Abstract
In many carcinomas, infiltrating macrophages are commonly found closely associated with tumour cells but little is known concerning the nature or significance of adhesion molecules involved in these cellular interactions. Here we demonstrate in primary human breast cancers that sialoadhesin (Sn), a macrophage-restricted adhesion molecule, is frequently expressed on infiltrating cells that often make close contact with breast carcinoma cells. To determine whether Sn could act as a specific receptor for ligands on breast cancer cell lines, binding assays were performed with a recombinant form of the protein fused to the Fc portion of human immunoglobulin G1 (IgG1) (Sn-Fc). Sn-Fc was found to bind specifically and in a sialic acid-dependent manner to the breast cancer cell lines MCF-7, T47.D and BT-20 both in solid- and solution-phase binding assays. To investigate the nature of the sialoglycoproteins recognized by Sn on breast cancer cells, MCF-7 cells were labelled with [6-3H]glucosamine. Following precipitation with Sn-Fc, a major band of ≈240 000 MW was revealed, which was shown in reprecipitation and Western blotting experiments to be the epithelial mucin, MUC1.
INTRODUCTION
Macrophages are commonly found as infiltrating cells in breast cancer.1 Although their precise functions are unclear, one contemporary view is that macrophages are an important source of cytokines and angiogenic factors that could be causally related to the survival, vascularization and eventual spread of a primary tumour.2–4 However, a large body of evidence, particularly using experimental models, supports the view that if macrophages are activated appropriately, they can also display potent tumoricidal activities that result in reduced tumour cell growth and survival (reviewed in 5).
Currently, little is known of the nature of cell–cell interaction molecules involved in these varied processes. Such interactions are likely to be important not only for the entry and localization of macrophages within tumours but also for influencing the migratory behaviour and metastasis of tumour cells.6 Intimate cell–cell contact between macrophages and tumour cells can also be a requirement for efficient killing by tumoricidal macrophages via cell surface expressed tumour necrosis factor-α (TNF-α).7
Sialoadhesin (Sn) is a macrophage-restricted lectin-like receptor with 17 extracellular immunoglobulin-like domains that can function as a cell adhesion molecule.8 The membrane distal immunoglobulin-like domain binds specifically to oligosaccharides terminating in Neu5Acα2,3Gal,9 a glycosidic linkage that is commonly found on O- and N-linked glycans present on the surface of mammalian cells.10 The interaction of Sn with cells of the haemopoietic lineages has been well characterized using both the native protein11,12 as well as a soluble recombinant form of the receptor in which the first three extracellular immunoglobulin domains were fused to the Fc portion of human immunoglobulin G (IgG) (designated Sn-Fc).8,10–13 Sn appears to bind selectively to immature and mature granulocytes in vitro12 and in vivo,14 suggesting that one of its functions is to mediate biologically relevant interactions between macrophages and myeloid cells. The interaction of Sn with other cells, particularly carcinoma cells, has been less well documented.
Here, we investigate the interaction of Sn with breast cancer cells. Our results demonstrate that Sn is expressed on macrophages that naturally infiltrate primary breast cancers and that Sn-Fc can mediate high level, sialic acid-dependent binding of breast cancer cells. Avidity capture experiments with lysates of breast cancer cells reveal selective binding of Sn to the membrane mucin, MUC1, suggesting that Sn–MUC1 interactions could occur in vivo and perhaps play a role in disease progression.
MATERIALS AND METHODS
Materials
Unless otherwise specified, all reagents and chemicals were purchased from Sigma Chemicals (Poole, UK or St Louis, MO). 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein acetoxymethyl ester (BCECF-AM) was purchased from Molecular Probes Inc. (Eugene, OR). Vibrio cholerae sialidase was from Calbiochem (La Jolla, CA). 6-[3H]glucosamine and ECL™was from Amersham (Little Chalfont, UK). Microtitre plates (Immulon 3) were from Dynatech (Chantilly, VA). Nitrocellulose membranes for Western blotting were purchased from Schleicher and Schuell (Dassel, Germany).
Cell lines and plasmids
COS-1, MCF-7, BT-20 and T47.D cell lines were obtained from the Imperial Cancer Research Fund Cell Bank, Clare Hall, UK. MCF-7 cells were grown in RPMI+10% fetal calf serum (FCS)+10 μg/ml insulin; BT-20 cells in minimal essential medium (MEM)+15% FCS+10 μg/ml insulin; T47.D cells in Dulbecco’s modified minimal essential medium (DMEM)+10% FCS.
Antibodies and Fc-proteins
The mouse antihuman MUC1 monoclonal antibodies (mAbs) HMFG115 and SM316 react with epitopes in the tandem repeat of MUC117,18 and were provided by the Imperial Cancer Research Fund Hybridoma Unit, Clare Hall, UK. This was also the source of the mouse antihuman CD44 mAb, P2A1. Polyclonal antiserum, CT1, is directed to a synthetic peptide corresponding to the 17 COOH-terminal amino acids in the cytoplasmic tail of human MUC1.19 The mouse antihuman Sn mAb, HSn1 and an affinity-purified rabbit polyclonal antihuman Sn were prepared as described.20 Affinity-purified rabbit antimouse CD22 polyclonal antibodies were prepared as described.10 Biotinylated rabbit antimouse IgG, alkaline phosphatase conjugated mouse antirabbit IgG and alkaline phosphatase, antialkaline phosphatase (APAAP) conjugates were purchased from Dako (High Wycombe, UK). The avidin-biotinylated peroxidase ABC kit was from Vector Laboratories (Peterborough, UK). Fc-proteins were purified from supernatants of transiently transfected COS-1 cells using protein A–sepharose, as described in detail previously.10
Immunoperoxidase staining of breast cancer sections
Frozen sections of breast tissue were fixed in acetone for 30 min at room temperature. Sections were blocked with 20% normal rabbit serum for 10 min and MUC1 was stained with the mouse mAb HMFG1 for 1 hr at room temperature, using tissue culture supernatant. After washing in phosphate-buffered saline (PBS) containing 0·1% bovine serum albumin (PBSA), the sections were incubated with biotinylated rabbit antimouse IgG for 45 min, washed with PBSA, the bound biotin detected using the ABC kit and developed with diaminobenzidine to give a brown colour. The sections were then incubated for 1 hr with affinity-purified rabbit polyclonal antiserum to Sn at 2 μg/ml, washed with PBSA and incubated for 45 min with alkaline phosphatase conjugated mouse antirabbit IgG. After further washing in PBSA, the staining was visualised with alkaline phosphatase, antialkaline phosphatase and the colour developed with Fast Red. Appropriate controls, including omission of each primary antibody, showed that binding of anti-MUC-1 and anti-Sn antibodies was specific under these conditions.
Solid phase binding assays
Fc chimeras at varying concentrations were adsorbed for 3 hr at 37° to wells of microtitre plates that had been coated overnight with goat antihuman IgG, as described.10 Cells (107 in 1 ml RPMI-1640+20 mm HEPES) were labelled at 37° for 30 min with 10 μg BCECF-AM dye and washed three times in PBS+0·25% BSA. Immediately prior to binding assays, cells were resuspended in PBS+0·25% BSA+5 mm ethylenediamine tetra-acetic acid (EDTA) at a concentration of 4×106/ml, and 50 μl aliquots were added to the precoated wells of microtitre plates containing 50 μl of the same buffer. After 1 hr at 37°, 50 μl of 0·25% glutaraldehyde in PBS were added and bound cells were fixed for 5 min at room temperature with gentle shaking. The nonadherent cells were removed by three washes in PBS and fluorescence was measured using a Cytofluor™ 1350 (Millipore, Watford, UK) with an excitation wavelength of 485 nm and an emission wavelength of 530 nm. The percentage of cells binding in each well was determined from measurements of the fluorescent signals before and after washing the plates.
Fluorescence-activated cell sorting (FACS) analysis
Adherent cells were detached from culture flasks in PBS+5 mm EDTA and washed with cold PBS+0·2% BSA+10 mm sodium azide. Cells were then incubated in the same buffer with Fc chimeras at a concentration of 50 μg/ml for 30 min at 4°, washed, and then incubated with fluoroscein isothiocyanate (FITC)-conjugated goat antihuman IgG for 30 min at 4°. After two further washes, stained cells were fixed in PBS with 2% formaldehyde. To determine whether binding of Sn-Fc to the cell lines was sialic acid-dependent, cells were treated with 0·01 U/ml sialidase for 3 hr in DMEM+20 mm HEPES prior to staining. The cells were analysed by flow cytometry on a Becton Dickinson FACScan.
Metabolic labelling and immunoprecipitations
MCF-7 cells (106) were labelled overnight at 37° with 10 μCi/ml [6-3H]glucosamine in DMEM without glucose, supplemented with 5% dialysed FCS and 0·5% normal FCS. All subsequent steps were performed at 4°. Labelled cells were lysed at a concentation of 0·5–1×107 cells/ml in 50 mm Tris–HCl pH 8·0 containing 1% NP-40, 150 mm NaCl, 5 mm EDTA; 1 mm phenylmethylsulphonyl fluoride (PMSF), 0·5 mg/ml leupeptin, 0·2 mg/ml aprotonin. Insoluble material was pelleted at 10 000 g for 10 min. The lysate was precleared for 30 min with 50 μl of protein A–sepharose and lysate fractions corresponding to 106 cells were incubated overnight with 10 μg Sn-Fc, CD22-Fc or neural cell adhesion molecule (NCAM-Fc) or 5 μg anti-MUC1 antibodies. 50 μl of protein A–sepharose beads were added and after 1 hr incubation, the resin was washed three times with wash buffer (50 mm Tris–HCl pH 8·0 containing 0·1% NP-40, 150 mm NaCl, 5 mm EDTA, 0·1 mm PMSF) and bound material eluted with non-reducing sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) loading buffer by boiling for 3 min. Samples were resolved on 7·5% SDS–PAGE and detected by fluorography. For re-precipitation experiments, bound material was eluted at 100° in 40 μl 1% SDS followed by addition of 960 μl of lysis buffer to dilute the SDS. 10 μg of the Fc proteins or 5 μg IgG were added and precipitations performed as above.
Immunoblotting
Using cell lysates prepared as described, proteins were separated on 7·5% reducing polyacrylamide gels and electrophoretically transferred overnight at 4° to nitrocellulose membranes at 0·3 A, 50 V. All subsequent steps were performed at room temperature. Membranes were blocked for 1 hr in PBS+0·1% Tween 20+3% skimmed milk, incubated for 1 hr with mAbs directed against either MUC1 or CD44 at a concentration of 1·0 μg/ml, washed and incubated for 1 hr with peroxidase-conjugated goat antimouse IgG. After washing, bound proteins were detected by enhanced chemiluminescence using the ECL™ detection system, according to the manufacturer’s protocol.
RESULTS
Expression of Sn on macrophages in breast cancer lesions
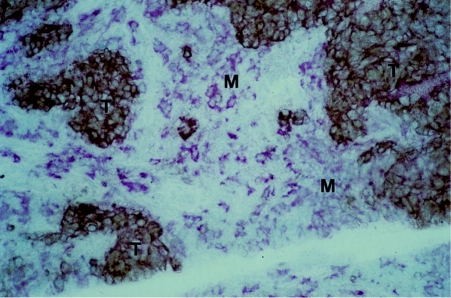
To investigate the expression of Sn in human breast cancer, frozen sections taken from up to 60 cases of primary breast cancer were stained using the antihuman Sn mAb, HSn1. In virtually all cases, Sn-positive cells were seen to be abundantly present throughout the stroma (Fig. 1). The Sn-positive cells resembled macrophages morphologically and were labelled by a pan-macrophage anti-CD68 mAb, EBM/11, in serial sections (not shown). To investigate the relationship between Sn-positive macrophages and breast cancer cells, double labelling experiments were performed using the anti-MUC1 mAb HMFG1 to detect breast cancer cells and an affinity-purified rabbit polyclonal antibody to detect Sn. MUC1 is an abundantly expressed membrane mucin of breast cancer cells as well as normal epithelia.21 The double-labelling experiments clearly revealed that the the Sn-positive macrophages are commonly found in close association with MUC1-positive breast carcinoma cells (Fig. 1). Although the rabbit antimouse Ab used to detect MUC-1 could theoretically react with the antirabbit used to detect Sn, control experiments showed that the binding of anti-MUC1 and anti-Sn was specific under the conditions used (not shown).
Figure 1.
Immunocytochemical double-staining of primary human breast cancer revealing close proximity of tumour cells and infiltrating Sn-positive macrophages. Frozen sections of breast tissue were stained by an indirect immunoperoxidase procedure using the HMFG1 anti-MUC-1 mAb and developed with diaminobenzidine to give a brown colour. The sections were then stained by an indirect alkaline phosphatase method using affinity-purified rabbit anti-Sn IgG and developed with Fast Red. Islands of brown MUC1-positive tumour cells (T) are surrounded by pink Sn-positive macrophages (M) (×250).
Binding of breast cancer cell lines to Sn-Fc and CD22-Fc
We next examined the ability of Sn to bind to the breast cancer cell lines MCF-7, T47.D and BT-20 by solid phase binding assays and by FACS analysis. These experiments were performed using a soluble chimaeric form of mouse Sn in which the first three extracellular domains were fused to the hinge and constant regions of human IgG1 (designated Sn-Fc). In solid-phase binding assays (Fig. 2), in which Fc-proteins were immobilized to plastic, all the cell lines bound at high levels to Sn-Fc, whereas a much lower level of binding was observed with CD22-Fc which specifically recognizes Neu5Ac(Gc)α2,6Gal in glycoproteins.13 No binding was observed with NCAM-Fc used as a negative control. In FACS assays, soluble Sn-Fc was found to bind specifically to MCF-7 cells (Fig. 3). Binding of Sn-Fc to cell lines in both the solid- and soluble-phase binding assays was sialic acid-dependent as pretreatment of the cells with sialidase completely abolished binding (Fig. 2b and Fig. 3).
Figure 2.

Solid-phase binding assays of Sn-Fc to breast cancer cell lines. Fc-proteins were immobilized to plastic wells, cells added and unbound cells washed off after 30 min. (a) Binding of breast cancer cell lines to Sn-Fc compared with CD22-Fc or NCAM-Fc used as a negative control. (b) Sialidase-treated MCF-7 cells bind to Sn-Fc at background levels. Bars show mean±1 SD of triplicate values. Results shown are representative of three experiments performed.
Figure 3.
FACS analysis of Sn-Fc binding to MCF-7 cells. Cells were incubated with either Sn-Fc or NCAM-Fc as a control, washed, and binding detected with antihuman IgG-FITC. Sn-Fc shows strong binding which is reduced to background levels after sialidase treatment of the MCF-7 cells.
Sn-Fc precipitates MUC-1 from MCF-7 cell lysates
To identify the nature of sialoglycoproteins recognized by Sn-Fc, precipitations were performed with lysates of MCF-7 cells labelled overnight with 6-[3H]glucosamine. Metabolic interconversion results in incorporation of the label into N-acetyl glucosamine, N-acetyl galactosamine and sialic acids, making it possible to identify both N- and O-glycosylated molecules.22 Following addition of protein A–sepharose, Sn-Fc precipitated a major broad band at ≈220–240 000 MW, which was not observed with CD22-Fc or NCAM-Fc used as a negative control (Fig. 4a).
Figure 4.
Precipitation of glycoproteins from 6-[3H]glucosamine-labelled MCF-7 cells. MCF-7 cells were labelled overnight with 6-[3H]glucosamine, lysed and precipitated with the indicated reagents. (a) Sn-Fc specifically precipitates high-molecular-weight material that comigrates with glycoproteins recognized by anti-MUC1 mAb SM3. (b) Lysates from MCF-7 cells labelled with 6-[3H]glucosamine were immunoprecipitated with anti-MUC1 mAb SM3 (left track) and eluates were reprecipitated with either Sn-Fc or NCAM-Fc. Sn-Fc specifically precipitates both allelic forms of MUC1.
The electrophoretic properties of the Sn-Fc precipitate were reminiscent of the MUC1 molecule which is well- labelled by 6-[3H]glucosamine.23 To test this possibility, 6-[3H]glucosamine-labelled MCF-7 cell lysates were immunoprecipitated with the SM3 anti-MUC1 mAb. MUC1 appeared as two glycoprotein bands of ≈220 000 and 240 000 MW, which comigrated with the material precipitated by Sn-Fc (Fig. 4a). These two bands correspond to the allelic forms of MUC1 expressed by MCF-7 cells.24
To test directly whether Sn-Fc could precipitate MUC1, reprecipitation experiments were performed (Fig. 4b). Following immunoprecipitation with anti-MUC1 mAb the eluate was reprecipitated with either Sn-Fc or NCAM-Fc. Sn-Fc quantitatively reprecipitated the two allelic forms of MUC1, whereas no bands were observed with NCAM-Fc, used as a negative control. These results show that Sn-Fc binds to MUC1 in a specific manner.
Further evidence that Sn-Fc precipitates MUC1 specifically was obtained in Western blots (Fig. 5). Lysates of MCF-7 cells were subjected to precipitation with Sn-Fc or CD22-Fc, eluted and western blots probed with anti-MUC1 mAb or an anti-CD44 mAb as a negative control. As shown in Fig. 5, MUC1 antigen was specifically precipitated by Sn-Fc, but not by CD22-Fc.
Figure 5.
Western blot analysis of glycoproteins precipitated by CD22-Fc or Sn-Fc. Lysates prepared from 6-[3H] glucosamine-labelled MCF-7 cells were precipitated with either CD22-Fc or Sn-Fc, proteins separated by SDS-PAGE and transferred to nitrocellulose membranes. The blots were probed with either anti-MUC1 mAb SM3 or anti-CD44 mAb P2A1. Sn-Fc precipitates glycoproteins that are recognized specifically by the anti-MUC1 but not by the anti-CD44 mAb. The nature of the faint band detected with anti-MUC-1 following precipitation with CD22-Fc is unknown.
DISCUSSION
The results of this study provide evidence that MUC-1 is a specific counter-receptor for the macrophage receptor, sialoadhesin. Using recently developed antibodies to human Sn,20 we first demonstrated that this receptor is present on the majority of infiltrating macrophages in many cases of breast cancer and that at least some of these macrophages are in close contact with MUC1-positive breast cancer cells. In vitro binding assays with a recombinant form of the receptor revealed that Sn can mediate high-level, sialic acid-dependent binding to breast cancer cell lines. Finally, we found that Sn-Fc is able to specifically precipitate the MUC1 molecule from lysates of the MCF-7 breast cancer cell line. It should be noted that, although a murine form of Sn-Fc was used in the present experiments, our recent characterization of human Sn has revealed that the human and mouse proteins exhibit indistinguishable binding specificities (unpublished observations). Thus, it is very likely that the binding properties described here for murine Sn-Fc reflect those that would occur with the human protein expressed by macrophages in human tumours.
MUC1 is an extended molecule because of extensive O-glycosylation and an extracellular domain which is made up of exact tandem repeats of 20 amino acids, each of which contains potential glycosylation sites.24 A size polymorphism is observed due to the presence of different numbers of tandem repeats (30–100 depending on the allele), but the smallest allele with around 25 repeats is still likely to encode a protein several times longer than other membrane mucins such as leukosialin or P-selectin glycoprotein ligand-1 (PSGL-1).25–27 Mucin-like membrane glycoproteins carrying multiple sialylated O-glycans can have a potentially antiadhesive function because of their extended structure and the negative charge that they carry. The MUC1 molecule has in fact been reported to exhibit an antiadhesive function27,28 that could, in the case of a cancer cell, affect metastatic progression. However, where a mucin can, through a carbohydrate structure, be recognized by a lectin on another cell, cell–cell adhesion is potentially enhanced, as has been well documented in the case of the selectin family of cell adhesion molecules (reviewed in 29). We speculate from the data presented here that the extended structure of the MUC1 polypeptide could act as a particularly effective scaffolding for presententation of oligosaccharide chains to Sn, thereby promoting cell–cell interactions between tumour cells and infiltrating macrophages.
Studies with normal and malignant breast cells have shown that the O-glycans added to MUC1 in normal cells are extended by core 2, branched structures while shorter chains are added to the mucin produced by breast cancer cell lines,23,30,31 resulting in the exposure of normally cryptic peptide epitopes. Although both forms of the glycoprotein carry O-glycans with terminal Neu5Acα2,3Gal (a preferred ligand for Sn10), the α2,3 sialyltransferase which adds sialic acid to Galβ1,3GalNAc (directly linked to serine or threonine), is dramatically upregulated in the breast cancer cells32 (and unpublished observations), resulting in an increase in the sialylated disaccharide carried on MUC1.23 Because of the aberrant glycosylation seen in breast cancer cells, the cancer mucin is antigenically distinct from the normal mucin and various formulations based on MUC1 are being tested in immunotherapy trials in the clinic. The interaction of MUC1 expressing cancer cells with the effector cells of the immune system is therefore particularly relevant. The data regarding the interaction of MUC1 with T cells are somewhat conflicting in that both inhibition of the interaction,33 enhancement of the interaction,34 and induction of cytotoxic T cells reactive with MUC1-expressing cells have been reported.35 Specific receptors on T cells have not been identified although intracellular adhesion molecule-1 (ICAM-1) has been reported to bind MUC1.36
While the present experiments indicate that Sn–MUC1 interactions are likely to occur in vivo, their biological significance remains to be determined. Interestingly, the degree of Sn-positive cell infiltration was positively correlated with disease severity in 33 cases analysed so far (unpublished observations). Sn has been shown to function as a non-phagocytic macrophage adhesion molecule for a range of cell types, including erythroid, myeloid and lymphoid cells.10,11,14,37,38 The physiological consequences of Sn-dependent adhesion are presently unclear, but it could influence, either positively or negatively, various macrophage effector functions such as phagocytic uptake of opsonized or apoptotic cells, antigen presentation,39 and cytotoxic activity that depends on membrane–membrane contact of macrophages with target cells. In addition, it is possible that Sn on macrophages can interact with soluble forms of MUC-1, which can be readily detected in the serum of breast cancer patients,40 possibly as a result of proteolytic cleavage from the cell surface.41 This circulating MUC1 could block Sn binding sites throughout the body and hence affect macrophage function at sites distal from the tumour itself. Future studies using mutant mice that are deficient in either MUC1,42 Sn or in both may shed light on these possibilities.
Acknowledgments
We are grateful to the Imperial Cancer Research Fund who supported this work.
Abbreviations
- BCECF-AM
2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein acetoxymethyl ester
- mAb
monoclonal antibody
- Sn
sialoadhesin
- Sn-Fc
recombinant form of mouse sialoadhesin, containing the first three N-terminal immunoglobulin-like domains fused to the Fc portion of human IgG1
References
- 1.O’sullivan C, Lewis CE. Tumour-associated leucocytes: friends or foes in breast carcinoma. J Pathol. 1994;172:229. doi: 10.1002/path.1711720302. [DOI] [PubMed] [Google Scholar]
- 2.O’Sullivan C, Lewis CE, Harris AL, Mcgee JO. Secretion of epidermal growth factor by macrophages associated with breast carcinoma. Lancet. 1993;342:148. doi: 10.1016/0140-6736(93)91348-p. [DOI] [PubMed] [Google Scholar]
- 3.Lewis CE, Leek R, Harris A, Mcgee JO. Cytokine regulation of angiogenesis in breast cancer: the role of tumor-associated macrophages. J Leukocyte Biol. 1995;57:747. doi: 10.1002/jlb.57.5.747. [DOI] [PubMed] [Google Scholar]
- 4.Leek RD, Lewis CE, Whitehouse R, Greenall M, Clarke J, Harris AL. Association of macrophage infiltration with angiogenesis and prognosis in invasive breast carcinoma. Cancer Res. 1996;56:4625. [PubMed] [Google Scholar]
- 5.Mantovani A, Bottazzi B, Colotta F, Sozzani S, Ruco L. The origin and function of tumor-associated macrophages. Immunol Today. 1992;13:265. doi: 10.1016/0167-5699(92)90008-U. [DOI] [PubMed] [Google Scholar]
- 6.Owen MR, Sherratt JA. Pattern formation and spatiotemporal irregularity in a model for macrophage–tumour interactions. J Theor Biol. 1997;189:63. doi: 10.1006/jtbi.1997.0494. [DOI] [PubMed] [Google Scholar]
- 7.Decker T, Lohmann-Matthes ML, Gifford GE. Cell-associated tumor necrosis factor (TNF) as a killing mechanism of activated cytotoxic macrophages. J Immunol. 1987;138:957. [PubMed] [Google Scholar]
- 8.Crocker PR, Mucklow S, Bouckson V, et al. Sialoadhesin, a macrophage sialic acid binding receptor for haemopoietic cells with 17 immunoglobulin-like domains. EMBO J. 1994;13:4490. doi: 10.1002/j.1460-2075.1994.tb06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nath D, Van Der Merwe PA, Kelm S, Bradfield P, Crocker PR. The amino-terminal immunoglobulin-like domain of sialoadhesin contains the sialic acid binding site. J Biol Chem. 1995;270:26184. doi: 10.1074/jbc.270.44.26184. [DOI] [PubMed] [Google Scholar]
- 10.Kelm S, Pelz A, Schauer R, et al. Sialoadhesin, MAG and CD22 define a new family of sialic acid-dependent adhesion molecules of the immunoglobulin superfamily. Curr Biol. 1994;4:965. doi: 10.1016/s0960-9822(00)00220-7. [DOI] [PubMed] [Google Scholar]
- 11.Crocker PR, Kelm S, Dubois C, et al. Purification and properties of sialoadhesin, a sialic acid-binding receptor of murine tissue macrophages. EMBO J. 1991;10:1661. doi: 10.1002/j.1460-2075.1991.tb07689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crocker PR, Freeman S, Gordon S, Kelm S. Sialoadhesin binds preferentially to cells of the granulocytic lineage. J Clin Invest. 1995;95:635. doi: 10.1172/JCI117708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelm S, Schauer R, Manuguerra J-C, Gross H-J, Crocker PR. Modifications of cell surface sialic acids modulate cell adhesion mediated by sialoadhesin and CD22. Glycoconj J. 1994;11:576. doi: 10.1007/BF00731309. [DOI] [PubMed] [Google Scholar]
- 14.Crocker PR, Werb Z, Gordon S, Bainton DF. Ultrastructural localization of a macrophage-restricted sialic acid binding hemagglutinin, SER, in macrophage–hematopoietic cell clusters. Blood. 1990;76:1131. [PubMed] [Google Scholar]
- 15.Burchell J, Durbin H, Taylor-Papadimitriou J. Complexity of expression of antigenic determinants, recognized by monoclonal antibodies HMFG-1 and HMFG-2, in normal and malignant human mammary epithelial cells. J Immunol. 1983;131:508. [PubMed] [Google Scholar]
- 16.Burchell J, Gendler S, Taylor-Papadimitriou J, et al. Development and characterization of breast cancer reactive monoclonal antibodies directed to the core protein of the human milk mucin. Cancer Res. 1987;47:5476. [PubMed] [Google Scholar]
- 17.Burchell J, Taylor-Papadimitriou J, Boshell M, Gendler S, Duhig T. A short sequence, within the amino acid tandem repeat of a cancer- associated mucin, contains immunodominant epitopes. Int J Cancer. 1989;44:691. doi: 10.1002/ijc.2910440423. [DOI] [PubMed] [Google Scholar]
- 18.Price MR, Clarke AJ, Robertson JF, O’sullivan C, Baldwin RW, Blamey RW. Detection of polymorphic epithelial mucins in the serum of systemic breast cancer patients using the monoclonal antibody, NCRC-11. Cancer Immunol Immunother. 1990;31:269. doi: 10.1007/BF01740933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pemberton L, Taylor-Papadimitriou J, Gendler SJ. Antibodies to the cytoplasmic domain of the MUC1 mucin show conservation throughout mammals. Biochem Biophys Res Commun. 1992;185:167. doi: 10.1016/s0006-291x(05)80971-4. [DOI] [PubMed] [Google Scholar]
- 20.Steiniger B, Barth P, Herbst B, Hartnell A, Crocker PR. The species-specific structure of microanatomical compartments in the human spleen: strongly sialoadhesin-positive macrophages occur in the perifollicular zone, but not in the marginal zone. Immunology. 1997;92:307. doi: 10.1046/j.1365-2567.1997.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girling A, Bartkova J, Burchell J, Gendler S, Gillett C, Taylor-Papadimitriou J. A core protein epitope of the polymorphic epithelial mucin detected by the monoclonal antibody SM-3 is selectively exposed in a range of primary carcinomas. Int J Cancer. 1989;43:1072. doi: 10.1002/ijc.2910430620. [DOI] [PubMed] [Google Scholar]
- 22.Cummings RD. Structural characterisation of N-glycans obtaind from metabolically-radiolabelled glycoproteins. In: Fukuda M, Kobata A, editors. Glycobiology a Practical Approach. Oxford: IRL Press; 1993. p. 243. [Google Scholar]
- 23.Lloyd KO, Burchell J, Kudryashov V, Yin BWT, Taylor-Papadimitriou J. Comparison of O-linked carbohydrate chains in MUC-1 mucin from normal breast epithelial cell lines and breast carcinoma cell lines. Demonstration of simpler and fewer glycan chains in tumor cells. J Biol Chem. 1996;271:33 325. doi: 10.1074/jbc.271.52.33325. [DOI] [PubMed] [Google Scholar]
- 24.Gendler SJ, Lancaster CA, Taylor-Papadimitriou J, et al. Molecular cloning and expression of human tumor-associated polymorphic epithelial mucin. J Biol Chem. 1990;265:15286. [PubMed] [Google Scholar]
- 25.Cyster JG, Shotton DM, Williams AF. The dimensions of the T lymphocyte glycoprotein leukosialin and identification of linear protein epitopes that can be modified by glycosylation. EMBO J. 1991;10:893. doi: 10.1002/j.1460-2075.1991.tb08022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li F, Erickson HP, James JA, Moore KL, Cummings RD, Mcever RP. Visualization of P-selectin glycoprotein ligand-1 as a highly extended molecule and mapping of protein epitopes for monoclonal antibodies. J Biol Chem. 1996;271:6342. doi: 10.1074/jbc.271.11.6342. [DOI] [PubMed] [Google Scholar]
- 27.Wesseling J, Van Der Valk SW, Hilkens J. A mechanism for inhibition of E-cadherin-mediated cell–cell adhesion by the membrane-associated mucin episialin/MUC1. Mol Biol Cell. 1996;7:565. doi: 10.1091/mbc.7.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ligtenberg MJ, Buijs F, Vos HL, Hilkens J. Suppression of cellular aggregation by high levels of episialin. Cancer Res. 1992;52:2318. [PubMed] [Google Scholar]
- 29.Crocker PR, Feizi T. Carbohydrate recognition systems: functional triads in cell–cell interactions. Curr Opin Struct Biol. 1996;6:679. doi: 10.1016/s0959-440x(96)80036-4. [DOI] [PubMed] [Google Scholar]
- 30.Hull SR, Bright A, Carraway KL, Abe M, Hayes DF, Kufe DW. Oligosaccharide differences in the DF3 sialomucin antigen from normal human milk and the BT-20 human breast carcinoma cell line. Cancer Commun. 1989;1:261. [PubMed] [Google Scholar]
- 31.Hanisch FG, Stadie TR, Deutzmann F, Peter-Katalinic J. MUC1 glycoforms in breast cancer-cell line T47D as a model for carcinoma-associated alterations of O-glycosylation. Eur J Biochem. 1996;236:318. doi: 10.1111/j.1432-1033.1996.00318.x. [DOI] [PubMed] [Google Scholar]
- 32.Brockhausen I, Yang JM, Burchell J, Whitehouse C, Taylor-Papadimitriou J. Mechanisms underlying aberrant glycosylation of MUC1 mucin in breast cancer cells. Eur J Biochem. 1995;233:607. doi: 10.1111/j.1432-1033.1995.607_2.x. [DOI] [PubMed] [Google Scholar]
- 33.Van De Wiel-Van Kemenade E, Ligtenberg MJ, De Boer AJ, et al. Episialin (MUC1) inhibits cytotoxic lymphocyte–target cell interaction. J Immunol. 1993;151:767. [PubMed] [Google Scholar]
- 34.Bohm CM, Mulder MC, Zennadi R, et al. Carbohydrate recognition on MUC1-expressing targets enhances cytotoxicity of a T cell subpopulation. Scand J Immunol. 1997;46:27. doi: 10.1046/j.1365-3083.1996.d01-91.x. [DOI] [PubMed] [Google Scholar]
- 35.Jerome KR, Barnd DL, Bendt KM, et al. Cytotoxic T-lymphocytes derived from patients with breast adenocarcinoma recognize an epitope present on the protein core of a mucin molecule preferentially expressed by malignant cells. Cancer Res. 1991;51:2908. [PubMed] [Google Scholar]
- 36.Regimbald LH, Pilarski LM, Longenecker BM, Reddish MA, Zimmermann G, Hugh JC. The breast mucin MUCI as a novel adhesion ligand for endothelial intercellular adhesion molecule 1 in breast cancer. Cancer Res. 1996;56:4244. [PubMed] [Google Scholar]
- 37.Morris L, Crocker PR, Fraser I, Hill M, Gordon S. Expression of a divalent cation-dependent erythroblast adhesion receptor by stromal macrophages from murine bone marrow. J Cell Sci. 1991;99:141. doi: 10.1242/jcs.99.1.141. [DOI] [PubMed] [Google Scholar]
- 38.Van Den Berg TK, Breve JJ, Damoiseaux JG, et al. Sialoadhesin on macrophages: its identification as a lymphocyte adhesion molecule. J Exp Med. 1992;176:647. doi: 10.1084/jem.176.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Umansky V, Beckhove P, Rocha M, Kruger A, Crocker PR, Schirrmacher V. A role for sialoadhesin-positive tissue macrophages in host resistance to lymphoma metastasis in vivo. Immunology. 1996;87:303. [PMC free article] [PubMed] [Google Scholar]
- 40.Burchell J, Wang D, Taylor-Papadimitriou J. Detection of the tumour-associated antigens recognized by the monoclonal antibodies HMFG-1 and 2 in serum from patients with breast cancer. Int J Cancer. 1984;34:763. doi: 10.1002/ijc.2910340605. [DOI] [PubMed] [Google Scholar]
- 41.Boshell M, Lalani EN, Pemberton L, Burchell J, Gendler S, Taylor-Papadimitriou J. The product of the human MUC1 gene when secreted by mouse cells transfected with the full-length cDNA lacks the cytoplasmic tail. Biochem Biophys Res Commun. 1992;185:1. doi: 10.1016/s0006-291x(05)80946-5. [DOI] [PubMed] [Google Scholar]
- 42.Spicer AP, Rowse GJ, Lidner TK, Gendler SJ. Delayed mammary tumor progression in Muc-1 null mice. J Biol Chem. 1995;270:30093. doi: 10.1074/jbc.270.50.30093. [DOI] [PubMed] [Google Scholar]