant than the alternative exon-containing mRNAs. However, mutant analysis revealed that the levels of these alternatively spliced mRNAs may be subject to regulation. Northern blots probed with cDNAs containing the alternative exon shows that one of the temperature-sensitive mutants we screened, emb-14 (g43) (19), contains much higher levels of the exon-included SR mRNAs than wild-type animals (Fig. 2 A and B, lanes 1 and 2). This result also is supported by RT-PCR of mRNA from g43 and wild-type animals using oligonucleotides that allow for detection of both SRp30b messages (Fig. 3, lanes 1 and 2). The sizes of the resulting RT-PCR products are consistent with the DNA sequence of SRp30b alternative cDNAs. The results indicate that the exon-included SRp30b mRNA represents approximately 0.6% of the total SRp30b mRNA in wild-type and 15% in g43 animals.
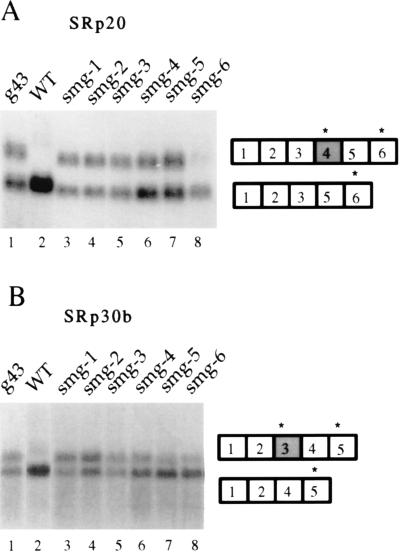
Figure 2.
Expression of SRp20 and SRp30b mRNAs in g43, wild-type, and smg mutant embryos. Total RNA was probed with cDNAs from the (A) SRp20 and (B) SRp30b genes. Lane 1: emb-14 (g43). Lane 2: Wild type (N2). Lane 3: smg-1 (r861). Lane 4: smg-2 (e2008). Lane 5: smg-3 (ma117). Lane 6: smg-4 (ma116). Lane 7: smg-5 (r860). Lane 8: smg-6 (r896). (Left) Northern blots. (Right) mRNAs.
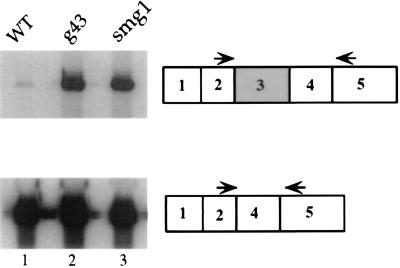
Figure 3.
RT-PCR detection of alternatively spliced SRp30b mRNAs in wild-type and mutant animals. Regions of SRp30b were amplified by PCR after reverse transcription of total mRNA from N2 (lane 1), g43 (lane 2), and smg-1 (r861) (lane 3), using the primers designated by arrows.
Several lines of evidence suggest that g43 contains an allele of a smg gene. First, g43 worms have protruding vulvae and abnormal male copulatory structures, phenotypes exhibited by all smg mutants (14). Second, g43 suppresses the paralyzed phenotype of unc-54 (r293) (A. K. Hopper, personal communication), another phenotype common to all smg mutants (14). Third, g43 maps to chromosome I near the smg-1 and smg-5 genes (19, 14). The fact that alleles of smg genes all have very similar phenotypes led us to test whether the molecular SR phenotype was a general smg phenomenon. Northern blot analysis indicates that representative alleles of six smg mutants express higher levels of the alternatively spliced messages to the same extent as g43 (Fig. 2 A and B). The smg-6 allele shows a much weaker effect, which is consistent with its weaker effect in other smg assays (15). RT-PCR of mRNA isolated from smg-1 mutant animals using oligonucleotides that detect both SRp30b mRNAs also supports the conclusion that g43 contains an allele of a smg gene (Fig. 3, lane 3).
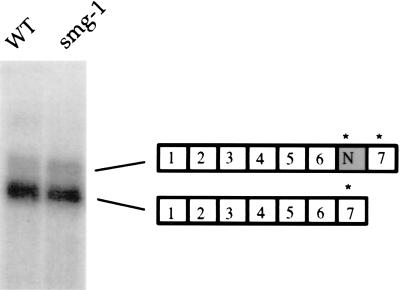
To determine whether the presence of an alternative stop codon containing exon in an mRNA always reveals a smg-dependent effect we assayed the expression of the catalytic subunit of the cAMP-dependent protein kinase (CeCat). Like SR genes the CeCat gene expresses two messages, one of which contains an alternative exon that introduces an alternative stop codon (25). Northern blot analysis shows that the relative ratio of the two alternative CeCat mRNAs does not vary between wild-type and a smg-1 mutant (Fig. 4). Indistinguishable results were obtained for g43 (data not shown). The failure of smg to affect the level of these alternate mRNAs indicates that smg acts on a specific subset of mRNAs.
Figure 4.
cAMP-dependent protein kinase catalytic subunit (CeCat) expression is the same in wild-type and smg-1 animals. Poly(A)+ RNA was probed with a fragment of the CeCat gene. (Right) The alternatively spliced mRNAs. ∗ indicate alternative stop codons.
DISCUSSION
We report that mutations in smg genes have dramatic and specific effects on the level of naturally occurring alternatively spliced mRNAs from the SRp20 and SRp30b genes. Most prominent is the increase in abundance of the SR mRNAs that contain alternative exons in smg mutants. Though we find a slight decrease in the level of the exon-skipped SRp30b and SR20 alternative mRNA in smg mutants, the absolute amount of mRNA remains approximately constant (less than a 2-fold change). These findings indicate that in wild-type nematodes smg activity may be involved in regulating the levels of the exon-included SRp20 and SRp30b mRNAs.
Previously smg genes were shown to have effects only on mutant genes. The finding that smg can effect the regulation of expression of wild-type genes raises the possibility that smg gene products play a role in processes in which RNA turnover is likely to be important, such as cell cycle progression and degradation of maternal mRNAs during early embryonic development. Because smg mutants affect the levels of SR protein mRNAs and because SR proteins participate widely in regulating alternative pre-mRNA splicing (30, 31), smg activity could have additional indirect effects on gene expression.
The alternative exons in SRp20, SRp30b, and CeCat all introduce upstream stop codons. Because this is a naturally occurring event we refer to it as an alternative stop codon and not a premature stop codon, a term used to describe nonsense mutations. The levels of alternative stop codon-containing mRNAs are known to be independently regulated and important for such processes as cell adhesion (32) and erythropoeisis (33). That smg genes affect the levels of exon-included alternative stop codon-containing mRNAs from SR genes, but not from CeCat, suggests either that some mRNAs are subject to regulation at the level of stability and others are not or that additional smg-independent degradation mechanisms exist to regulate the levels of alternative stop codon-containing mRNAs.
Though smg gene products may effect splicing, previous work on unc-54 (15) favors the possibility that they effect turnover. We propose that significant amounts of both alternatively spliced SRp20 and SRp30b mRNAs are produced and smg activity destabilizes the exon-included mRNA. This would account for the low level of the exon-included message in wild-type animals and its increase in smg mutants. This work raises the possibility that independent regulation of the stability of alternatively spliced mRNAs may contribute to alternative pre-mRNA processing.
Acknowledgments
We thank Phil Anderson for strains. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center (University of Minnesota, St. Paul), which is funded by the National Institutes of Health National Center for Research Resources. We thank Alan Coulson for grids of genomic DNA clones. We thank Jim Manley for communicating unpublished results. M.M. is supported by Training Grant 2T32HD07183–16 from the National Institutes of Health. K.S.H. is supported by Grant GM1659–03 from the National Institutes of Health. This work was funded by National Institutes of Health Grant GM48435–01A2 to M.B.R.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: RT-PCR, reverse transcription–PCR; RRM, RNA recognition motifs.
References
- 1.Breitbart R E, Andreadis A, Nadal-Ginard B. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- 2.Baker B S. Nature (London) 1989;340:521–524. doi: 10.1038/340521a0. [DOI] [PubMed] [Google Scholar]
- 3.Maniatis T. Science. 1991;251:33–34. doi: 10.1126/science.1824726. [DOI] [PubMed] [Google Scholar]
- 4.McKeown M. Annu Rev Cell Biol. 1992;8:133–155. doi: 10.1146/annurev.cb.08.110192.001025. [DOI] [PubMed] [Google Scholar]
- 5.Surdej P, Riedl A, Jacobs-Lorena M. Annu Rev Genet. 1993;28:263–282. doi: 10.1146/annurev.ge.28.120194.001403. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg M E, Belasco J G. In: Control of Messenger RNA Stability. Belasco J, Brawerman G, editors. London: Academic; 1993. pp. 199–218. [Google Scholar]
- 7.Theodorakis N G, Cleveland D W. In: Control of Messenger RNA Stability. Belasco J, Brawerman G, editors. London: Academic; 1993. pp. 219–238. [Google Scholar]
- 8.Brown B D, Zipkin I D, Harland R M. Genes Dev. 1993;7:1620–1631. doi: 10.1101/gad.7.8.1620. [DOI] [PubMed] [Google Scholar]
- 9.Muhlrad D, Parker R. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 10.Treisman R. Cell. 1985;42:889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- 11.Wisdom R, Lee W. Genes Dev. 1991;5:232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, Ruiz-Echevarria M J, Quan Y, Peltz S W. Mol Cell Biol. 1995;15:2231–2244. doi: 10.1128/mcb.15.4.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leeds P, Wood J M, Lee B, Culbertson M R. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pulak R, Anderson P. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 16.Andrews C, Hopper A K, Hall B D. Mol Gen Genet. 1976;144:29–37. doi: 10.1007/BF00277300. [DOI] [PubMed] [Google Scholar]
- 17.Vijayraghavan U, Company M, Abelson J. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 18.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cassada R, Isnenghi E, Culotti M, von Ehrensrein G. Dev Biol. 1981;84:193–205. doi: 10.1016/0012-1606(81)90383-3. [DOI] [PubMed] [Google Scholar]
- 20.Sulston J, Hodgkin J. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 587–606. [Google Scholar]
- 21.Xie W Q, Rothblum L I. Biotechniques. 1991;11:325–327. [PubMed] [Google Scholar]
- 22.Ausubel F M, Brent R, Kingston R E, Moore D D, Seideman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. I. New York: Wiley; 1992. pp. 4.5.1–4.5.5. and 4.9.1–4.9.4. [Google Scholar]
- 23.Rupp R A, Weintraub H. Cell. 1991;38:317–331. doi: 10.1016/0092-8674(91)90545-a. [DOI] [PubMed] [Google Scholar]
- 24.Zahler A M, Lane W S, Stolk J A, Roth M B. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 25.Gross R E, Bagchi S, Lu X, Rubin C S. J Biol Chem. 1990;265:6896–6907. [PubMed] [Google Scholar]
- 26.McCombie W R, Andrews M D, Kelley J M, Fitzgerald M G, Utterback T R, Kahn M, Dubnick M, Kerlavage A R, Venter J, Fields C. Nat Genet. 1992;1:124–131. doi: 10.1038/ng0592-124. [DOI] [PubMed] [Google Scholar]
- 27.Waterston R, Martin C, Craxton M, Huynh C, Coulson A, Hillier L, Durbin R, Green P, Shownkeen R, Halloran N, Metzstein M, Hawkins T, Wilson R, Berks M, Du Z, Thomas K, Thierry-Mieg J, Sulston J. Nat Genet. 1992;1:114–123. doi: 10.1038/ng0592-114. [DOI] [PubMed] [Google Scholar]
- 28.Ge H, Zuo P, Manley J L. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- 29.Zuo P, Manley J L. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng X, Mount S M. Mol Cell Biol. 1995;15:6273–6282. doi: 10.1128/mcb.15.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 32.Gower H J, Barton H, Elsom V L, Thompson J, Moore S E, Dickson G, Walsh F S. Cell. 1988;55:955–964. doi: 10.1016/0092-8674(88)90241-3. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura Y, Komatsu N, Nakauchi H. Science. 1992;257:1138–1141. doi: 10.1126/science.257.5073.1138. [DOI] [PubMed] [Google Scholar]