Abstract
Melanoma‐specific cytotoxic T lymphocytes (CTL) can be generated from peripheral blood lymphocytes (PBL) by mixed lymphocyte–tumour cell cultures. Analysis of CTL precursor frequencies in peripheral blood of melanoma patients is generally used for immunomonitoring purposes to evaluate vaccination efficacy. At present, it is unclear whether PBL‐derived CTL generated in vitro are indicative of an anti‐tumour immune response in vivo. Three tumour‐specific human leucocyte antigen (HLA)‐B/C‐restricted CTL clones were derived from peripheral blood of a melanoma patient immunized with interleukin‐7 (IL‐7) gene‐modified tumour cells. CTL clones differing in their T‐cell receptor‐γ (TCRγ) rearrangement produced interferon‐γ, IL‐4 and/or IL‐10. On the basis of their unique TCRγ gene rearrangements clone‐specific primers were generated for detection of clone‐specific DNA by polymerase chain reaction. One CTL clone (E5) of the three was found to be selectively expanded in one of seven metastases obtained at autopsy, as determined by Southern blot hybridization. However, the presence of E5 in only one of seven metastases at death indicates that the in vivo accumulation of the specific CTL clone was not sufficient to contain tumour progression. Nevertheless, our data support the proposition that analysis of anti‐tumour activity of PBL‐derived CTLs may reflect an anti‐tumour immune response in vivo.
Introduction
Human malignant melanoma in its advanced state has no hope for cure.1 Nevertheless, the immune system is believed to play an important role in the host defence against melanoma, with spontaneous tumour regression occurring in a minority of patients.2,3 Melanomas are frequently characterized by a lymphocyte infiltration that, in some cases, has been associated with a good prognosis,4 thus suggesting that an anti‐tumour immune response can occur naturally. Tumour‐infiltrating lymphocytes (TILs) expressing a restricted set of T‐cell receptor (TCR) V‐genes have been shown to accumulate at the tumour site.5 Selective expression of TCRV‐gene subfamilies in tumour lesions strongly suggests an antigen‐induced proliferation of oligoclonal T cells in response to tumour‐associated antigenic peptides.6–8 In line with this observation, it has independently been reported by two groups that a melanoma‐specific cytotoxic T‐cell clone derived from TILs was expanded in two different metastases of the same patient.9,10
Given the proposed importance of the immune system in fighting melanoma, various forms of active specific immunotherapies, including peptide vaccination, dendritic cell‐based therapy11 and gene therapy,12,13 have recently been applied in melanoma patients. For immunomonitoring purposes, the analysis of cytotoxic T lymphocyte (CTL) precursor frequencies in peripheral blood of melanoma patients is generally performed to evaluate vaccination efficacy, in as much as that anti‐melanoma cytotoxic CD8+ CTL can often be generated from peripheral blood lymphocytes (PBL) by mixed lymphocyte–tumour cultures (MLTC),14 and that peripheral blood can readily be obtained from virtually all melanoma patients undergoing immunotherapy. However, the important question as to whether the in vitro sensitized PBL‐derived CTL are truly representative of the cytotoxic T cells accumulating in vivo within tumour lesions, remains unanswered.
In the present study, we have investigated the in vivo clonal expansion of three anti‐melanoma CTL clones derived from peripheral blood of a melanoma patient immunized with interleukin‐7 (IL‐7) gene‐modified autologous tumour cells. The human leucocyte antigen (HLA) restriction element and tumour‐specific cytokine secretion by these CTL were also examined.
Materials and methods
Patient characteristics and melanoma specimens
The patient (J.L.) under study was a 51‐year‐old woman suffering from a primary malignant melanoma at the back of her left shoulder. Fifteen months after surgical excision of the primary tumour, distant metastases were detected. After having failed to respond to various chemoimmunotherapies, she was subsequently immunized with gene‐transduced autologous tumour cells secreting IL‐7 with mixed response.12 Peripheral blood mononuclear cells were isolated before vaccination and 2 weeks after the third vaccination and were stored in liquid nitrogen until use. Metastatic lesions from different sites (axilla, inguina, spleen) were obtained at autopsy.
Tumour cell lines
An autologous melanoma cell line (UKRV‐MEL‐6a, referred to hereafter as Mauto) was established from an accessible cutaneous melanoma metastasis as described.12 Seven additional melanoma cell lines (UKRV‐MEL‐15a, UKRV‐MEL‐21a, UKRV‐MEL‐7, UKRV‐MEL‐17, UKRV‐MEL‐23, UKRV‐MEL‐29, UKRV‐MEL‐19a, referred to as M1–M7, respectively) derived from different patients, along with the natural killer (NK)‐sensitive target K562 and an autologous Epstein–Barr virus (EBV)‐immortalized B‐cell line, were used as targets in cytotoxicity assays. All cell lines were grown in RPMI‐1640 medium supplemented with 10% fetal calf serum (FCS), 2 mm glutamine (Seromed, Berlin, Germany) and 100 U/100 µg/ml penicillin/streptomycin (Seromed).
Generation of T‐cell lines
A limiting dilution assay was carried out as described.15 Briefly, cryopreserved peripheral blood mononuclear cells (PBMC) were stimulated at 10 000, 5000, 2500, 1250, 625 and 312 cells per microwell with 1 × 104 mitomycin C (MMC)‐inactivated autologous melanoma cells in 200 µl RPMI‐1640 medium supplemented with 10% heat‐inactivated pooled human AB serum (Sigma, Deisenhofen, Germany), 2 mm glutamine, 100 U/100 µg/ml penicillin/streptomycin, 25 mm HEPES, and 20 U/ml recombinant human IL‐2 (rhIL‐2). The microcultures were restimulated at days 7, 14 and 21 and were initially screened for cytotoxic activity against autologous melanoma cells and K562 at day 28, as previously described.12 The tumour‐reactive CTL lines obtained were restimulated weekly (2 × 105–3 × 105/well) with MMC‐inactivated autologous melanoma cells (5 × 104) and autologous EBV‐B cells (2 × 105–3 × 105) as feeders in 2 ml in 24‐well plates.
Immunofluorescence analysis
The phenotypic analysis of the T‐cell lines was performed by direct immunofluorescence using monoclonal antibodies (mAb) towards CD3, CD8, TCRαβ, CD4 and CD56 (all from Immunotech, Hamburg, Germany). In brief, 3 × 105 T cells in 100 µl of Ca2+Mg2+‐free phosphate‐buffered saline (PBS), containing 1% bovine serum albumin and 0·1% sodium azide, were incubated with 5 µl of mAb for 30 min on ice. Cells were washed twice and analysed on an EPICS XL flow cytometer (Coulter Electronics, Krefeld, Germany). The percentage of positive cells was calculated. Isotype‐matched irrelevant antibodies [immunoglobulin G1 (IgG1) and IgG2a] served as a negative control.
Cytotoxicity assay
The cytotoxic activity of T‐cell lines against melanoma cells, K562, or autologous EBV‐B cells was measured by a 6‐hr lactate dehydrogenase (LDH)‐release assay, as previously described.12 Briefly, 5000 viable target cells in triplicate were co‐cultured with various amounts of effector cells [effector to target (E : T) ratios ranging between 20 : 1 and 2·5 : 1] in 200 µl assay medium (phenol red‐free RPMI‐1640 medium supplemented with 3% FCS) in U‐bottom microwell plates (Nunc, Wiesbaden, Germany). Following 6‐hr incubation at 37° in 5% CO2, 100 µl/well supernatant was collected and LDH activity in the supernatant was immediately measured using a commercially available detection kit for LDH (Boehringer Mannheim, Mannheim, Germany). LDH activity present in the assay medium alone served as background control and was subtracted from all values. The percentage of specific LDH‐release was calculated with the following formula:
 |
Effector or target spontaneous release was obtained by incubating effector or target cells with assay medium alone, respectively. Maximum release of LDH was determined by incubating target cells in the presence of 1% Triton X‐100 (Sigma Chemical Co., St. Louis. MO). The spontaneous release of target cells was always < 15% of the maximum LDH‐release in all experiments.
In order to test the cross‐reactive cytotoxicity of anti‐melanoma T‐cell clones, a panel of allogeneic melanoma cell lines was employed in the LDH‐release assay at a fixed E : T ratio of 5 : 1.
Analysis of HLA restriction elements
Inhibition of T‐cell‐mediated tumour lysis was assessed by preincubating tumour cells with neutralizing mAbs including W6/32 (anti‐HLA‐class I heavy chains, monomorphic);16 MA2.1 (anti‐HLA‐A2)17 and B1.23.2 (anti‐HLA‐B, ‐C)18 (all ascitic fluids kindly provided by Dr Coulie; Brussels, Belgium). Briefly, 1 × 104 tumour cells were pretreated in triplicate with different mAbs (at a 1 : 20 dilution) in 50 µl for 60 min at 37°. Then, effector cells were added to give a final volume of 200 µl/well. E : T ratios ranged from 3 : 1 to 0·3 : 1. The cytotoxicity was determined in a 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) assay as described.12 Standard deviations were < 5%. Inhibition of cytolysis by blocking antibody was calculated as:
Control lysis was defined as lysis in the absence of blocking antibodies at a given E : T ratio. Unspecific effects of the ascites on target cells were tested by coincubation for 1 hr at 37° before the assay in order to exclude an antibody‐dependent mechanism of cell lysis in the absence of T cells.
Cytokine release
At least 8 days after the last stimulation, T cells were washed and stimulated (3 × 105 cells) with autologous melanoma cells (5 × 104) in a final volume of 2 ml in 24‐well plates. Control wells contained melanoma cells with medium alone. Supernatants were harvested 24‐hr later and were stored at – 20° until use. All cytokine measurements were performed using commercially available enzyme‐linked immunosorbent assay (ELISA) kits: IL‐4, interferon‐γ (IFN‐γ), and IL‐10 ELISA kits were obtained from Immunotech, Hamburg, Germany, whereas the transforming growth factor‐β1 (TGF‐β1) ELISA kit was purchased from Genzyme, Cambridge, MA.
Analysis of clone‐specific junctional sequences
From each T‐cell line (A2, D3, E5), DNA was isolated as described.19 Briefly, the cell pellet was redissolved in a digestion buffer [0·2 m Tris–Cl, pH 8, 10 mm ethylenediamine tetraacetic acid (EDTA), 1% sodium dodecyl sulphate (SDS), 1 mg/ml Proteinase K]. After incubation at 55° for 24 hr, DNA was extracted by phenol/chloroform, precipitated in ethanol and resuspended in 100 µl of distilled and autoclaved water.
For identification of the clone‐specific TCRγ junctional sequences, ≈ 1 µg of DNA was used as a template for polymerase chain reaction (PCR) amplification reactions using primers annealing to conserved regions of TCRγ genes as described.20 Primer Varicons anneals between nucleotides 272 and 290 of TCRγ variable (V) genes and Primer Jointcons anneals between nucleotides 45 and 62 of TCRγ joint genes.21 The PCR reaction mixture consisted of 3 U Taq polymerase (Boehringer Mannheim), a 1 × reaction buffer as supplied by the manufacturer, 300 ng of each primer, 200 µm of each dNTP, and ≈ 1 µg of DNA in a volume of 50 µl overlaid with mineral oil. Using a Perkin Elmer Cetus DNA Thermal Cycler, the samples were exposed to 40 cycles of 94° for 1 min, 55° for 30 seconds, 72° for 30 seconds. The PCR products were subjected to DNA sequence analyses using an automated Applied Biosystems DNA sequencer (Dr Metzger, Vaterstetten, Germany). With knowledge of these sequences, clone‐specific primers (Primer Nspec), complementary to the junctional region of each clone were designed and their sequences are shown in Table 4.
Table 4.
Clone‐specific primers (Primer Nspec) designed individually for junctional regions of each T‐cell clone
| A2 | Nspec | 5′ | −TGT | TCC | ACT | GCC | AAA | GAG | TTG | 3′ |
| D3 | Nspec | 5′ | CCA | CTG | CCA | AAG | AGT | CAC | GAA | 3′ |
| E5 | Nspec | 5′ | ACT | GCC | AAA | GAG | TTT | CTT | ATA | 3′ |
Generation of clone specific hybridization probes
DNA from each CTL line was extracted as described above and used as a template for PCR amplification reaction. PCR was carried out using clone‐specific primers (see Table 4) in conjunction with primer Vari 8 or Vari 2 in 100 µl of a reaction mixture containing 5 µl of DNA, 200 µm of each dNTP, 1 µm of each primer, and 2·5 U of Taq DNA polymerase (Boehringer Mannheim) on a DNA thermal cycler (PTC‐200 DNA Engine, MJ Research, Inc., Boston, MA). Amplification was performed as described above. Negative controls included reactions without DNA. Ten microlitres of amplified products was analysed by agarose gel electrophoresis. The amplified DNA products were labelled with non‐radioactive digoxigenin‐11‐dUTP (DIG‐11‐dUTP) (Boehringer Mannheim), as indicated by the manufacturers. The DIG‐labelled TCR probes were used for hybridization, as described below.
Southern blot hybridization
In order to assess the presence of any of the characterized CTL lines in tumour lesions in vivo, DNA was extracted from seven formalin‐fixed, paraffin‐embedded metastases of the patient obtained at autopsy, as described.22 DNA from each specimen was used as a template for PCR amplification reactions using one of the Nspec primers in conjunction with a primer annealing to the V‐gamma‐8 gene (Primer V‐gamma‐8: 5′ CTT CCT GTA GAA AAT GCC GTC 3′) or V‐gamma‐2 gene (Primer V‐gamma‐2: 5′ CTT GCT GAA GGA AGT AAC GGC 3′), respectively. The primers V‐gamma‐8 and V‐gamma‐2 can be regarded as ‘semi‐specific’, since they anneal to DNA from all lymphocytes which use the V‐gamma‐2 or V‐gamma‐8 segment for rearrangement. However, in conjunction with a N‐specific primer, only DNA from the characterized clones will be amplified.
The PCR products were size separated by 2% agarose gel electrophoresis for 1‐hr and blotted onto a nylon membrane (Boehringer Mannheim). Membranes were prehybridized at 40·6° in a volume of 20 ml hybridization solution (DIG Easy Hyb, Boehringer Mannheim) for 60 min. Hybridization was performed by incubating membranes at 40·6° overnight in 5 ml hybridization solution with 5 ng/ml of DIG‐labelled individual TCR probes which were obtained by PCR using the respective T‐cell lines as a template and the respective N‐specific primers (see Table 4). After stringency washes, the hybridized blots were submitted to immunological chemiluminescence detection using anti‐digoxigenin antibody conjugated to alkaline phosphatase and CSPD®, ready‐to‐use (Boehringer Mannheim), as indicated by the manufacturer.
Results
Derivation and characterization of tumour‐specific T‐cell clones
Fifty‐five T‐cell lines were generated from peripheral blood of a melanoma patient (J.L.), as described in the Materials and Methods. Phenotypic analyses performed 8 weeks after initiation of culture revealed that all T‐cell lines established expressed CD3+ CD56– markers as well as TCRα and ‐β chains (data not shown). Eight out of the 55 T‐cell lines showed predominantly CD8+ populations (> 80% CD8+ cells), whereas 11 of the 55 expressed mainly CD4 markers (> 80% CD4+ cells); the remaining 36 T‐cell lines contained mixed populations of CD4+ and CD8+ cells (data not shown). Of the 55 T‐cell lines, 21 (six containing predominantly CD8+ populations, 15 were mixtures of CD4+ and CD8+ populations) displayed significant cytolytic activity against autologous melanoma cells, but failed to lyse autologous EBV‐B cells or NK‐sensitive K562 cells in multiple assays (Table 2, and data not shown).
Table 2.
Characterization of T‐cell lines by cytotoxicity and cytokine secretion
| Cytotoxicity against* | Cytokine secretion‡ | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T-cell lines | Mauto | K562 | EBV-B | M1 | M2 | M3 | M4 | M5 | M6 | M7 | IFN-γ (IU/ml) | IL-4 (pg/ml) | IL-10 (pg/ml) | TGF-β1 (ng/ml) |
| E5 | 70† | 0 | 0 | 10 | 4·4 | 0·6 | 0·9 | 3 | 0·8 | 1·4 | 26·8§ | 3519 | 2594 | 0 |
| A2 | 69·3 | 5·5 | 0 | 2·2 | 0·7 | 1·4 | 2·8 | 0·7 | 1·5 | 1·6 | 68·3 | 1200 | 2688 | 0 |
| D3 | 21 | 0 | 6·2 | ND¶ | ND | ND | ND | ND | ND | ND | 14·5 | 1842 | 531 | 0 |
Specificity and reactivity of each T‐cell line were determined by 6 hr LDH-release assays. Results are expressed as percentage lysis. Specific lysis for each T-cell clone was tested at least three times and representative experiments are shown for an E : T ratio of 5 : 1.
Percentage lysis given as boldface was significantly different from background lysis.
3 × 105 CD8+ CTL were stimulated with 5 × 104 autologous tumour cells in a final volume of 2 ml. After 24 hr supernatants were harvested and assayed for cytokine content by ELISAs.
Underlined values indicate results obtained after dilution of supernatants in order to obtain values within standard curve of ELISAs (0·08–25 IU/ml for IFN‐γ; 5–1000 pg/ml for IL‐4; 5–2000 pg/ml for IL‐10).
ND, not done.
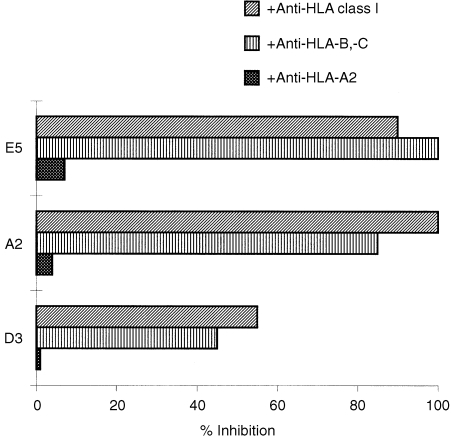
We initially focused on six CTL lines containing predominantly CD8+ populations and expanded them in the presence of autologous EBV‐B as feeder cells. However, three CTL lines could not be maintained beyond 4 months and were no longer available for further analyses. The remaining CTL lines (termed A2, E5, D3), characterized by a good expansion rate, were eligible for further analyses. The results presented in Table 2 clearly showed the specific lysis of autologous tumour cells (UKRV‐Mel‐6a) by three CTL lines. The recognition of tumour cells by these CTL was restricted by HLA‐B or ‐C molecules, in as much as that pretreatment of tumour cells with neutralizing antibodies towards HLA class I or HLA‐B/C strongly inhibited CTL cytolysis, whereas anti‐HLA‐A2 mAb (MA2.1) had no effect (Fig. 1). Since patient J.L. was homozygous for the HLA‐B, ‐C locus (Table 1), it is therefore conceivable that the precise restriction molecule was B60, B62, or Cw3. Examination of cross‐reactivity of these CTL with a panel of allogeneic melanoma cell lines with defined HLA‐type (Table 1) and the known tumour antigen expression revealed no significant cytolysis of allogeneic tumour cells tested (Table 2).
Figure 1.
Inhibition of the specific cytotoxicity of three T‐cell lines towards autologous melanoma cells by mAbs. Melanoma cells were preincubated with mAbs directed to HLA‐class I (W6/32), HLA‐B, ‐C (B1.23.2) and HLA‐A2 (MA2.1) at a 1 : 20 dilution of ascites for 60 min at 37° prior to the addition of the effectors (E5, A2 and D3 T‐cell lines). Cytotoxicity was determined in a MTT assay at an E : T ratio of 3 : 1. Percentage of inhibition was calculated as described in the Materials and Methods.
Table 1.
HLA-type of autologous and seven allogeneic melanoma cell lines
| UKRV-Mel-6a | (Mauto) | A1, A2 | B60, B62 | Cw3 | DR4, DR13 |
| UKRV‐Mel‐15a | (M1) | A2, A11 | B22, B75 | Cw3, Cw1 | DR1, DR15 |
| UKRV‐Mel‐21a | (M2) | A2, A9 | B44 | Cw4 | DR7, DR11 |
| UKRV-Mel-7 | (M3) | A11, A32 | B7, B52 | Cw5 | DR1, DR2 |
| UKRV-Mel-17 | (M4) | A1, A3 | B62, B57, Bw4, Bw6 | Cw3, Cw6 | DR3, DR4, DR52, DR53 |
| UKRV-Mel-23 | (M5) | A1, A11 | B8, B35 | Cw4, Cw7 | DR1, DR17 |
| UKRV-Mel-29 | (M6) | A2 | B60, Bw4, Bw6, B38 | Cw3 | DR1, DR15, DQ5, DQ6 |
| UKRV‐Mel‐19a | (M7) | A2, A33 | B14, B55, Bw6 | Cw3, Cw8 | DR1, DQ15, DR51 |
Previous studies on human CD8+ T cells reacting to conventional antigens have shown that CD8+ T cells preferentially producing T helper type 2 (Th2)‐type cytokines IL‐4 and IL‐5 are less cytotoxic than non‐IL‐4/IL‐5‐producing cells.23 However, this was not observed for tumour‐specific CTL.24 Results presented in Table 2 showed that all three CTL lines upon stimulation with melanoma cells, produced high levels of IL‐4 (up to 3519 pg/ml) and/or IL‐10 (up to 2688 pg/ml), in addition to Th1‐type cytokine IFN‐γ (up to 68·3 IU/ml). Neither CTL lines produced TGF‐β (Table 2). While the functional role of Th2‐type cytokines in these anti‐tumour CTL remains unclear, kinetic studies suggest that IL‐4 and IL‐10 production by these cells did not result in impaired cytotoxic function (data not shown).
For molecular characterization of these anti‐tumour CTL lines, we examined the highly variable junctional region of rearranged TCR‐γ genes (Table 3). DNA sequencing after PCR amplification revealed different junctional regions. In clone A2, the variable gamma‐8 segment rearranged with a joint segment and no nucleotides were inserted at the junction, in contrast to D3 where different breakpoints were used and where six nucleotides were interposed. In clone E5, the variable gamma‐2 segment rearranged with the joint segment at different breakpoints and no nucleotides were inserted at the junction (Table 3).
Table 3.
Junctional sequences of rearranged TCRγ genes of three individual CTL clones
| V8 | Joint | |||||||||||
| A2 | ACG | TGA | CTC | TGG | GGT | C | / | AAC | TCT | TTG | GCA | G |
| V8 | N | Joint | ||||||||||
| D3 | TGC | CAC | CTG | GGA | / | TTC | GTG | / | AC | TCT | TTG | GCA |
| V2 | Joint | |||||||||||
| E5 | CTG | GGA | CGGG | / | TAT | AAG | AAA | CTC | T |
Detection of tumour‐specific CTL clones in metastatic lesions by Southern blot hybridization
In order to detect T‐cell accumulation at tumour sites, DNA isolated from seven metastases obtained at autopsy, was used as a template for PCR using TCR clone‐specific primers in conjunction with semi‐specific V‐gamma primers (Vari 2 or Vari 8). After blotting the PCR products onto nylon membranes, hybridization was performed with DIG‐labelled TCR probes which were generated from DNA of the original T‐cell clones (D3, E5, A2) using the identical sets of primers. As shown in Fig. 2, a hybridization signal specific for E5 was identified in DNA from the inguinal lymph node of the patient (lane 4), but not in any other lesions. However, clone‐specific DNA for A2 or D3 was not detected in any of the metastatic lesions analysed (data not shown).
Figure 2.
Hybridization signal of an E5‐specific probe designed for its unique TCRγ gene rearrangement by Southern blot. DNA extracted from seven metastases of the patient (lanes 1–7: right axillary lymph node; left axillary lymph node; left axillary lymph node; left inguinal lymph node; right inguinal lymph node; spleen; spleen, respectively.) obtained at autopsy was used as a template for PCR amplification reactions using Primer E5‐Nspec and Primer V‐gamma‐2. The PCR products were electrophoresed for 1 hr in a 2% agarose gel and transferred on to a nylon membrane. Hybridization was performed by incubating the membrane at 40·6° overnight in 5 ml hybridization solution with 5 ng/ml of a DIG‐labelled TCR probe which was obtained from the E5 T‐cell clone by PCR amplification using primer E5‐Nspec and primer V‐gamma‐2.
Discussion
Melanoma is thought to be a highly immunogenic tumour to which multitypes of active specific immunotherapy have recently been applied. As a readout system, analysis of anti‐melanoma CD8+ CTL response has widely been employed, since CD8+ CTL are most effective in recognizing and destroying the tumour and that such effectors can often be elicited when T cells from PBL or TILs are co‐cultured with autologous tumour cells.14,25
Previous efforts at analysing TIL‐derived anti‐tumour CTL have indicated the in vivo existence of a melanoma antigen‐driven tumour‐specific CTL response. Mackensen et al.26,27 isolated an anti‐tumour CTL clone from a spontaneously regressive melanoma lesion that in vitro displayed HLA‐B14‐restricted cytolytic activity towards autologous tumour cells. With the analysis of TCR expression and immunohistochemistry, they found that this in vitro cultured anti‐melanoma CTL clone could be detected in vivo in the tumour area, thus providing an indication of a local adaptive immune response which is clinically associated with tumour regression. Sensi and co‐workers28 have demonstrated selective expansion of an HLA‐A2‐restricted TIL‐derived CTL clone even in an advanced metastatic melanoma lesion. More recently, two groups9,10 independently reported in vivo accumulation of a TIL‐derived anti‐melanoma CTL line in various metastases after vaccination with tumour cells. In these cases, however, clonal expansion of anti‐tumour CTL failed to mediate objective tumour regression. Nevertheless, these studies provided evidence that an in vivo selection of anti‐melanoma cytotoxic T cells occurs and that such CTL may circulate and accumulate at different tumour sites.
Having considered that peripheral blood is easily attainable and can serve as a consistent source for generation of tumour‐specific CTL, many current studies with melanoma vaccines have used peripheral blood as a source of CTL generation. However, whether analysis of PBL‐derived CTL response to melanoma antigens accurately reflects the in vivo immune status, remains undefined. Relevant to this, it would be important to determine whether PBL‐derived CTL lines or clones had selectively been expanded in vivo at the tumour sites. In the present study, we have tested three HLA‐B/C‐restricted CTL clones generated from PBL of a melanoma patient for in vivo accumulation using PCR and Southern blot hybridization techniques. One CTL clone (E5) with highly cytotoxic activity against autologous melanoma cells in vitro, was detected in one out of the seven metastatic lesions analysed (Fig. 2). Unfortunately, the antigen specificity of this clone has not yet been established. However, autologous EBV‐B cells transduced with full‐length cDNA of gp100 and tyrosinase (two melanoma antigens known to be expressed on autologous tumour cells, as determined by reverse transcription–PCR) did not induce cytokine release from E5 CTL, suggesting that this CTL clone recognizes unidentified shared melanoma antigens.
The accumulation of clone E5 in patient with far‐advanced disease failed to contain the tumour progression, consistent with previous observations from TIL‐derived CTL lines.9,10,28 This might be due to tumour‐induced CTL dysfunction, apparent immunosuppression, including production of TGF‐β,12 or down‐regulated expression of melanoma antigens and/or HLA class I and TAP (transporter associated with antigen processing),29,30 preventing tumour recognition by CTL. On the basis of these observations, we assumed that the anti‐tumour CTL response would be more pronounced in and relevant to primary tumour.
Since after long‐term culture only three CTL clones were available for in vivo analysis, it is uncertain whether these clones are representative of the in vivo anti‐melanoma responses. However, the demonstration that in vitro‐propagated, PBL‐derived, anti‐melanoma CTL do accumulate in vivo within the tumour, suggests the occurrence of in vivo selection of anti‐melanoma cytotoxic T cells. Taken together, the current study supports the proposition that analysing PBL during immunization is meaningful and that PBL‐derived CTL may truly reflect an in vivo anti‐tumour immune response. However, further studies are needed to confirm these initial observations.
Acknowledgments
This work was supported by the DFG (Scha 422/6–2). The authors are grateful to Mrs Antje Sucker and Helga Kemmer for their excellent technical assistance. The authors are also thankful to Mrs. Margaret Vazansky for her editing of the manuscript.
Glossary
Abbreviations
- LDH
lactate dehydrogenase
- MLTC
mixed lymphocyte–tumour cultures
- MTT
3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide
- PBL
peripheral blood lymphocytes
- TILs
tumour‐infiltrating lymphocytes
References
- 1.Johnson TM, Smith Ii JW, Nelson BR, Chang A. Continuing medical education: Current therapy for cutaneous melanoma. J Am Acad Dermatol. 1995;32:689. doi: 10.1016/0190-9622(95)91443-9. [DOI] [PubMed] [Google Scholar]
- 2.Nathanson L. Spontaneous regression of malignant melanoma: a review of the literature on incidence, clinical features, and possible mechanisms. Conference on spontaneous regression of cancer. Natl Cancer Inst Monogr. 1976;44:67. [PubMed] [Google Scholar]
- 3.Bodurtha AJ. Spontaneous regression of malignant melanoma. In: Clark WH, Goldman LI, Mastrangelo JM, editors. Human Malignant Melanoma. New York: Grune & Stratton; 1979. p. 227. [Google Scholar]
- 4.Clark WH, Elder DE, Guerry DP, et al. Model predicting survival in stage I melanoma based on tumor progression. J Natl Cancer Inst. 1989;81:1893. doi: 10.1093/jnci/81.24.1893. [DOI] [PubMed] [Google Scholar]
- 5.Sensi M, Parmiani G. Analysis of TCR usage in human tumors: a new tool for assessing tumor‐specific immune responses. Immunol Today. 1995;16:588. doi: 10.1016/0167-5699(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 6.Nitta T, Oksenberg JR, Rao NA, Steinman L. Predominant expression of T‐cell receptor Vα7 in tumor‐infiltrating lymphocytes of uveal melanoma. Science. 1990;249:672. doi: 10.1126/science.2382141. [DOI] [PubMed] [Google Scholar]
- 7.Weidmann E, Trucco M, Whiteside TL. Relevance of the T‐cell receptor for immunotherapy of cancer. Cancer Immunol Immunother. 1994;39:1. doi: 10.1007/BF01517174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferradini L, MacKensen A, Genevee C, et al. Analysis of T cell receptor variability in tumor‐infiltrating lymphocytes from a human regressive melanoma: Evidence for in situ T cell clonal expansion. J Clin Invest. 1993;91:1183. doi: 10.1172/JCI116278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sensi M, Farina C, MacCalli C, et al. Clonal expansion of T lymphocytes in human melanoma metastases after treatment with a hapten‐modified autologous tumor vaccine. J Clin Invest. 1997;99:710. doi: 10.1172/JCI119215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hishii M, Andrews D, Boyle LA, et al. In vivo accumulation of the same anti‐melanoma T cell clone in two different metastatic sites. Proc Natl Acad Sci USA. 1997;94:1378. doi: 10.1073/pnas.94.4.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nestle FO, Alijagic S, Gilliet M, et al. Vaccination of melanoma patients with peptide‐ or tumor lysate‐pulsed dendritic cells. Nat Med. 1998;4:328. doi: 10.1038/nm0398-328. [DOI] [PubMed] [Google Scholar]
- 12.Möller P, Sun Y, Dorbic T, et al. Vaccination with IL‐7 gene‐modified autologous melanoma cells can enhance the anti‐melanoma lytic activity in peripheral blood of patients with a good clinical performance status – A clinical phase I study. Br J Cancer. 1998;77:1907. doi: 10.1038/bjc.1998.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y, Jurgovsky K, Möller P, et al. Vaccination with IL‐12 gene‐modified autologous melanoma cells: preclinical results and a first clinical phase I study. Gene Ther. 1998;5:481. doi: 10.1038/sj.gt.3300619. [DOI] [PubMed] [Google Scholar]
- 14.Boon T, Cerottini J‐C, Van Den Eynde B, Van Der Bruggen P, Van Pel A. Tumor antigens recognized by T lymphocytes. Annu Rev Immunol. 1994;12:337. doi: 10.1146/annurev.iy.12.040194.002005. [DOI] [PubMed] [Google Scholar]
- 15.Coulie PG, Somville M, Lehmann F, et al. Precursor frequency analysis of human cytolytic T lymphocytes directed against autologous melanoma cells. Int J Cancer. 1992;50:289. doi: 10.1002/ijc.2910500220. [DOI] [PubMed] [Google Scholar]
- 16.Brodsky FM, Parham P. Monomorphic anti‐HLA‐A,‐B, ‐C monoclonal antibodies detecting molecular subunits and combinatorial determinants. J Immunol. 1982;128:129. [PubMed] [Google Scholar]
- 17.McMichael AJ, Parham P, Rust N, Brodsky FM. A monoclonal antibody that recognizes an antigenic determinant shared by HLA‐A2 and B17. Hum Immunol. 1980;1:121. doi: 10.1016/0198-8859(80)90099-3. [DOI] [PubMed] [Google Scholar]
- 18.Rebai N, Malissen B. Structural and genetic analyses of HLA class I molecules using monoclonal xenoantibodies. Tissue Antigens. 1983;22:107. doi: 10.1111/j.1399-0039.1983.tb01176.x. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A Laboratory Manual. Cold Spring Harbor NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Volkenandt M, Soyer HP, Kerl H, Bertino JR. Development of a highly specific and sensitive molecular probe for detection of cutaneous lymphoma. J Invest Dermatol. 1991;97:137. doi: 10.1111/1523-1747.ep12479308. [DOI] [PubMed] [Google Scholar]
- 21.Lefranc MP, Forster A, Baer R, Stinson MA, Rabbitts TH. Diversity and rearrangement of the human T cell rearranging γ genes: nine germ‐line variable genes belonging to two subgroups. Cell. 1986;45:237. doi: 10.1016/0092-8674(86)90388-0. [DOI] [PubMed] [Google Scholar]
- 22.Volkenandt M, McNutt NS, Albino AP. Sequence analysis of DNA from formalin‐fixed, paraffin‐embedded human malignant melanoma. J Cutan Pathol. 1991;18:210. doi: 10.1111/j.1600-0560.1991.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 23.Maggi E, Giudizi MG, Biagiotti R, et al. Th2‐like CD8+ T cells showing B cell helper function and reduced cytolytic activity in human immunodeficiency virus type 1 infection. J Exp Med. 1994;180:489. doi: 10.1084/jem.180.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacCalli C, Mortarini R, Parmiani G, Anichini A. Multiple sub‐sets of CD4+ and CD8+ cytotoxic T‐cell clones directed to autologous human melanoma identified by cytokine profiles. Int J Cancer. 1994;57:56. doi: 10.1002/ijc.2910570111. [DOI] [PubMed] [Google Scholar]
- 25.Rosenberg SA. Development of cancer immunotherapies based on identification of the genes encoding cancer regression antigens. J Natl Cancer Inst. 1996;88:1635. doi: 10.1093/jnci/88.22.1635. [DOI] [PubMed] [Google Scholar]
- 26.MacKensen A, Ferradini L, Carcelain G, et al. Evidence for in situ amplification of cytotoxic T‐lymphocytes with antitumor activity in a human regressive melanoma. Cancer Res. 1993;53:3569. [PubMed] [Google Scholar]
- 27.MacKensen A, Carcelain G, Viel S, et al. Direct evidence to support the immunosurveillance concept in a human regressive melanoma. J Clin Invest. 1994;93:1397. doi: 10.1172/JCI117116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sensi M, Salvi S, Castelli C, et al. T cell receptor (TCR) structure of autologous melanoma‐reactive cytotoxic T lymphocyte (CTL) clones: tumor‐infiltrating lymphocytes overexpress in vivo the TCR beta chain sequence used by an HLA‐A2‐restricted and melanocyte‐lineage‐specific CTL clone. J Exp Med. 1993;178:1231. doi: 10.1084/jem.178.4.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee KH, Panelli MC, Kim CJ, et al. Functional dissociation between local and systemic immune response during anti‐melanoma peptide vaccination. J Immunol. 1998;161:4183. [PubMed] [Google Scholar]
- 30.Hicklin DJ, Marincola FM, Ferrone S. HLA class I antigen downregulation in human cancers: T‐cell immunotherapy revives an old story. Mol Med Today. 1999;5:178. doi: 10.1016/s1357-4310(99)01451-3. [DOI] [PubMed] [Google Scholar]