Abstract
Immunoglobulin (IgM) is found in various states of covalent polymerization (μL)n, where n is typically 8, 10, or 12. The usual form of IgM of bony fish is tetrameric (8 μL units) as compared to the pentameric form (10 μL units) observed in cartilaginous fish and mammals. Two hypotheses were tested in this study. First, that the length of the μ-chain C terminus following Cys575 determines whether an IgM polymerizes as a tetramer or as a pentamer. This was tested by examining the covalent polymerization state of mouse IgM mutated to contain a series of μ-chain C-termini from bony and cartilaginous fish. The results proved this hypothesis wrong: mouse IgM bearing the C-terminal sequence of shark, salmon and cod μ-chain behaved identically to native mouse IgM, forming predominantly (μL)10 and (μL)12 forms. The second hypothesis was that an additional Cys residue near the C terminus of the μ-chain is responsible for the multiple covalent structures seen in IgM of the channel catfish. The addition of a catfish C terminus to the mouse μ-chain resulted, as predicted, in the production of a series of covalently bonded forms, with the major species being (μL)4. When a Ser-Cys unit was removed from the catfish C terminus added to the mouse μ-chain, this resulted in production of IgM indistinguishable in structure from that of wild-type mouse IgM.
INTRODUCTION
The assembly of polymeric immunoglobulin M (IgM) is a complex process that, despite substantial study, remains incompletely understood.1–10 The formation of stable polymeric mammalian IgM is considered11 to proceed in stages: the association of heavy and light chains to form μL ‘halfmers’ is followed by the subsequent assembly of halfmers into monomers (μL)2 and so-on up to the full-size pentameric (μL)10 or hexameric (μL)12 IgM molecule. However, other modes of assembly are possible, such as formation of an H-chain dimer, followed by the association of two L-chains to give (μL)2.11 The intramolecular heterogeneity of antigen-binding sites in IgM (wherein half the antigen-binding sites of certain IgM molecules are of high affinity and half are of low affinity3) has been suggested to arise as a result of the utilization of alternative pathways of IgM assembly.3 Whatever the pathway of assembly of IgM, the formation of disulphide bonds between polypeptides should be preceded by non-covalent interactions that bring two cysteines into the correct orientation for disulphide-bond formation. The non-covalent interactions in mammalian IgM are weak,12 and mutation (to Ser) of the three Cys residues involved in μ–μ disulphide bonding results in the exclusive production of μL halfmers.9 Identical results were seen when these same Cys residues were reduced and alkylated.9 Of the three Cys residues involved in μ–μ disulphide bond formation, the subterminal one (Cys575; position 627 in the numbering system of Kabat et al.13) appears to be the most important, in that it is both necessary and sufficient for the formation of IgM polymers, although in some experiments significant proportions of hexameric IgM (μL)12, were found. In contrast Cys337 and Cys414 were neither necessary nor sufficient for the formation of IgM polymers.9 J-chain binds to Cys575 and participates in the polymerization of mammalian IgM, although evidence suggests that it may act primarily to direct the assembly of IgM as the (μL)10 pentamer.
The IgM of bony fish (teleosts) is distinctive in several respects. First, its major polymeric form is the (μL)8 tetramer. Second, significant amounts of monomeric (μL)2 IgM can be found normally in the blood of some species. Third, the polymeric IgM of many bony fishes is held together to some degree by non-covalent interactions: upon exposure to denaturation under non-reducing conditions, the polymeric IgM dissociates into a variety of smaller forms.18–20 Particularly interesting in this respect is the IgM of the channel catfish, Ictalurus punctatus. The serum IgM of this species is a typical (μL)8 tetrameric form, but upon exposure to sodium dodecyl sulphate (SDS) it dissociates into a series of subunits containing from one to eight halfmers, i.e. of structure (μL)1–8. The presence of two Cys residues near the C terminus of the catfish μ-chain is unusual, and might explain this phenomenon if intrasubunit, rather than intersubunit, disulphide bonds were to be formed by these Cys residues.22 The competition for formation of intra- and inter-μ-chain disulphide bonds between Cys627 and Cys629 results in each IgM polymer being held together by a mixture of covalent and non-covalent interactions.22
The reason that the IgM of bony fish is tetrameric (μL)8, rather than pentameric (μL)10, is unknown. One suggestion is that it is caused by the structure of the C-terminal region of the μ-chains. Specifically, the presence of 2–5 residues C-terminal to Cys575 has been hypothesized to induce packing problems in the centre of the molecule that result in the adoption of a tetrameric structure.16
The study reported here was undertaken to test two hypotheses. First, that extending the C terminus of the mouse μ-chain beyond Cys575 using the C termini of teleost μ-chains, would induce mouse IgM to assemble as a (μL)8 tetramer. Second, that the additional Cys residue in the C terminus of the catfish μ-chain is responsible for the unusual covalent structure of the IgM in this species.
MATERIALS AND METHODS
Constructs
The parent plasmids used for mutagenesis were pR-Sp6 and pR-Sp6(S414), kindly provided by Dr Marc Shulman (University of Toronto, Canada). These plasmids7 express neomycin resistance and contain the mouse μ gene from Sp6/HL which secretes a dinitrophenyl (DNP)/trinitrophenyl (TNP)-specific μ/κ IgM. pR-Sp6 contains the wild-type μ sequence, whereas pR-Sp6(S414) is a mutant in which Cys414 has been converted to Ser.7 Mutagenesis was carried out by polymerase chain reaction (PCR; Excite kit, Stratagene, La Jolla, CA) on the Bam HI/KpnI fragment containing all of exons 3 and 4 and most of exon 2, cloned in pBluescript (pBS; Stratagene), as illustrated in Fig. 1. In each PCR reaction, primer 1175 (Table 1) was paired with one of the species-specific mutagenic primers (1174, 1197–1199, 1201). Synthetic oligonucleotides used for mutagenesis (Table 1) were synthesized in the Medical University of South Carolina Nucleic Acid Synthesis Facility. Each PCR product was gel-purified, treated with DpnI, cut with EcoRI, ligated and transformed into Escherichia coli (XL1-Blue MRF’, Stratagene). The sequences of the mutant clones were checked for accurate introduction of the new sequence, and for the absence of additional unwanted mutations. The BstEII/Kpn I fragment containing the mutated segment of exon 4 was then cloned back into the parental pR-Sp6 and pR-Sp6(S414) plasmids by the scheme shown in Fig. 1. Table 2 shows the amino acid sequences introduced into the C terminus of the mouse μ-chain by the mutagenesis procedure. The sequence of μ-chains of cod (Gadus morhua), salmon (Salmo salar), channel catfish (Ictalurus punctatus) and horned shark (Heterodontus francisci ) were taken from database entries with the following accession numbers, respectively; X58870, S48652, M27230 and S01853.
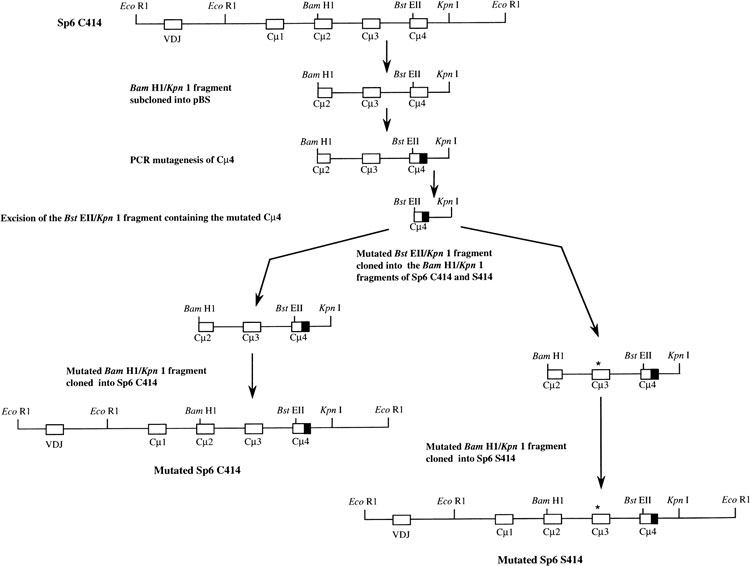
Figure 1.
Introduction of mutations into the 3′ coding region of the mouse μ gene. The Bam HI/KpnI fragment of the μ gene was the target for the introduction of mutations (shaded) into the Cμ4 exon. Following the confirmation of sequence the Bst EII/Kpn I fragment was cloned back into the parental Sp6(C414) and Sp6(S414) genes as shown. The asterisk (*) indicates Ser 414.
Table 1.
Table Primers for PCR mutagenesis of the mouse μ gene

The mouse sequence is shown in italics, the introduced (mutant) sequence is shown underlined, and the EcoRI site is shown in bold.
Table 2.
Table Sequences of the C-terminal region of the mutated μ-chains
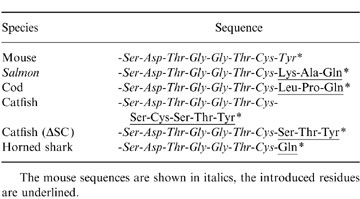
The mouse sequences are shown in italics, the introduced residues are underlined.
Cells and tissue culture
The X-10 cell line,9 a derivative of Sp6/HL which secretes only light chain (a kind gift of Dr Marc Shulman, University of Toronto) and the Sp6 parental cell line (a gift of Dr Jan Andersson, Uppsala University, Sweden) were grown in Dulbecco’s modified Eagle’s minimal essential medium (F-DMEM) supplemented with 15% fetal calf serum (Gibco-BRL, Grand Island, NY), gentamycin (50 μg/ml) and 50 μm 2-mercaptoethanol (2-ME) at 37°, in 5% CO2–95% air. For electroporation, X-10 cells were washed once in serum-free F-DMEM and cells (2 × 106 or 4 × 106) were suspended in 0·4 ml of the same medium and electroporated with 10 μg or 20 μg (respectively) of KpnI-linearized plasmid. Electroporation was carried out using the Gene Pulser™ (Bio-Rad Laboratories Richmond CA), using 2-mm gap cuvettes, a capacitance of 960 μF and 180 V, which gave a time constant of 6·5 ms. After electroporation the cells were distributed into two 96-well tissue-culture plates in F-DMEM/15% serum, and at 24 hr selection was initiated with G418 (Gibco) at an effective concentration of 0·6 mg/ml. Wells containing G418-resistant cells were tested for the production of DNP-specific IgM by antigen-capture enzyme-linked immunosorbent assay (ELISA). Limiting dilution was used to derive clones from each transfection. G-418-resistant clones were established for each construct except the Cys414 μ-chain with the horned shark C terminus.
ELISA
To test IgM production a capture assay was used in which the first antibody (bound to the 96-well plate) was goat IgG anti-mouse μ-chain (Sigma BioSciences, St. Louis, MO), and the second antibody was alkaline phosphatase-conjugated goat anti-mouse μ-chain (Sigma). The substrate was 100 μl/ well of p-nitrophenylphosphate (1 mg/ml in 0·85 m diethanolamine/HCl, pH 9·8, 0·5 m MgCl2) and the assay was read, at 30 min, at 414 nm on the Multiskan® MCC/340 (Labsystems, Sweden). To determine DNP-specific antibody, the same antigen capture assay was used, but with bovine serum albumin–DNP (BSA–DNP)23 bound to the 96-well plate. For quantitative analysis of IgM, 106 cells were pelleted, washed once in fresh 2-ME-free F-DMEM with 15% serum, resuspended in 1 ml of the same medium and incubated for 12 hr at 37° in 5% CO2–95% air. Mouse IgMκ (Sigma) was used as the standard for calibration of this assay.
35S-methionine labelling, immunoselection and gel analysis
Cells were washed once in serum-free, 2-ME-free and methionine-free F-DMEM (SVA, Uppsala, Sweden), and resuspended in this medium at 106 cells/ml. The cells were incubated in a 24-well plate, at 1 ml/well, with the addition of 50 μCi 35S-labelled methionine (>1000 Ci/mm, Amersham Sweden, Stockholm) per well, and incubated at 37° in 5% CO2–95% air, for 12 hr. After incubation the cells were pelleted (13 000 g for 5 min at room temperature) and the supernatant was collected. IgM was affinity purified by incubating the supernatant with 50 μl of DNP–BSA-coupled to CNBr-activated Sepharose (Pharmacia Biotech, Uppsala, Sweden) overnight at 4° with mixing. The Sepharose was then washed three times with 0·15 m NaCl, 0·05 m Tris–HCl, pH 7·5, 0·5% Tween-20, and bound proteins were eluted with SDS–polyacrylamide gel elctrophoresis (PAGE) sample buffer for 20 min. SDS–PAGE was performed in slab gels using the system described by Laemmli,24 with a 3% stacking gel and a 3·5% resolving gel for unreduced samples, and a 5% stacking gel, 10% resolving gel for reduced samples. Labelled proteins were visualized and quantified after drying of the gel by use of a GS-250 Molecular Imager (BioRad). Each non-reduced gel contained a lane in which, as a standard, the 35S-labelled IgM antibody secreted by Sp6 cells (a mixture of pentameric (μL)10 and hexameric (μL)12 molecules7) was run. Unlabelled Sp6 antibody, purified by affinity chromatography on DNP–BSA Sepharose, was also used as a standard. Mouse Sp6 IgM and mouse TEPC 183 IgM [Sigma, which yields predominantly (μL)10 with a minor (μL)12 component], and catfish IgM, which yields a ladder of eight bands, of structure (μL)1–8, were run simultaneously with the 35S-labelled samples, but on a parallel gel, and silver stained.
RESULTS
Secretion of IgM
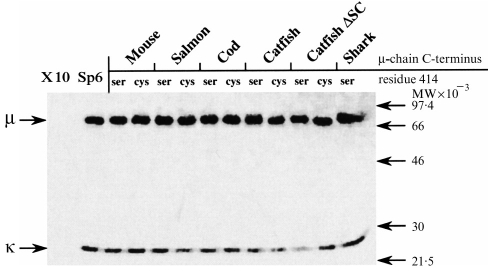
Stably transfected cloned lines were established for each of the constructs (except Cys414, horned shark C terminus), and the concentration of IgM in the medium following a 12-hr incubation was determined by ELISA (Table 3). The IgM produced by these cell lines and bound to DNP-Sepharose contained, as anticipated, μ and L chains when analysed under reducing conditions (Fig. 2). Some minor variations were observed in the mobilities of the μ-chains secreted by the stably transfected cells, and were attributed to effects of the introduced terminal residues.
Table 3.
Table Secretion of IgM by stably transfected cell lines
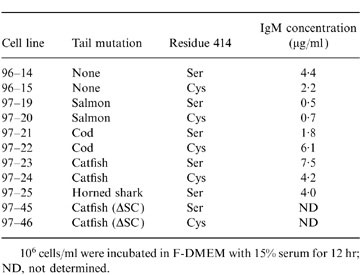
106 cells/ml were incubated in F-DMEM with 15% serum for 12 hr; ND, not determined.
Figure 2.
Analysis, by SDS–polyacrylamide slab gel electrophoresis under reducing conditions, of 35S-labelled DNP-binding antibody produced by transfected and untransfected cell lines. X-10 is the parental cell line producing κ chains only, and Sp6 is the parental cell line producing both κ-and wild-type mouse μ-chains. Other lanes show the analysis of mutant mouse antibody produced by transfection of X-10 cells with a mouse μ gene in which the C-terminal sequence has been altered. The name of the species from which the introduced μ-chain C-terminal sequence was taken (Table 2) is indicated above the lanes. The presence of Cys or Ser at position 414 of the μ-chain is also indicated above each lane. The positions of standards (phosphorylase b, serum albumin, ovalbumin, carbonic anhydrase and trypsin inhibitor) are shown by arrows.
Covalent structure of the secreted IgM
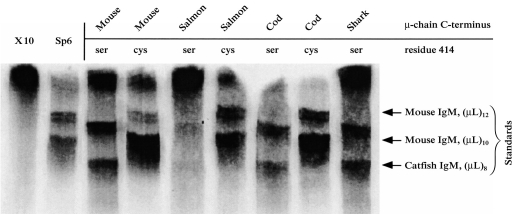
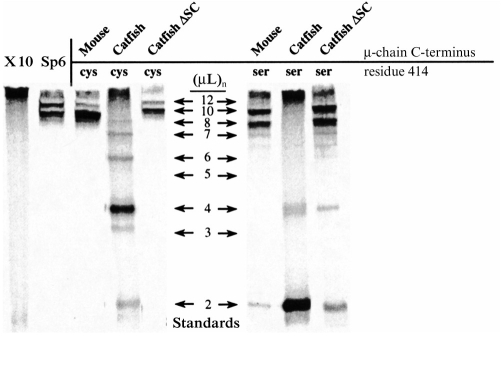
The nature of the covalent bonding in the IgM secreted by each of the cell lines was determined by SDS–PAGE analysis (under non-reducing conditions) of the 35S-labelled DNP-binding antibodies (Fig. 3). The negative control cell line X-10 (which secretes only κ light chain) did not yield any bands that were resolvable as IgM, although radioactivity remained at the top of the gel. Such material was seen in other samples (Fig. 3), but did not appear in every case. Although the nature of this material is unknown, it did not react with antibody to mouse IgM in Western blot (data not shown). The control cell line Sp6 secreted antibody which was resolved into two major bands. The antibody secreted by Sp6 consists of a slower-migrating hexameric (μL)12 IgM and a faster-migrating, broader band of pentameric (μL)10 IgM. The heterogeneity in the IgM as revealed in this electrophoretic analysis (see also refs 7,9) may reflect different patterns of intramolecular disulphide bonding, but other explanations, such as heterogeneity of glycosylation, cannot be ruled out. The X-10 cells transfected with the wild-type mouse μ gene produced IgM with an essentially identical polymerization pattern to that seen with Sp6; i.e. predominantly (μL)10 pentamer, but with some hexameric (μL)12 IgM. The cells transfected with the Ser414 mouse μ gene secreted IgM that resolved into two major bands (Fig. 3). The mobility of these bands, in comparison to the standards, was interpreted as indicating the secretion of pentameric (μL)10 and tetrameric (μL)8 IgM. This interpretation is consistent with prior analyses of the IgM expressed from this Ser414 mouse μ construct, which (by ultracentrifugal and electron microscopic analysis7) showed it to consist of pentamers and tetramers. The structure of the IgM produced by cells transfected with the genes encoding mouse μ-chains with C-terminal sequences of salmon and cod μ-chains was indistinguishable from that produced in the case of the wild-type mouse μ sequence (Fig. 3). Similarly, the IgM produced by cells transfected with the Ser414 mouse μ genes with salmon, horned shark and cod C-terminal sequences was also indistinguishable from the IgM expressed from the parental mouse Ser414 μ gene (Fig. 3). The cells transfected with the Ser414 construct bearing the salmon C-terminal sequence expressed IgM only weakly (Fig. 3) for reasons that are not understood. In contrast to the results seen with the mouse μ genes with C-terminal sequences of cod, salmon and shark μ-chains, the IgM expressed by cells transfected with a Cys414 mouse μ gene bearing the C-terminal sequence of catfish μ-chains showed an unusual covalent structure (Fig. 4). The predominant form was the dimer (μL)4 although other polymerized forms were visible. The mobilities suggest that they contained two, four, six, seven and eight covalently bonded μL pairs. When the Ser414 μ gene with the catfish μ-chain C terminus was expressed in X-10 cells, the predominant covalent form was the monomer (μL)2. Small amounts of dimer (μL)4 were also visible, but there were no detectable polymers larger than dimers (Fig. 4). The deletion of Ser-Cys from the constructs with the catfish C-terminal sequence (the ‘catfish ΔSC’ constructs) had a marked effect: it resulted in the expression of IgM essentially indistinguishable from that seen with the wild-type mouse μ gene (cf. Figs 3 and 4). This was the case with both the Cys414 and Ser414 constructs, although small amounts of monomer (μL)2 and dimer (μL)4 were seen with the S414 gene, in addition to the substantial (μL)10 pentamer and (μL)8 tetramer bands.
Figure 3.
Analysis, by SDS–PAGE under non-reducing conditions, of 35S-labelled DNP-binding antibody produced by transfected and untransfected cell lines. X-10 is the parental cell line producing κ-chains only, and Sp6 is the parental cell line producing both κ- and wild-type mouse μ-chains. Other lanes show the analysis of mutant mouse antibody, produced by transfection of X-10 cells with a mouse μ gene in which the C-terminal sequence has been altered. The name of the species from which the introduced μ-chain C-terminal sequence was taken (Table 2) is indicated above each lanes. The presence of Cys or Ser at position 414 of the μ-chain is also indicated above each lane. The mobility of the IgM standards [mouse IgM (TEPC 183 and Sp6) and catfish IgM] with eight, 10, or 12 ‘halfmer’ (μL) units is indicated by arrows.
Figure 4.
Analysis, by SDS–PAGE under non-reducing conditions, of 35S-labelled DNP-binding antibody produced by transfected and untransfected cell lines. X-10 is the parental cell line producing μ-chains only, and Sp6 is the parental cell line producing both κ- and wild-type mouse μ-chains. Other lanes show the analysis of mutant mouse antibody, produced by transfection of X-10 cells with a mouse μ gene in which the C-terminal sequence has been altered. The name of the species from which the introduced μ-chain C-terminal sequence was taken (Table 2) is indicated above the lanes. The mobilities of the IgM proteins used as standards are indicated by arrows and labelled according to the number of (μL) units they contain. The standards were mouse IgM [TEPC 183 and Sp6 with 10 and 12 (μL) subunits] and catfish IgM with two to eight (μL) subunits.
DISCUSSION
This study examined the contribution of the C-terminal sequence of the μ-chain to the covalent structure of IgM in two contexts. The first was the hypothesis16 that the C-terminal extension seen with μ-chains of teleost fish is responsible (by interfering with packing in the centre of the IgM molecule) for the tetrameric structure of their IgM. The second was the hypothesis22 that the additional Cys residue in the C terminus of the catfish μ-chain is responsible for the unusual covalent structure of this species’ IgM. The present results clearly show that the first hypothesis is wrong. Adding the C-terminal sequence of cod, salmon or catfish μ-chains to the mouse μ-chain did not induce a teleost-like (μL)8, tetrameric structure in the resulting IgM. In fact, mouse IgM with the C-terminal μ sequence from salmon and cod was indistinguishable in structure from that obtained with the wild-type sequence. Thus, alternative explanations for the tetrameric structure of teleost IgM, involving perhaps the Pro residues near the C terminus, must now be considered. Previous studies utilizing the tailpieces of mammalian μ- and α chains have also shown that the final polymerized structures of multimeric immunoglobulins depend on factors other than the tailpiece alone. Interestingly, it was only with the C-terminal sequence from catfish that the covalent structure of the resulting IgM differed from that seen with the mouse μ-chain. In this case, a variety of structures were seen and interpreted as containing from two to eight μL units. It is not clear if single μL units were also present, as these would not have been resolved on the SDS–PAGE gels that were used. However, these results showed that introduction of five C-terminal amino acids of the catfish μ-chain into the mouse sequence resulted in the production of an IgM whose covalent structure was very similar to that of the native catfish IgM.22 This result permitted the testing of the hypothesis that the additional Cys residue in the C terminus of the catfish μ-chain is responsible for this unusual covalent structure.22 The wild-type sequence of the catfish μ-chain C terminus is Cys-Ser-Cys-Ser-Thr-Tyr. From this, Ser-Cys was deleted, to give the sequence Cys-Ser-Thr-Tyr. The structure of the IgM bearing this C-terminal sequence was indistinguishable from that formed with the wild-type mouse μ. It seems most reasonable that the effects, on IgM polymerization, of removing a Ser-Cys unit from the catfish μ-chain C terminus can be attributed to the Cys, rather than to the Ser residue. Thus, the results presented here strongly support the suggestion22 that the second Cys residue in the C terminus of the μ-chain is responsible for the unusual covalent structure of catfish IgM.
The results of the present study also shed some light on the role of Cys414 in the polymerization of IgM. The following conclusions can be drawn in comparing the covalent IgM structures obtained with the full catfish C-terminal sequence in either the Cys414 or Ser414 constructs (Fig. 4). First, the major covalent form seen with catfish wild-type/Cys414 is dimer, 4 μL units, whereas with catfish wild-type/Ser414 the major form is monomer (2 μL units). Second, with catfish wild-type/Cys414, larger covalent forms containing six, seven and eight μL units are observed, whereas with the catfish wild-type/Ser414 construct, significant amounts of polymers larger than dimers (μL)4, are not seen. Thus, the role of Cys414 in the polymerization of IgM varies,9 depending upon the context in which it is assessed.
Acknowledgments
We thank Dr Marc Shulman for gifts of cells and plasmids, and for helpful advice, and Professor Jan Andersson for the gift of cells. This work was supported by grants from the Swedish Council for Forestry and Agricultural Research (No. 40.0484/96), and the National Science Foundation (MCB-9406249). G. Warr was in receipt of a Visiting Research Fellowship from Uppsala University and A. Getahun was a student in the ERASMUS exchange programme.
REFERENCES
- 1.Brewer JW, Randall TD, Parkhouse RME, Corley RB. Mechanism and subcellular localization of secretory IgM polymer assembly. J Biol Chem. 1994;269:17338. [PubMed] [Google Scholar]
- 2.Bornemann KD, Brewer JW, Beck-Engeser GB, Corley RB, Haas IG, Jäck H-M. Roles of heavy and light chains in IgM polymerization. Proc Natl Acad Sci USA. 1995;92:4912. doi: 10.1073/pnas.92.11.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pascual D, Clem LW. Ligand binding by murine IgM antibodies: Intramolecular heterogeneity exists in certain, but not all, cases. Mol Immunol. 1988;25:87. doi: 10.1016/0161-5890(88)90094-6. [DOI] [PubMed] [Google Scholar]
- 4.Niles MJ, Matsuuchi L, Koshland ME. Polymer IgM assembly and secretion in lymphoid and nonlymphoid cell lines: Evidence that J chain is required for pentamer IgM synthesis. Proc Natl Acad Sci USA. 1995;92:2884. doi: 10.1073/pnas.92.7.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis AC, Roux KH, Shulman MJ. On the structure of polymeric IgM. Eur J Immunol. 1988;18:1001. doi: 10.1002/eji.1830180705. [DOI] [PubMed] [Google Scholar]
- 6.Davis AC, Shulman MJ. IgM – Molecular requirements for its assembly and function. Immunology Today. 1989;10:118. doi: 10.1016/0167-5699(89)90244-2. [DOI] [PubMed] [Google Scholar]
- 7.Davis AC, Roux KH, Pursey J, Shulman MJ. Intermolecular disulfide bonding in IgM: effects of replacing cysteine residues in the μ heavy chain. EMBO J. 1989;8:2519. doi: 10.1002/j.1460-2075.1989.tb08389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis AC, Collins C, Yosimura MI, D’agastaro G, Shulman MJ. Mutations of mouse μ H chain which prevent polymer assembly. J Immunol. 1989;143:1352. [PubMed] [Google Scholar]
- 9.Wiersma EJ, Shulman MJ. Assembly of IgM. Role of disulfide bonding and noncovalent interactions. J Immunol. 1995;154:5265. [PubMed] [Google Scholar]
- 10.Wiersma EJ, Chen F, Bazin R, et al. Analysis of IgM structures involved in J chain incorporation. J Immunol. 1997;158:1719. [PubMed] [Google Scholar]
- 11.Baumal R, Scharff MD. Synthesis, assembly and secretion of mouse immunoglobulin. Transplant Rev. 1973;14:163. doi: 10.1111/j.1600-065x.1973.tb00106.x. [DOI] [PubMed] [Google Scholar]
- 12.Solheim BG, Harboe M. Reversible dissociation of reduced and alkylated IgM subunits to half subunits. Immunochemistry. 1972;9:623. doi: 10.1016/0019-2791(72)90248-0. [DOI] [PubMed] [Google Scholar]
- 13.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of proteins of immunological interest. Bethesda: U. S. Department of Health and Human Services, Public Health Service, National Institute of Health; 1991. [Google Scholar]
- 14.Wilson MR, Warr GW. Fish immunoglobulins and the genes that encode them. Annu Rev Fish Dis. 1992;2:201. [Google Scholar]
- 15.Warr GW. The immunoglobulin genes of fish. Dev Comp Immunol. 1995;19:1. doi: 10.1016/0145-305x(94)00052-h. [DOI] [PubMed] [Google Scholar]
- 16.Pilström L, Bengtén E. Immunoglobulin in fish – genes, expression and structure. Fish Shellfish Immunol. 1996;6:243. [Google Scholar]
- 17.Clem LW, McLean WE. Phylogeny of immunoglobulin structure and function. VII. Monomeric and tetrameric immunoglobulins of the margate, a marine teleost fish. Immunology. 1975;29:791. [PMC free article] [PubMed] [Google Scholar]
- 18.Warr GW. Immunoglobulin of the toadfish, Spheroides glaber. Comp Biochem Physiol. 1983;76B:507. doi: 10.1016/0305-0491(83)90284-5. [DOI] [PubMed] [Google Scholar]
- 19.Lobb CJ, Clem LW. Distinctive subpopulations of catfish serum antibody and immunoglobulin. Mol Immunol. 1983;20:811. doi: 10.1016/0161-5890(83)90077-9. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi K, Hara A, Takano K, Hirai H. Studies on subunit components of immunoglobulin M from a bony fish, the chum salmon (Oncorhynchus keta) Mol Immunol. 1982;19:95. doi: 10.1016/0161-5890(82)90251-6. [DOI] [PubMed] [Google Scholar]
- 21.Lobb CJ. Covalent structure and affinity of channel catfish anti-dinitrophenyl antibodies. Mol Immunol. 1985;22:993. doi: 10.1016/0161-5890(85)90087-2. [DOI] [PubMed] [Google Scholar]
- 22.Ghaffari SH, Lobb CJ. Cloning and sequence analysis of channel catfish heavy chain cDNA indicate phylogenetic diversity within the IgM immunoglobulin family. J Immunol. 1989;142:1356. [PubMed] [Google Scholar]
- 23.Rittenberg MB, Pratt KL. Antitrinitrophenyl (TNP) plaque assay. Primary response of BALB/c mice to soluble and particulate immunogen. Proc Soc Exp Biol Med. 1969;132:575. doi: 10.3181/00379727-132-34264. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Fazel S, Wiersma EJ, Shulman MJ. Interplay of J chain and disulfide bonding in assembly of polymeric IgM. Int Immunol. 1997;9:1149. doi: 10.1093/intimm/9.8.1149. [DOI] [PubMed] [Google Scholar]
- 26.Hsu E. The variation in immunoglobulin heavy chain constant regions in evolution. Sem Immunol. 1994;6:383. doi: 10.1006/smim.1994.1048. [DOI] [PubMed] [Google Scholar]
- 27.Smith RIF, Coloma MJ, Morrison SL. Addition of a μ-tailpiece to IgG results in polymeric antibodies with enhanced effector functions including complement-mediated cytolysis by IgG 4. J Immunol. 1995;154:2226. [PubMed] [Google Scholar]
- 28.Sørensen V, Rasmussen IB, Norderhaug L, Natvig I, Michaelsen TE, Sandlie I. Effect of the IgM and IgA secretory tailpieces on polymerization and secretion of IgM and IgG. J Immunol. 1996;156:2858. [PubMed] [Google Scholar]